Principal Components
advertisement

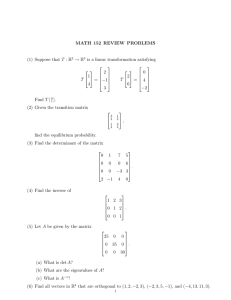
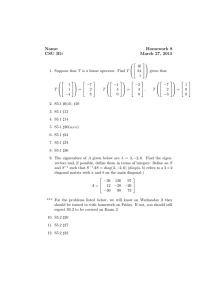
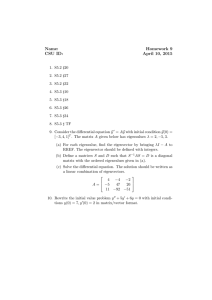
Determinant of a matrix |A| Examples: a b ad bc , c d 2 2 (2 )(3 ) 2 2 5 4 ( 1)( 4) 1 3 Eigenvalues of A: All for which |A-I|=0 2 2 has eigenvalues 1 and 4 (see above) 1 3 Example: A Facts: Determinants and eigenvalues (may be complex) exist for all square matrices. If |A|=0 then 0 is an eigenvalue and A has no inverse. Eigenvectors: For each eigenvalue , there exists a column vector X = (x1, x2)’ for which X’X=1 and AX = X Example: 2 2 1 1 4 2 2 x 4 x 1 so 1 3 x x 2 1 1/ does any multiple of it. Normalizing, we see that 1/ 2 1 1 4 and so 3 1 1 1 satisfies AX = X as 1 2 is one eigenvector. 2 2/ 5 2 2 1 1 2 2 1 1 is the 1 2 2 x 1 x 0.5 so 1 and 1/ 5 1 3 x x 1 3 0.5 0.5 other eigenvector. Note that any multiple of an eigenvector will also satisfy AX=X. Example: symmetric correlation matrix 1 1 1 1 (1 ) and 1 1 1 1 1 (1 ) so eigenvectors are 1 1 1 1/ 2 1/ 2 , 1/ 2 1/ 2 for the special case of a 2x2 correlation matrix. What are the eigenvalues? ___ ____ Now collect the 1 0 1 1 1 ' . Note that V V and we have shown that 2 1 1 0 1 0 1 1 V V . Multiplying through on the left by the transpose of V we see that 1 0 1 0 1 1 V V so if X1 and X2 are centered and scaled (mean 0, variance 1) random 1 1 0 eigenvectors in a matrix V = P1 x1 ( x1 x 2 ) / 2 P1 , the variance matrix of is V P2 x2 ( x1 x 2 ) / 2 P2 0 1 1 x1 the expected value of V x1 x2 V which is V V . This is a big deal 1 1 0 x2 variables with correlation then if since it means that we have converted correlated variables x1 and x2 to uncorrelated variables P1 and P2 by taking linear combinations of x1 and x2 based on the eigenvectors of the (x1 , x2) correlation matrix. The variance of P1 is seen to be 1+ and that of P2 is seen to be 1- which are the eigenvalues of the correlation matrix. By convention the eigenvalues are listed in descending order which imposes an order on the eigenvectors as well. The linear combinations P1 and P2, when applied to observed data, are the principal components corresponding to variables X1 and X2. In general, a kxk correlation matrix has k eigenvalues which are variances of the principal components and there are k associated eigenvectors which describe the directions in the k-dimensional data space in which the principal component axes point. Suppose we have an nxk data matrix X of n observations on k random variables. Suppose also that we have centered and scaled the data matrix so that the sample variance covariance matrix S has all 1s on the diagonal (in other words S is the correlation matrix of the centered and scaled data). This means that 1 1 X X Z Z S where Z X i.e. X n 1Z . The diagonal elements of S=Z’Z are all n 1 n 1 1. We can find matrices L (nxk) and R (kxk) such that Z=LDR’ where D (kxk) is a diagonal matrix, L’L=I and R’R=I. This is called the singular value decomposition of Z and the elements of D are called singular values. This implies that ZR=LD where D is a diagonal matrix. Note that ZR consists of k linear combinations of the columns of Z and hence of X. The ith linear combination uses the ith column of R to provide the weights in the linear combination. Note also that LD consists of linear combinations of the columns of L but since D is diagonal these are just multiples of the columns of L which are orthogonal to each other because L’L=Ik. Notice that R’SR=R’Z’ZR = D’L’LD=D’D=D2. Now D2 is diagonal, implying that the columns of ZR are orthogonal to each other and the sums of squares of the columns of ZR are the diagonal elements of D2. Multiplying both sides of R’Z’ZR =D2 by R on the left gives Z’ZR =SR= RD2 so it is no surprise that R is the matrix of eigenvectors of the sample correlation matrix S. Multiplying the left and right sides of this by (n-1) we see that similarly X’XR = R (n-1)D2 =R(D0)2 where (D0)2 =(n-1)D2 is diagonal, consisting of eigenvalues associated with the eigenvectors of X’X (columns of R) which we see are also the eigenvectors of S. Since the eigenvalues of S are variances of the centered and scaled data points, the elements of D0= n 1D are the corresponding standard deviations along the principal component axes. Note that X=LD0R’ is the singular value decomposition of X just as LDR’ is the singular value decomposition of Z. The principal components are the columns of P where P=LD0, a matrix whose columns are scalar multiples (D0) of orthogonal columns (L). We have shown these same principal components can be computed as linear combinations of the columns of X, namely P=XR. The elements of D0 are called singular values of X and are standard deviations along the principal component axes. Similarly the elements of D02 are variances along the principal component axes. Now replace L by its first few columns, say k’<k. Then the new LD has just k’ columns (k’ principal components). These principal components are also a new XR where the new R is just the first k’ columns of the old R. The new version of LD0R’ is the best rank k’ approximation to all the elements of X in the sense of least squares. That is, if you sum and square the elements of X minus the new LDR’ this is smaller than that of any other approximation to X that you can get by taking linear combinations of the columns of an nxk’ matrix. In a regression on k input variables we might get close to the same predictions by regressing on just k’ principal components. Because these are orthogonal we might then get some nice mathematical properties for the regression calculations but note that to get the principal components in the first place we need to measure all k of the input variables. If we have 5 X variables we might get an idea of what the 5 dimensional data scatterplot looks like by plotting the first 2 or 3 principal components by graphical methods we have already seen. Furthermore if we want to cluster the 5 dimensional vectors we might want to cluster based on just the first few principal components. Here is a small example in which we have two wraps A and B for some packages of frozen food we are transporting. Upon arrival at the destination we measure the temperatures of each box top and bottom for a two dimensional plot. You will see the directions of the principal component axes in the (x1,x2) space and the differences in variance along the two principal component axes. The second program shows the math worked out above. Run princomp.sas Run princomp2.sas