Hollie's slides
advertisement

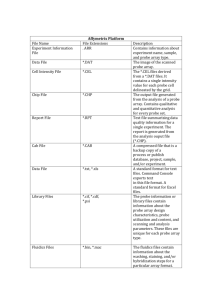
Microarrays A snapshot that captures the activity pattern of thousands of genes at once. Custom spotted arrays Affymetrix GeneChip Practical Applications of Microarrays Gene Target Discovery By allowing scientists to compare diseased cells with normal cells, arrays can be used to discover sets of genes that play key roles in diseases. Genes that are either overexpressed or underexpressed in the diseased cells often present excellent targets for therapeutic drugs. Pharmacology and Toxicology Arrays can provide a highly sensitive indicator of a drug’s activity (pharmacology) and toxicity (toxicology) in cell culture or test animals. This information can then be used to screen or optimize drug candidates prior to launching costly clinical trials. Diagnostics Array technology can be used to diagnose clinical conditions by detecting gene expression patterns associated with disease states in either biopsy samples or peripheral blood cells. Microarray Platforms •Oligonucleotide-based arrays •25mers spotted on a glass wafer, Affymetrix GeneChip arrays •Custom spotted 50-80mers generated from known sequences. •cDNA •Inserts from cDNA libraries •PCR products generated from gene specific or universal primers GeneChip Instrument System ® Fluidics Station Scanner made by Hewlett-Packard Computer Workstation GeneChip Probe Array ® GeneChip Probe Arrays ® GeneChip Probe Array Hybridized Probe Cell Single stranded, fluorescently labeled DNA target Oligonucleotide probe 1.28cm * * * * * 24µm Each probe cell or feature contains millions of copies of a specific oligonucleotide probe Over 250,000 different probes complementary to genetic information of interest Image of Hybridized Probe Array Synthesis of Ordered Oligonucleotide Arrays Light (deprotection) Mask OOOOO TTOOO HO HO O O O T– Substrate Light (deprotection) Mask CATAT AGCTG TTCCG TTCCO TTOOO C– Substrate REPEAT Probe Tiling Strategy Gene Expression (25-mer) Gene Expression Tiling Strategy [ ] [ ] [ [ Uninduced ] ] [ ] [ ] Induced 40 separate hybridization events are involved in determining the presence or absence of a transcript 80 separate hybridization events are involved determining differential gene expression of a transcript between two samples Starting material for Microarrays Platform Affymetrix Poly (A)+ mRNA Total RNA ~2 mg ~10 mg Spotted arrays Poly (A) + Total RNA ~0.4 – 2 mg 10 -100 mg Agilent Bioanalyzer 2100 Total RNA Fragmented cRNA Experimental Design Biotin - labeled cRNA transcript Cells B + Poly (A) RNA Or Total RNA IVT cDNA Biotin-UTP Biotin-CTP B B B B B B Fragment heat, Mg2+ B Hybridize B B B B Wash & Stain Scan (8 minutes) (75 minutes) (16 hours) Biotin - labeled cRNA fragments Add Oligo B2 & Staggered Spike Controls .DAT file and .CEL file .CHP file Absolute Analysis Normalization and Scaling Non-biological factors can contribute to the variability of data in many biological assays, therefore it is important to minimize the non-biological differences. Factors that may contribute to variation include: •Amount and quality of target hybridized to array •Amount of stain applied •Experimental variables The data can be normalized from: •a limited group of probe sets •all probe sets Thus the normalization of the array is multiplied by a Normalization Factor (NF) to make its Average Intensity equivalent to the Average Intensity of the baseline array. Normalization and Scaling Average intensity of an array is calculated by averaging all the Average Difference values of every probe set on the array, excluding the highest 2% and lowest 2% of the values. Background Calculation: Measure of non-specific fluorescence attributed to hybridization conditions and sample. Defaults - 16 sectors Horizontal (HZ) : 4 Vertical (VZ) : 4 Background • Probe Cells with the lowest 2 % intensity values for each sector are averaged. Probe Array • This value is subtracted from all cell intensities in each sector before further analysis. Signal Noise Noise results from small variations in the digitized signal observed by the scanner as it samples the probe array’s surface. The level of the noise is calculated by the software, and then used as one of the criteria to determine the significance of differences between PM and MM probe cells, and differences in probe set intensities across two probe arrays. Noise Calculation: Q Pixel to pixel variation determined from background Q 1/ N stdevi iallbgcells Total # of background cells - lowest 2% for each sector. Noise for each sector of a given probe array pixeli SF NF Total # of pixels in a feature i Standard deviation of the intensities of the pixels making up feature i Normalization Factor Scaling Factor What determines a positive or negative probe pair? Positive Probe Pair 1) PM - MM > SDT and 2) PM / MM > SRT PM is more than MM. Yes, this probe pair is detecting a signal. Negative Probe Pair 1) MM - PM > SDT and 2) MM / PM > SRT PM and MM are similar. More MM than PM. No differential signal Signal is not specific to detected. targeted sequence. Statistical Difference Threshold PM - MM > SDT • Calculated by the software based on the noise (Q): SDT = (Q) x (SDTmult) SDTmult (multiplier) is set by default to 2.0 when the single SAPE staining protocol is used (usually with 50m feature arrays), and 4.0 when the antibody amplification protocol is used (usually with 24m feature arrays). (SDTmult) can be modified by user: - increasing makes the analysis more stringent; decreasing less stringent Statistical Ratio Threshold PM / MM > SRT • SRT is set by user – increasing makes the analysis more stringent; decreasing less stringent • Default SRT value is 1.5 – an SRT threshold value of 1.5 means that the intensity of the PM must be 50% greater than MM (after background subtraction) to meet criteria Probe Pairs in Average Used in calculation of Log Average Ratio and Average Difference Pairs in Average A “Trimmed” probe set prevents outlier probe pairs (extremely positive or negative) from inclusion in calculations for Log Average Ratio (and Average Difference) 8 probe pairs or fewer: Greater than 8 probe pairs: • All probe pairs are • Super Scoring takes used place Super Scoring Used in calculation of Log Average Ratio and Average Difference A mean and a standard deviation are calculated for the intensity differences among an entire probe set. A filter is then applied to each member of the probe set. Probe pairs outside of the number of standard deviations set in the parameters are excluded from the calculations of Log Average Ratio and Average Difference STP is the parameter for setting the number of standard deviations used in Super Scoring. Default Setting is 3 (excludes everything outside of 99.7% of the mean) Positive Fraction Number of positive probe pairs/total pairs used 15/20 = 0.75 Log Average Ratio Log Avg = 10 * [ log (PM /MM)]/(# Probe Pairs in Average) An average of the log of the intensity ratios is calculated for each probe set from the pairs in average and multiplied by 10. Positive/Negative Ratio Ratio of positive probe pairs to Negative probe pairs in a probe set Pos/Neg = 18/2 = 9 Average Difference Avg Diff = (PM - MM) / Pairs in Average Average difference is calculated by taking the difference between PM and MM of every probe pair and averaging the differences over the entire probe set. Average difference correlates with expression level Average Difference is not used in the Absolute Call Decision Matrix Absolute Call Decision Matrix - Absolute Analysis Threshold Values Present Marginal 4.0 0.43 1.3 Max 3.0 0.33 0.9 Min Absent Pos/Neg Ratio Positive Log Avg Ratio Fraction Calls must be in the Present bin in order for quantification metrics to be informative Comparison Analysis Increased Probe Pairs & Decreased Probe Pairs Increased Probe Pair Neither Increased or Decreased Decreased Probe Pair Increased Probe Pair (PM - MM)exp - (PM - MM)base > Change threshold (CT) And [(PM - MM)exp - (PM - MM)base] / (PM - MM)base > (PCT)/100 Probe Set on Baseline Probe Array Probe Set on Experimental Probe Array Increased Probe Pairs Compares changes in relative intensity between two probe pairs on two probe arrays, not positive/negative probe pair changes Decreased Probe Pair (PM - MM)base - (PM - MM)exp > Change threshold (CT) And [(PM - MM)base - (PM - MM)exp] / (PM - MM)base > (PCT)/100 Probe Set on Baseline Array Probe Set on Experimental Array Decreased Probe Pair Compares changes in relative intensity between two probe pairs on two probe arrays, not positive/negative probe pair changes Thresholds used in comparison analysis Change Threshold (CT) • The CT can be calculated in either of two ways: • Calculated by the software, based on the SDTs of the two probe arrays being compared • Calculated as the product of a parameter called CT multiplier (CTmult) and Q. SDT 2 SDT 2 - applied if input field is blank exp bl CT OR CTmult max Qexp , Qbl - applied by setting CTmult CTmult is a default setting (80) or can be set by the user Percent Change Threshold (PCT) User Defined (default 80); means a probe pair must change 80% Increase or Decrease Ratio Increase Ratio = # Increased probe pairs / # probe pairs used Decrease Ratio = # Decreased probe pairs / # probe pairs used Probe Set on Baseline Array Probe Set on Experimental Array 10 Increased Probe Pairs / 20 = 0.5 Compares changes in relative intensity between two probe pairs on two probe arrays, not positive/negative probe pair changes Max (Increase/PP used),(Decrease/PP used) Calculates the number of probe pairs that have changed in a certain direction. Increase/PP used = number of increased probe pairs/number of probe pairs used Decrease/PP used = number of decreased probe pairs/number of probe pairs used Max Inc & Dec = Max (0.95, 0.05) = 0.95 This larger of the values will be used in the decision matrix, which determines whether each transcript’s expression level has changed between baseline and experimental. Increase/Decrease Ratio Ratio of increase probe pairs over decreased probe pairs Increased: 6 Decreased:1 Inc/Dec = 6/1 = 6 Dpos - Dneg Ratio Dpos-Dneg Ratio = positive change - negative change / # pp used Positive Change = # Positive Probe Pairsexp - # Negative Change = # Negative Probe Pairsexp - Positive Probe Pairsbase # Negative Probe Pairsbas • Dpos - Dneg Ratio flags and excludes probe sets that change in two directions () within one transcript. It also accounts for changes in the neither bin. Probe Set on Baseline Array 7 Positive PP Example: 3 Negative PP 10 Neither Probe Set on Experimental Array 14 Positive PP 4 Negative PP Positive Change = (14 - 7) = 7 Negative Change = (4 - 3) = 1 2 Neither Dpos -Dneg Ratio = (7 - 1) / 20 = 0.3 Log Average Ratio Change • Log Average is recomputed for each probe set based on probe pairs used in both the baseline and experimental probe arrays. Log Avg Ratio Change = Log Avgexp - Log Avgbase Example: Probe Set on Baseline Array Probe Set on Experimental Array 2 probe pairs Not in 1 probe pair Not in Average Average Total = 3 probe pairs Not in Average Log average is recomputed for each probe set to take into account any probe pairs that have been dropped (not in average) or masked Differential Call Comparison Analysis Threshold Values - Increase No Change Decrease 4.0 0.33 0.2 0.9 Max 3.0 0.43 0.3 1.3 Min Inc/Dec Ratio Inc/Total Dpos/Total Log Avg Ratio Calls must be in the Increase or Decrease bin in order for quantification metrics to be informative Average Difference Change • Average Difference is recomputed for each probe set based on common probe pairs to take into account any probe pairs that have been dropped (not in average) or masked Avg Diff Change = Avg Diffexp - Avg Diffbase Average difference change correlates to changes in expression level Average Difference Change is used in Fold Change calculations, but not used in the Comparison Call Decision Matrix B = A When an “ * ” is present in this column, it signifies that In the baseline array, this transcript was absent. Example: Absolute call A B = A * Diff Call I Define not significant, slight increase from A baseline to A experimental A * D not significant, slight decrease from A baseline to A experimental P * I IMPORTANT, increase in gene expression, A baseline to P exp. Fold Change: Measure of the relative change in mRNA expression levels between experiments. FC = Avg Diff Change (exp-base) (recomputed) max[ min (Avg Diff exp,Avg Diff base), QM * Qc] Lesser of the two values if (QM x QC) of either array is greater than the average difference of the transcript in either the baseline or experimental, the fold change is calculated using the Noise. In this case the fold change is preceded by a (~) and considered an approximation. Defined by the library file +1 or -1 AvgDiffexp AvgDiffbase AvgDiffexp AvgDiffbase Greater of the scaled or normalized Qexp or Qbase Sort Score Based on a calculation that basically multiplies Fold Change and Average Difference Change. The larger the Sort score, the more further away the values are from the noise. Example: Avg. Diff (baseline): 10 Avg. Diff (experimental) :100 Avg. Diff Change : 90 Fold change:10 Avg. Diff (baseline):100 Avg. Diff (experimental): 1000 Avg. Diff Change :900 Fold Change: 10 The fold change in both experiments is 10; however the Sort Score will be approximately 10 times larger in experiment #2 than #1, due to higher average difference change. A fold change with a high sort score means that the average difference change is relatively large. Data Analysis Save as *.txt file and import into other statistical software programs Data Visualization: Data visualization is an important technique for gaining a fundamental understanding of results of a microarray experiment. you can detect outliers/anomalies, overall trends, clusters, correlations with the following visual techniques. • 1-D Profile Plots - e.g. time series response data • Histogram / Frequency Plots - analyze distribution of gene expression data • Star Plots - signature analysis of gene expression profiles • Intensity Plots – color genes by gene expression across all exp’ts at once • Scatter Plots – allow you to visualize high-dimensional microarray data Integration: Integrated into your environment by reading files in standard formats and writing the results out in standard formats. • Import flat files, comma or tab separated Formats, or URL’s •Import from ODBC Data Source •Tiles Saved in Portable Comma Separated (.csv) Format •Automate Via Embedded Tcl Scripting Language •Link to Other Applications by Selecting Data in Spreadsheet or in Graphics. Data Processing: •Analytical Spreadsheet can Handle Millions of Rows or Columns •Scaling & Normalization (e.g. standardize, log-scale, log & linear scale, power) •Sort rows by Value or by Similarity to Prototype (find genes most similar to specified prototype) •Missing Data Handling (e.g. analysis, casewise deletion, imputation) Exploratory Analysis: New, unexpected discoveries are most easily made during the exploratory analysis stage. •Cluster analysis – identify genes with similar expression profile •Principal Components Analysis – visually and numerically analyze the correlation inherent in the data (similarity of genes, of experiments) •Multidimensional Scaling – visually analyze similarity of genes, tissues, or time points using any one of 20 measures of similarity. •Linear, Non-linear correlation – find significantly correlated genes, tissues, or time points. •Parametric & Nonparametric tests (e.g. chi-square, t-test, anova, kolmogorov-smirnov) – genes that are significantly different •Correspondence Analysis – measure the correspondence between (for example) a cluster analysis grouping and a known functional class of genes. •Randomization Experiments & Permutation Tests – evaluate likelihood of chance More Analysis Features Correlations •Find Genes with expression profiles similar to a chosen Gene •Find Expression Profiles similar to a drawn Profile •Multiple Ways to Define 'Similar' in the 'Find Similar' Search Quantitative Restrictions •Filter data by degree of expression or x-fold comparisons across experiments. Find Interesting Genes Function Pathways •Identify Potential New Candidates Modeling: Modeling tools allow us to make use of discoveries to build predictions on new, unknown data. For example, we can build a neural network (or other predictive model) capable of accurately categorizing tissue type or condition, based on just one, two, or a few genes alone. •Variable Selection – find the few genes needed to represent a profile for a particular phenotype. •Neural Networks – build a neural network to categorize new samples based on the gene expression of the few selected genes. •Discriminant Analysis (linear, quadratic) - build a statistical model to categorize new samples based on the gene expression of few selected genes. •Automated Model Validation - use cross-validation, jackknife, or bootstrap to estimate the accuracy of your sample identification model •Save Models for Prediction of new observations – save your predictive model for actual use. Cluster and Tree View Microarray Process Indirect labeling Simple, highly sensitive technique requires less starting RNA, and creates evenly labeled DNA without dye bias. •Uniform incorporation of fluorescent dyes produces more reliable signals •High sensitivity to detect lowcopy signals •Requires only 10 to 20 µg of total RNA or 0.4 to 1 µg of polyA RNA Clontech Atlas™ Glass Fluorescent Labeling Kit Stratagene FairPlay™ Microarray Labeling Kit Products used for spotting Easy-To-Spot™ Products (Incyte Genomics) •Every clone is sequence-verified prior to PCR • PCR products are purified to remove excess salts, unincorporated nucleotides, primers, and particulates • Quality controlled production process with failure rate1 of less than 10% • 8,734 PCR products from sequenced-verified clones from the UniGene database from NCBI, average length is greater than 500 nucleotides •Between 1-3 ug of DNA per well. Enough to fabricate 500 to 1,000 arrays • Corresponding clones available for purchase for further research Array Ready Oligo set (Operon Technologies) Complete Yeast Genome Oligo Set • Optimized 70-mer oligonucleotides for each of the 6,307 open reading frames (ORFs) of Saccharomyces cerevisiae from the Saccharomyces Genome Database (SGD) at Stanford University •The amount of sample provided with each set is sufficient to print between 2000 and 6000 slides, depending on the printing procedure used. Human Genome Oligo Set •This Array-Ready Oligo Set™ contains arrayable 70-mers representing 13,971 well-characterized human genes from the UniGene database. This database is located at the National Center for Biotechnology Information. •All 70-mer oligonucleotides in the Human Genome Oligo Set were designed from the representative sequences in the UniGene database, Hs build #119. The set also contains 29 controls. GeneMachine Omni Grid Arrayer Printing Pin Axon GenePix4000A Scanner • 10mm pixel size • Simultaneously scans array slides at two wavelengths • User-selectable laser power • User-selectable focus poisitions GenePix Pro Features • Auto Align Before Auto Align After Auto Align GenePix Pro Features •Feature Viewer P = pixel intensity F = feature intensity B = background intensity Rp = ratio of pixel intensities Rm = ratio of means mR = median of ratios rR = regression ratio GenePix Pro Features •Feature Pixel Plot GenePix Pro Features •Histogram GenePix Pro Features •Scatter Plot GenePix Pro Features • Results Spotted glass slide microarrays Advantages Low cost per array Custom gene selection Any species Competitive hybridization Open architecture Disadvantages Clone management Clone cost Quality control Affymetrix GeneChip system Advantages Stream line production Large number of genes and ESTs/chip Several number of species Disadvantages System cost GeneChip cost Propietary system Limits on customizing http://genome-www4.stanford.edu/cgi-bin/SMD/source/sourceSearch The Stanford Online Universal Resource for Clones and ESTs (SOURCE) compiles information from several publicly accessible databases, including UniGene , dbEST , SwissProt , GeneMapp99, RHdb , GeneCards and LocusLink. The mission of SOURCE is to provide a unique scientific resource that pools publicly available data commonly sought after for any clone, GenBank accession number, or gene. SOURCE was specifically designed to facilitate analysis of the large data sets that biologists can now produce using genome-scale experimental approaches. Choose organism: Choose search option: Enter a search term: Use a wildcard character (*) at the end and/or beginning of the term to broaden your search.Choose type of information to display: GeneReport: Gene Information (limited to those in UniGene) CloneReport: cDNA Clone Information (limited to those in dbEST) GeneCards Database http://207.123.190.10/ Dragon Database • annotate microarray data sets with biological information • search for genes or proteins that have shared characteristics • compare the genes represented on multiple array technologies Dragon View • view annotated microarray data Dragon Map • explore the map of human gene expression Challenges in analyzing Microarray Data •Amount of DNA in spot is not consistant •Spot contamination •cDNA may not be proportional to that in the tissue •Low hybridization quality •Measurement errors •Spliced variants •Outliers •Data are high-dimensional “multi-variant” •Biological signal may be subtle, complex, non linear, and buried in a cloud of noise •Normalization •Comparison across multiple arrays, time points, tissues, treatments •How do you reveal biological relationships among genes? •How do you distinguish real effect from artifact? Factors to consider in designing microarray experiments •Need to do lots of control experiments-validate method •Do replicate spotting, replicate chips, and reverse labeling for custom spotted chips •Do pilot studies before doing “mega chip” experiments •Don’t design experiment without replication; nothing will be learned from a single failed experiment •Design simple (one-two factor) experiments, i.e. treatment vs. untreatment •Understand measurement errors •In designing Databases; they are useful ONLY if quality of data is assured •Involve statistical colleagues in the design stages of your studies Once you have identified an interesting expression pattern, what comes next? •With some arrays it is possible to purchase clones of interest for further experimentation. •Confirm that the particular clone you now have in your hand shows the expression pattern so indicated by the array, quantitating individual mRNA species. •RT-PCR, Relative, quantitative RT-PCR uses an internal standard to monitor each reaction and allow comparisons between different reactions to be made. • Competitive RT-PCR --a competition between a known amount of a template and an unknown target. •Northern analysis