Scott Hollingsworth
advertisement
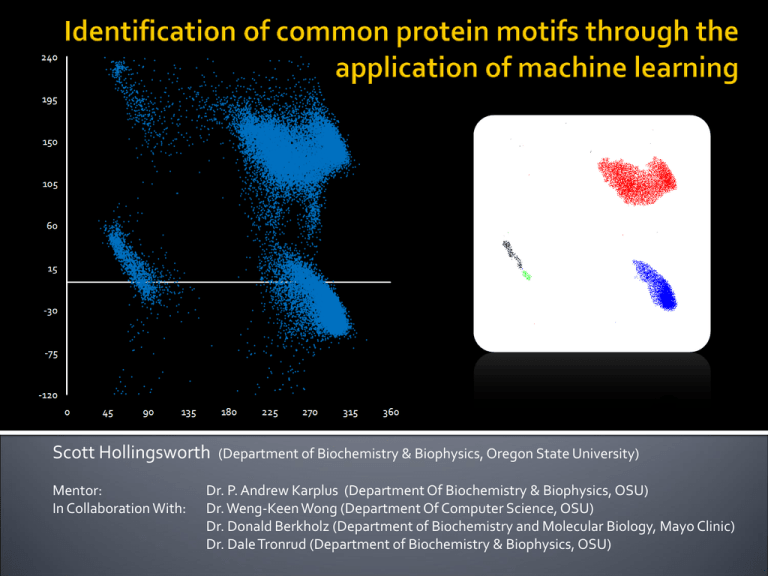
Scott Hollingsworth Mentor: In Collaboration With: (Department of Biochemistry & Biophysics, Oregon State University) Dr. P. Andrew Karplus (Department Of Biochemistry & Biophysics, OSU) Dr. Weng-Keen Wong (Department Of Computer Science, OSU) Dr. Donald Berkholz (Department of Biochemistry and Molecular Biology, Mayo Clinic) Dr. Dale Tronrud (Department of Biochemistry & Biophysics, OSU) Each protein has an individual structure Structure flows from function Understand structure, understand function Ptr Tox A Phi & Psi (φ, ψ) Phi and psi describe the conformation of the planar peptide (amino acid) in regards to other peptides One amino acid – two angles φ ψ Ramachandran Plot Voet, Voet & Pratt Biochemistry (Upcoming 4th Edition) 310 Helix Use of Protein Geometry Database (PGD) to identify linear group existence (i.e. α-helix, β-sheet, πhelix…) Simple repeating structures Methods: manual searches Hollingsworth et al. 2009. “On the occurrence of linear groups in proteins.” Protein Sci. 18:1321-25 α-Helix Linear groups are only part of the picture Not all common protein motifs are repeating structures Many have changing conformations Goal of this research: Identify all common motifs in proteins Too complex for manual searches Enter machine learning Form of artificial intelligence Can identify clusters within a dataset Cluster – significant grouping of data points Visual example… Topographical map of Oregon Data value: Elevation Mt. Hood (11,239 Feet) Mt. Jefferson (10,497 Feet) Three Sisters (10,358-10,047 Feet) Highest points (Individual peaks) Topographical map of Oregon Data value: Elevation Highest points (Individual peaks) Topographical map of Oregon Data value: Elevation TUALATIN HILLS OCHOCO STRAWBER RIES PAULINA MTS MAHOGANY MTS JACKASS MTS SISKIYOUS ( KALAMAT H ) HART MTN TROUT CREEK MTS Mountain ranges (Broad patterns) φ ψ Similar approach with our data 2-Dimensional Example α-helix β Abundance PII αL ψ Similar approach with our data 2-Dimensional Example φ Complications… Our Data: 4-dimensional dataset 4D to 2D distance conversions What has and hasn’t been observed? No definitive source Abundance / Peak Heights Machine learning programs can identify both previously documented and unknown common motifs and their abundances 1) Create and prep datasets with resolution of at least 1.2Å or higher, 1.75Å or higher 2) Run cuevas 3) Analyze identified clusters Automated process using Python to remove bias 4) Analyze context of motifs 2D-visual example of cuevas clustering Goal: Definitive list of the most common protein motifs In order of abundance “Everest” Method Locate “highest” peak first ▪ Bad pun : “Mt. Alpha-rest” Locate second highest peak Locate third……. Identifying motifs Search for peaks while looking for ranges Results: Definitive list of common protein motifs in order of abundance The list… Points Per Circle r=10 Degree2 5644 247 173 147 125 117 88 55 51 43 40 36 35 34 31 31 30 29 24 20 20 20 19 17 15 14 11 10 9 9 8 8 7 6 6 6 6 6 18.07 0.7909 0.5540 0.4707 0.4003 0.3747 0.2818 0.1761 0.1633 0.1377 0.1281 0.1153 0.1121 0.1089 0.0993 0.0993 0.0961 0.0929 0.0769 0.0640 0.0640 0.0640 0.0608 0.0544 0.0480 0.0448 0.0352 0.0320 0.0288 0.0288 0.0256 0.0256 0.0224 0.0192 0.0192 0.0192 0.0192 0.0192 φi ψi φi+1 ψi+1 -63.4 -125.5 -69.9 -65.5 -70.4 -57.2 -88.3 -88.1 -91.8 93.5 -133.9 -82.4 54.9 -122.3 -136.1 65.3 82.6 56.7 78 -78.3 -96.6 50.5 -69.9 -129.1 53.7 -87.6 76.3 78.8 -138.5 92.8 -107.6 84.6 -85.8 -102.4 -77.9 83.8 57.1 -128.3 -42 132.4 157.4 -21.4 153.6 131 -2 1.3 -1.9 -0.1 164.3 -26.8 38.3 119.6 70.4 28.3 5.6 -133.5 0.5 116 0.9 49.9 -32.3 80.8 48 61 -169.3 171.1 165.7 165.9 16.8 8.1 71.8 -9 -8.6 -166.3 44.5 98.7 -64 -118 -61 -90.3 -60.4 82.4 -64.7 87.9 -58.4 -71.7 -62.2 -146.3 84.5 52.7 -65 -67.2 -103.1 -73.7 -67.5 -89.1 -133.8 -61.2 -129.8 -70.3 -118.9 -140.3 -61.4 -69.3 57.7 -62.5 80 -143 -83.1 92.6 86.7 -121.9 -152.5 56.7 -40.6 130.2 -36.3 1.5 143 -0.6 136.9 5.7 -42.5 146 -34.1 152.1 0.8 41 -19 140.8 137.5 -10.7 -43.1 -31.1 156.3 148.3 73.1 141.9 126.6 149.5 138.3 -29.6 -137.8 -35.7 -177 169.3 163.5 163.3 174.2 132.1 158.8 -133.3 Residue i i+1 α β PII α PII PII δ δ δ δL β δ αL β ζ αL δL PII` δL PII δ αL α ζ αL γ` PII` PII` β ε δ δL γ` δ δ PII` αL ζ α β α δ PII δL PII δL α PII α β δL αL α PII β δ α δ β PII ζ PII β β PII α PII` α PII` β PII ε ε β β PII` Cluster Size Motif Name 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 3 1 1 1 1 2 1 1 1 2 1 2 1 1 1 3 4 1 1 1 1 α-helix / 310-helix β-strand PII- Helix N-Cap / Capping Box Type I Turn# PII Type II Turn Type I Turn Cap Schellman Motif Reverse Type I Turn Reverse Type II Turn βα Turn Classic Beta Bulge‡ Type I` Turn β → αL ζ → αP† G1 Beta Bulge δL → β Type II` Turn δL → α Type VIa1 Turn (S) Classic Beta Bulge (S) Wide Beta Bulge (S) α → ζ† ζ → PII αL → β (S) γ` Turn PII` → PII PII` → α (S) β → PII` ε→α Reverse Type II` Turn δL → β γ` → PII δ→ε δ → ε (S) PII` → β αL → β ζ → PII` New Motif X X X X X X X X X X X X X X X X X X X Motif “shapes” Results: Each motif analyzed by plotting New insight into each motif’s of each motif range Understand the shape of the cluster/motif structure Context Comparisons Example Cluster Shape Type II Vs. Type II` Type II Vs. Type II` Hairpin turns 180° Turn Two Residues Defined as mirror images of each other φ Distributions show differences between the two structures Nearly four years in the making… ψ The results go on… Motif analysis ▪ Viral forming of “Pangea” Range and peak method sections ▪ Adapting cuevas for our data ▪ Python automation ▪ Identification of 310 Helix & Type I Turn 6D, 8D, 10D and 12D clustering ▪ Full helix caps, loops, halfturns… For full story, a manuscript for publication is being prepared: Hollingsworth et al. “The protein parts list: motif identification through the application of machine learning.”(Unpublished) Cuevas was successful in identifying both documented and undocumented motifs Previously described: Linear groups, helix caps, β-turns (& reverses), β-bulges, α-turns, loops, helix bends, π-structures… Numerous new motifs Successful from 4D through 20D Results form the “Protein Parts” List Comprehensive list of all common protein motifs found in proteins • • • • • • Dr. P. Andrew Karplus Dr. Weng-Keen Wong Dr. Donald Berkholz Dr. Dale Tronrud Dr. Kevin Ahern Howard Hughes Medical Institute