Final_Exam_Q1-3_solution_Dec152008.doc
advertisement

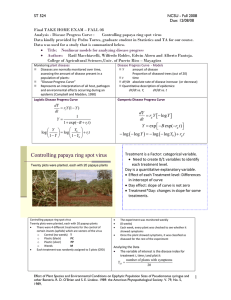
ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Final TAKE HOME EXAM – FALL 08 Analysis - Disease Progress Curve : Controlling papaya ring spot virus Data kindly provided by Pedro Torres, graduate student in Statistics and TA for our course. Data was used for a study that is summarized below. Title: Nonlinear models for analyzing disease progress Authors: Raúl Macchiavelli, Wilfredo Robles, Edwin Abreu and Alberto Pantoja. College of Agricultural Sciences,Univ. of Puerto Rico – Mayagüez Monitoring plant diseases Diseases are normally monitored over time, assessing the amount of disease present in a population of plants: “Disease Progress Curve” Represents an interpretation of all host, pathogen and environmental effects occurring during an epidemic (Campbell and Madden, 1990) Disease Progress Curve - Models Y amount of disease Proportion of diseased trees (out of 20) t time dY/dt absolute rate of disease increase (or decrease) Quantitative description of epidemics: dY/dt vs. Y, dY/dt vs. t Logistic Disease Progress Curve Gompertz Disease Progress Curve dY rl Y (1 Y ) dt 1 Y 1 exp( B rl t ) Y log 1 Y dY rg Y log Y dt Y exp B exp(rg t ) log log Y log log Y0 rg t Y0 rl t log 1 Y0 Treatment is a Factor: categorical variable. Need to create 0/1 variables to identify each treatment level. Day is a quantitative explanatory variable. Effect of each Treatment level: Differences in intercept of curve Day effect: slope of curve is not zero Treatment*Day: changes in slope for some treatments. Controlling papaya ring spot virus Twenty plots were planted, each with 20 papaya plants Controlling papaya ring spot virus Twenty plots were planted, each with 20 papaya plants There were 4 different treatments for the control of certain insects (aphids) which are vectors of the virus o Control (no weeds) T o Plastic (black) PC o Plastic (silver) PP o Weeds M Each treatment was randomly assigned to 5 plots (CRD) The experiment was monitored weekly (8 weeks) Each week, every plant was checked to see whether it showed symptoms Once the plant showed symptoms, it was classified as diseased for the rest of the experiment Analyzing the Data The variable of interest is the disease index for treatment i, time j and plot k: number of plants with symptoms Yijk 20 1 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Problems with traditional approach o Non normal distribution o Non linear models o Non constant Variances o Observations of the same tree in different weeks are dependent o Observations on the same plot may not be independent (contagion) Traditional analysis o Fit separate curves for each plot using linear regression o Compare slopes for different treatments Generalized linear models o The linear component is defined like in traditional linear models: i xi ' o A monotonic differentiable link function g describes how the expected value of y , E Y , related to the linear predictor: g ( i ) xi ' o The response variables y are independent and have a probability distribution from an exponential family. This implies that the variance of the response depends on the mean through a variance function V: var(Yi ) V ( i ) o The dispersion parameter is either assumed known (for example, for the binomial distribution, = 1) or it must be estimated to account for overdispersion. a) Nonlinear fitting of observed proportion of diseased plants (out of 20 plants per plot) using PROC NLIN in SAS Need to set up dummy variables for treatments Needs initial values of parameters Logistic Fit Yijk number trees diseased 20 E Yijk o i 1 day j 90 2i day j 90 * i log 1 E Yijk ij Yijk exp ij 1 exp ij eijk ij o i 1 day j 90 2i day j 90 * i oi 1i day ij 90 Normal 0, 2 eijk Gompertz Fit Yijk number trees diseased 20 log log E Yijk ij o i 1 day j 90 2i day j 90 * i Yijk exp exp ij eijk ij o i 1 day j 90 2i day j 90 * i oi 1i day ij 90 eijk Normal 0, 2 2 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 b) Estimation of parameters and treatment effect of Generalized Linear Model with PROC GENMOD in SAS and o o o o No need for initial estimates Use of CLASS statement sets up dummy variables (treatment effect) directly Use of CONTRAST statement allows comparing treatment effect, equality of slopes, etc. Use of ESTIMATE statement allows predictions, confidence intervals . Yijk number of diseased trees out of 20 per plot binomial 20, ij Yijk log ij 1 ij i 1day j 2i day j i E Yijk ij Var Yijk 20 ij 1 ij i = 1, 2, 3, 4 treatments j=1, 2, 3, 4, 5, 6, 7, 8 timepoints k = 1, 2, 3, 4, 5 blocks c) Note Non linear models with normal residuals (NLIN) do not take into account actual distribution or longitudinal nature. Because of contagion, Number of diseased trees (out of 20) is not a binomial random variable, variance may not correspond to a binomial random variable. E Yijk ij Var Yijk 20 ij 1 ij overdispersion parameter Non linear models fitting a binomial distribution with possibly overdispersion do not take into account longitudinal nature. Overdispersion parameter may be estimated as the square root of deviance divided by its degrees of freedom. If the ratio (deviance/d.f) is greater than 1 indicates that overdispersion is present. Use scale= deviance option in PROC GENMOD, MODEL statement, to fit a binomial with overdispersion. Standard errors and tests are adjusted to account for extra variation d) Estimation of the parameters of Generalized Linear Model with PROC NLINMIXED in SAS Repeated observations from the same plot are correlated, same random plot effect. Yijk number of diseased trees out of 20 per plot Yijk | uik binomial 20, ij ij log 1 ij uik E Yijk ij i 1day j 2i day j i uik Normal 0, 2 i = 1, 2, 3, 4 treatments j = 1, 2, 3, 4, 5, 6, 7, 8 timepoints k = 1, 2, 3, 4 blocks Var Yijk 20 ij 1 ij ij exp int i 1day j 2i day j i 1 exp int i 1day j 2i day j i No accounting for correlation between measurements within same tree. 3 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Questions PROC NLIN is used to fit two disease curves. Q1.a. Write down both estimated equations. Logistic Fitting Yijk number of diseased trees out of 20 per plot Yijk binomial 20, ij log ij 1 ij 0.2073 0.3215* id _ m 1.7844* id _ pc 2.3447 * id _ pp 0.0593* day j 0.00231* day j * id _ m 0.0425* day j * id _ pc 0.0535* day j * id _ pp Treatment=C log ij 1 ij 0.2073 0.0593* day j Treatment= M ij log 1 ij 0.2073 0.3215* id _ m 0.0593* day j 0.00231* day j * id _ m 0.2073 0.3215 0.0593* day j 0.00231* day j .1142 0.06161day j Treatment = PC log ij 1 ij 0.2073 1.7844* id _ pc 0.0593* day j 0.0425* day j * id _ pc 0.2073 1.7844 0.0593* day j 0.0425* day j 1.5771 0.1018* day j Treatment=PP log ij 1 ij 0.2073 2.3447 * id _ pp 0.0593* day j 0.0535* day j * id _ pp 0.2073 2.3447 0.0593* day j 0.0535* day j 2.1374 0.1128* day j Gompertz Fitting Yijk number trees diseased 20 log log E Yijk o i 1 day j 90 2i day j 90 * i 0.6125 0.2169* id _ m 1.3102* id _ pc 1.7687 * id _ pp 0.0404* day j 90 0.000993* day j 90 * id _ m 0.0332* day j 90 * id _ pc 0.0439* day j 90 * id _ pp 4 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Treatment C log log E Yijk 0.6125 0.0404* day j 90 Treatment M log log E Yijk 0.6125 0.2169* id _ m 0.0404* day j 90 0.000993* day j 90 * id _ m 0.6125 0.2169 0.0404* day j 90 0.000993 day j 90 0.3956 0.041393* day j 90 Treatment PC log log E Yijk 0.6125 1.3102* id _ pc 0.0404* day j 90 0.0332* day j 90 * id _ pc 0.6125 1.3102 0.0404* day j 90 0.0332* day j 90 0.6977 0.0736* day j 90 Treatment PP log log E Yijk 0.6125 1.7687 * id _ pp 0.0404* day j 90 0.0439* day j 90 * id _ pp 0.6125 1.7687 0.0404* day j 90 0.0439* day j 90 1.1562 0.0843* day j 90 5 ST 524 Solution Final Take Home Exam Q1.b. NCSU - Fall 2008 Due: 12/09/08 Calculate R2, measure of goodness of fit, Error SS R 2 100 1 % USS Total For Logistic Fitting 1.6916 R 2 100 1 96.97% 55.88 For Logistic Fitting 1.9131 R 2 100 1 96.58% 55.88 Q1.c. which model, GOMPERTZ or Logistic, shows better fit? Logistic fitting have a higher R-square, thus Logistic model is better fit for proportion of diseased plants. Note that model may be improved since the approximate 95% Confidence limits for parameters bm and rm includes value 0, which would indicate that there is no differences between the logistic curves for treatments M and Control. Also, it should be of interest to test whether both plastic treatments respond to same logistic curve, i.e., no differences between these two treatments; and whether there are differences between treatments M and C against the “plastic” treatments PP and PC. PROC GENMOD is used for a fitting the number of diseased trees within each plot as a binomial random variable with n=20 (trees) and the probability for a tree being diseased as a function of Treatment and Day. Full model fits four slopes (for linear time effect), one for each treatment and four separate intercepts (treatment effects). Q1.d. Would you recommend to adjust for overdispersion?. Yes, Deviance/DF = 280.1112/152 = 1.8428 is greater than 1, indicating overdispersion. Contrasts test ☼ whether slopes for plastic covers have same effects ☼ whether slopes for control and weedy condition are the same. ☼ whether average slope for “plastic” is equal to average slope for “nonplastic” ☼ whether effects of plastic covers are the same ☼ whether effects of control and weedy treatments are the same. ☼ whether average effect for “plastic” treatments is equal to average effect for “nonplastic” treatments Q1.e. Which model do you select, based on above results (PROC GENMOD)? Make reference to contrasts. Write down conclusions. Indicate limitations. After adjusting for overdispersion, Type III likelihood ratio test, in Table LR Statistics for Type 3 Analysis indicates that Treatments, Day and their interaction are highly significant (p-value < 0.0001). Contrasts show that there are not differences between main effects of the two plastic treatments (p=0.3434), No differences between between main effects of the M and C treatments (p =0.5173), significant differences between these two groups (p<0.0001) , and similarly for the day effect: no differences in linear slope between the two “plastic” treatments (p=0.4793), nor the two M and C treaments (p=0.8031), but significant differences between the linear day effect of these two groups (p<0.001). The recommended model should fit a common binomial model (curve) for the two “plastic” treatments, and a common binomial model (curve) for the two M and C treatments, with correction for overdispersion. 6 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Note: The scale parameter was estimated by the square root of DEVIANCE/DOF. LR Statistics For Type 3 Analysis Source Num DF Den DF F Value Pr > F Chi-Square Pr > ChiSq treatid 3 152 14.72 <.0001 44.15 <.0001 day 1 152 1113.09 <.0001 1113.09 <.0001 day*treatid 3 152 7.21 0.0001 21.62 <.0001 Contrast Results Contrast Num DF Den DF F Value Pr > F Chi-Square Pr > ChiSq Type PP=PC 1 152 0.90 0.3449 0.90 0.3434 LR T=M 1 152 0.42 0.5183 0.42 0.5173 LR (PPandPC= TandM) 1 152 43.18 <.0001 43.18 <.0001 LR rpc=rpp 1 152 0.50 0.4804 0.50 0.4793 LR rm=rt 1 152 0.06 0.8034 0.06 0.8031 LR (rpc and rpp) = (rm and rt) 1 152 21.32 <.0001 21.32 <.0001 LR PROC NLMIXED is used to fit a model taking into account the distribution of the number of diseased trees within a plot as a binomial random variable with parameter ij depending on the treatment and time of measurement. Random block effects are also included in model. Full model fits separate slopes and intercepts for each treatment group, while the second model fits two models with common intercept and slope for treatments T and M and separate common intercept and slope for treatment PP and PC. Q1.f. Which model do you select, make reference to contrasts. Limitations. Results from NLMIXED are similar to what we found in PROC GENMOD, significance of contrasts tested indicate that there should be two models for the logits, one modeling logits from the “plastic” treatments, and the other modeling logits from the M and C treatments. A limitation of this modeling is that the repeated measures structure of the residual variance has been modeled, although random effect associated with repetitions is included in the model, while GENMOD does not include either random effects or the repeated measures structure for the residual variation. 7 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Parameter Estimates Parameter Estimate Standard Error DF t Value Pr > |t| Alpha Lower Upper Gradient b0 0.3505 0.1498 159 2.34 0.0205 0.05 0.05468 0.6463 -0.0013 bm -0.2966 0.2102 159 -1.41 0.1602 0.05 -0.7119 0.1186 -0.00035 bpc -1.7488 0.2503 159 -6.99 <.0001 0.05 -2.2432 -1.2544 -0.00049 bpp -2.2418 0.2734 159 -8.20 <.0001 0.05 -2.7818 -1.7018 0.000131 r 0.06896 0.005835 159 11.82 <.0001 0.05 0.05744 0.08049 0.030399 rm 0.001911 0.008116 159 0.24 0.8141 0.05 -0.01412 0.01794 0.029448 rpc 0.03107 0.01009 159 3.08 0.0024 0.05 0.01114 0.05099 -0.00962 rpp 0.03934 0.01088 159 3.61 0.0004 0.05 0.01785 0.06083 0.006952 sigma2 0.3813 0.1122 159 3.40 0.0009 0.05 0.1597 0.6030 0.000135 Contrasts Label Num DF Den DF F Value Pr > F PP=PC 1 159 2.63 0.1065 T=M 1 159 3.77 0.0539 rpc=rpp 1 159 0.44 0.5077 PPandPC = TandM 1 159 79.45 <.0001 slopes: (T,M) = (PP,PC) 1 159 98.96 <.0001 Final model is Parameter Estimates Parameter Estimate Standard Error DF t Value Pr > |t| Alpha Lower Upper Gradient b0 0.2077 0.1088 159 1.91 0.0582 0.05 -0.00730 0.4226 -0.00242 bpp -1.8373 0.1883 159 -9.75 <.0001 0.05 -2.2093 -1.4653 -0.0008 r 0.07018 0.004330 159 16.21 <.0001 0.05 0.06163 0.07873 -0.03194 rpp 0.03384 0.007520 159 4.50 <.0001 0.05 0.01899 0.04869 -0.02421 sigma2 0.4212 0.1183 159 3.56 0.0005 0.05 0.1876 0.6548 0.000523 8 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Yijk number of diseased trees out of 20 per plot Yijk binomial 20, ij log ij 1 ij 0.2077 1.8373* id _ pp +0.0718* day j 90 0.03384* day j 90 * id _ pp Q1.g. Write down the model for the proportion of diseased trees in a plot receiving a silver plastic cover at day t. log ij 1 ij 0.2077 1.8373* id _ pp+0.07018* day j 90 0.03384* day j 90 * id _ pp silver plastic 0.2077 1.8373 0.07018 0.03384 * day j 90 1.6296 0.10402* day j 90 Q1.h. Interpret coefficients of model. -1.6296 is the predicted value for logit at t=0, which means that at t=0, i.e., at day 90, the predicted e1.6296 proportion of diseased plants per plot is 0.1639 , 16.39% at day = 90 1 e1.6296 0.10402 is the increase in the logit when day increases one unit, i.e., the daily rate of increase on logit is given by 0.10402, which indicates that the odds ratio is 1.1096, and a daily increase of 10.96% Q1.i. Write down the equation for the prediction of response in a plot with a silver plastic cover at day 85. And similarly for a weedy plot at day 85. log ij 0.2077 1.8373* id _ pp+0.07018* day j 90 0.03384* day j 90 * id _ pp 1 ij silver plastic ,t 85 0.2077 1.8373 0.07018 0.03384 * 85 90 1.6296 0.10402* 85 90 2.1497 silverplastict day 85 log ij 1 ij e2.1497 0.1044 , 10.44% 1 e2.1497 0.2077 0.07018* day j 90 silver plastic ,t 85 0.2077 0.07018* 85 90 0.2077 0.07018* 85 90 weedy day 85 0.1432 0.1432 e 0.4543 , 45.43% 1 e0.1432 9 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Question 2 Scientific paper: Effect of Plant Species and Environmental Conditions on Epiphytic Population Sizes of Pseudomonas syringae and other Bacteria. R. D. O’Brien and S. E. Lindow. 1989. the American Phytopathological Society. V. 79, No. 5, 1989, Question 2 will Refer only to Experiment 1 in the above paper. Please answer the following Description Q2.1. Objective 9 strains * 4 plant species 10 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 dry + high light 72 hrs Humid + low light 48 hrs Uncontrolled outdoor environment 12 days Sampling unit: 15-40 individual leaves per plant (15-20 g. fresh weigh per plant) Sampling periods: ☼ Before Inoculation ☼ Immediately Before Inoculation ☼ Immediately after inoculation ☼ After wet and dry growth chamber incubation ☼ Periodically during incubation of plants under field conditions. o 11 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Response Variable Bacterial Population size per gram fresh weight of leaves. Log-transformed population size Q2.3. Indicate What Are The Different Experimental Units, a. Main-Unit : a run of the experiment at each environment b. Sub-Unit: a 20-cm diameter pot , one plant per pot c. Sub-Sub Unit: a sample of 15-20 g fresh weigh leaves Q2.4. How Is A Block Defined? a. There were 3-4 replicates for each experimental condition, a block can be assumed a complete run of the experiment, thus there will be three to four runs (?) No information about blocks in text. Q2.5. What Are The Factors And Their Type: Random Fixed, a. Main-plot Factor: Environment: Humid+Dry; Humid+uncontrolled : Fixed b. Sub-plot Factor: i. Strain : Random (random sampled selected) ii. and Plant Species : Fixed c. Sub-sub-sub unit factor: Removal method : Fixed All Factors were considered fixed-effect factors. Counting Q2.6. Number Of Blocks: 3-4 Q2.7. Total Number Of Main-Units: 2 * (3-4) = 6-8 Q2.8. Total Number Of Sub-Units: 2*(9*4)*(3-4) = 216-288 Q2.9. Total Number Of Sub-Sub-Units: 2*2*(9*4)*(3-4) =432-576 Q2.10. How Many Main-Unit Within Each Block: 2 Q2.11. How Many Sub-Units Within Each Main-Unit: 9*4=36 Q2.2. 12 ST 524 Q2.12. Solution Final Take Home Exam How Many Sub-Sub-Units Within Each Sub-Unit: Each block should have 2 main units 2*9*4 = 72 sub units 2*2*9*4 = 144 sub-sub units NCSU - Fall 2008 Due: 12/09/08 2 Statistical Analysis Q2.13. Linear Model, based on above information. Yijk log colony size Yij Ei Bm aim S j Pk S * P jk E * S ij E * P ik E * S * P ijk b jkmi Rl E * R il S * R jl P * R kl S * P * R jkl E * S * R ijl E * P * R ikl E * S * P * R ijkl eijklm aim Normal 0, a2 b jkm i Normal 0, b2 Normal 0, e2 eijklm Q2.14. Present The ANOVA Table, Sources Of Variation, Df, Ms If Possible, Number of blocks = 4 MS Block DF r=4 3 4.51 E(MS) Fixed Effect 2 e2 2r b2 72r a2 144 block Environment 1 195.41 e2 2 b2 72 a2 Q E Error(a) 3 0.71 e2 2 b2 72 a2 Strain 8 6.51 e2 2 b2 Q S Plant Species 3 6.53 e2 2 b2 Q P Strain*Plant Sp 24 0.65 e2 2 b2 Q S * P Env*Strain 8 2.52 e2 2 b2 Q E * S Env*PlantSp 3 2.75 e2 2 b2 Q E * P Env*Strain*PlantSp 24 0.50 e2 2 b2 Q E * S * P Error(b) 210 Removal 1 12.23 e2 Q R Env*Rem 1 4.75 e2 Q E * R Strain*Rem 8 e2 Q S * R Plant Sp*Rem 3 e2 Q P * R Strain*Plant Sp*Rem 24 e2 Q S * P * R Env*Strain*Rem 8 e2 Q E * S * R Env*Plant Sp*Rem 3 e2 Q E * P * R Env*Strain*Plant Sp*Rem 24 e2 Q E * S * P * R Residual 216 e2 Total 575 Source e2 2 b2 13 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Random Effect: Strain and its interactions Source MS E(MS) Random Effects Block DF r=4 3 4.51 2 e2 2r b2 72r a2 144 block Environment 1 195.41 e2 2 b2 72 a2 Q E Error(a) 3 0.71 e2 2 b2 72 a2 Strain 8 6.51 e2 2 b2 64 S2 Plant Species 3 6.53 e2 2 b2 Q P Strain*Plant Sp 24 0.65 e2 2 b2 16 S3*P Env*Strain 8 2.52 e2 2 b2 32 E2*S Env*PlantSp 3 2.75 e2 2 b2 Q E * P Env*Strain*PlantSp 24 0.50 e2 2 b2 8 E2*S*P Error(b) 210 Removal 1 12.23 e2 Q R Env*Rem 1 4.75 e2 Q E * R Strain*Rem 8 e2 32 S2*R Plant Sp*Rem 3 e2 Q P * R Strain*Plant Sp*Rem 24 e2 8 S2*P*R Env*Strain*Rem 8 e2 16 E2*S*R Env*Plant Sp*Rem 3 e2 Q E * P * R Env*Strain*Plant Sp*Rem 24 e2 4 E2*S*P*R Residual 216 e2 Total 575 e2 2 b2 14 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Yijk log colony size Yij Ei Bm aim s j Pk s * P jk E * s ij E * P ik E * s * P ijk b jkmi Rl E * R il s * R jl P * R kl s * P * R jkl E * s * R ijl E * P * R ikl E * s * P * R ijkl eijklm aim Normal 0, a2 b jkmi Normal 0, b2 eijklm Normal 0, e2 sj Normal 0, s2 s * P jk Normal 0, s2*P E * s ij Normal 0, E2*s E * s * P ijk s * R jl s * P * R jkl E * s * P * R ijkl Normal 0, E2*s*P Normal 0, s2*R Normal 0, s2*P*R Normal 0, E2*s*P*R Sub-Sub-Plot Factor And Interactions Was Analyzed As A Randomized Complete Block Design. Indicate The Number Of Blocks That Should Be Considered. Each combination of BLOCK*E*S*P is considered a block when analyzing the sub-sub-plot factor. Thus there are r*2*9*4=72r (216 for r=3, or 288 for r=4) Q2.15. Compare Your ANOVA Table With Table 2. Experiment 1. If any discrepancies are observed, please explain them. 15 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 Whole plots = 2*4 = 8 df=8-1 = 7 Sub plots = 2*9*4 = 72 df=72-1=71-1missing=70 Error (b) = 423 From table: 210+216 = 426-3=423; Error (b) and Residual Error were combined Total = 575 – missing obs = 500 (75 obs missing) In second anova, for Removal, Error= 494 = 423 + 3 + 2*(8+3+24) –(1 + 1) , Strain, Plant and their interactions was pooled with Error Q2.16. Describe An Alternative Plan Of Statistical Analysis For The Described Model. Why the full model can not run? 16 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 1. Run a separate model for each Removal method Source DF r=4 MS E(MS) Block 3 2 e2 2r b2 72r a2 144 block Environment 1 e2 2 b2 72 a2 Q E Error(a) 3 e2 2 b2 72 a2 Strain 8 e2 2 b2 Q S Plant Species 3 e2 2 b2 Q P Strain*Plant Sp 24 e2 2 b2 Q S * P Env*Strain 8 e2 2 b2 Q E * S Env*PlantSp 3 e2 2 b2 Q E * P Env*Strain*PlantSp 24 e2 2 b2 Q E * S * P Residual 210 e2 2 b2 Total 287 2. The two removal methods are correlated, since they are measures taken from the same 1540 fresh individual leaves, create a new variable defined as the difference in population size between these two methods, and run model in 1. 3. Run analysis for each ENVIROMENT separate, include REMOVAL METHOD as repeated measures (pseudo-replication). 17 ST 524 Solution Final Take Home Exam NCSU - Fall 2008 Due: 12/09/08 4. Q3. This question ask you to write down a description of your research project, indicating Q3.1. Objective Q3.2. Response Variable Q3.3. Experimental design. Detailed description a. Indicate What Are The Different Experimental Units, i. Main-Unit ii. Sub-Unit (if any) iii. Sub-Sub Unit (if any) b. How Is A Block Defined? c. What Are The Factors And Their Type: Random Fixed, Q3.4. Present the Analysis of Variance table a. Sources of Variation (SOV) b. Degrees of Freedom c. Expected Mean Squares d. F test for each SOV Q3.5. What type of statistical tests do you plan to carried on results to answer your research questions: pairwise mean comparisons, contrasts, orthogonal polynomial contrasts, curve fitting, etc Q3.6. Do you have repeated measures, how do you plan to analyze them? References https://www.crops.org/publications/pdfs/CESGuide.pdf https://www.crops.org/publications/pdfs/cinstauthmans.pdf https://www.crops.org/publications/pdfs/jpr-instructions.pdf From North Dakota Agricultural Exp Station- Research Project Guidelines Procedures: This section is to provide a general design of the project. To begin, re-state each of the objective statements followed by a description of the procedures/methods for that objective. The procedure statements should show that the research needs and plans have been considered carefully and the proposed work has the potential to provide data and information which will permit accomplishing the objectives. While the details of the experimental design do not need to be specified, provide sufficient information to indicate that an appropriate design is planned. 18