g nin tu to
advertisement

CASI, 20th May 2005
National University of Ireland, Galway
Jochen Einbeck and John Hinde
parameters
Work supported by
Making the EM algorithm for NPML estimation less sensitive to tuning
i=1
k=1
=⇒ Nonparametric maximum likelihood (NPML).
effect distribution g(zi) is made.
with mass points zk and masses πk , where no parametric assumption about the random
i=1
The marginal likelihood can be approximated by a finite mixture (Laird, 1978)
( K
)
Z
n
n
Y
Y X
L(β, g(z)) =
f (yi|zi, β)g(zi) dzi ≈
f (yi|zk , β)πk
µi ≡ E(yi|zi, β) = h(ηi) ≡ h(x0iβ + zi).
Generalized linear model with random effect:
NPML estimation
k=1
K
X
πk f (y|zk , σk2)
simultaneously.
the number of components K
the masses πk ,
the mass points zk and the variances σk2,
Aim: Estimate
where f (y|zk , σk2) is a normal density with mean zk and standard deviation σk .
f (y|(zk , πk , σk )k=1,...,K ) =
structure of a finite Gaussian mixture, i.e.
Assume data Y1, . . . , Yn sampled from a population Y , which is presumed to have the
A special case: Fitting finite Gaussian mixtures
15000
Finite Gaussian mixture?
10000
25000
galaxy$velocity
20000
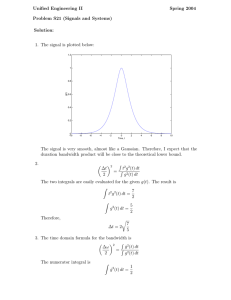
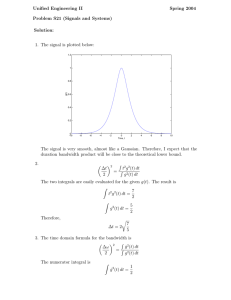
Recession velocities (in km/s) of 82 galaxies.
Example: Galaxy Data
30000
∂` − λ( πk − 1)
= 0,
∂πk
P
k=1
∂`
= 0,
∂σk
M-Step Update parameter estimates.
E-Step Adjust weights given current parameter estimates.
Starting points Select starting values zk0, πk0, and σk0, k = 1, . . . , K.
=⇒ can be solved by standard EM algorithm:
which turn out to be weighted versions of the single-distribution score equations.
∂`
= 0,
∂zk
and calculate the score equations
i=1
For fixed K, consider the log-likelihood
( K
)
n
X
X
`=
log
πk f (yi|zk , σk2) ,
Estimation
16.13 22.78 19.72 33.04
-2 log L:
380.9
0.423 0.043 1.721 0.626 0.922
Standard deviations:
0.085 0.024 0.512 0.342 0.037
Mixture proportions:
9.71
MASS1 MASS2 MASS3 MASS4 MASS5
Coefficients:
Set e.g. K=5:
0
20
EM Trajectories:
mass points
Application on Galaxy Data
35
30
25
20
15
10
60
EM iterations
40
80
1
n
P
(yi − ȳ)2.
the optimal mass points. This makes automatic starting point selection difficult.
The positions of the ’optimal’ starting points apparently ’have nothing to do’ with
The EM trajectories behave quite erratically in the first cycles, and tend to cross.
Finding the optimal solution requires a tedious grid search for tol.
where tol: scaling parameter, gk : Gauss-Hermite mass points, σ̂ =
zk0 = ȳ + tol ∗ σ̂ ∗ gk
Results depend heavily on the choice of starting points zk0, usually defined as
NPML estimates are ”impressively stable” (Aitkin, 1996) and reproducable.
The EM algorithm converges in every case.
Properties of current NPML implementations (as in GLIM 4)
successively until the likelihood ceases to fall.
There does not exist any automatic routine to estimate K. Usually, it is increased
(Richardson & Green, 1997).
”one of the things you do not know is the number of things that you do not know”
0
0
{ K, z10, . . . , zK
, π10, . . . , πK−1
, tol}.
– Set of tuning parameters to specify beforehand:
{K, z1, . . . , zK , π1, . . . , πK , σ1, . . . , σk }.
– Set of parameters to estimate:
General unsolved problem:
dj = 1 − (1 − tol)j ,
(0 < tol ≤ 1)
Optimal mass points are optimal starting points.
Reduces fluctuations and dependence on tol.
Damping has main effect in the first cycles.
i.e. d1 = tol and dj −→ 1 for j −→ ∞.
by the factor
Shrink estimated standard deviation σˆk of the mixture components in the j − th cycle
First Step: Damping the EM algorithm.
Possible improvements
0
20
40
80
EM iterations
0
61
No. iterations
93
60
0.45-0.82
opt. tol range
0.133-0.143
380.9
−2 log L
380.9
30
EM iterations
10
With damping
15
mass points
35
30
25
Without damping
mass points
10
20
35
30
25
20
15
10
50
20
25
velocity/1000
15
30
35
1
nh
i=1
K
³
.
10
20
25
velocity/1000
15
30
35
est. mixture components
Yi −y
h
´
components of a Gaussian mixture. But: Number of modes depends on bandwidth h.
Carreira & Perpiñan, 2003: The number of modes is a lower bound for the number of
10
estimated density
Idea: Consider density estimate fˆ(y, h) =
f
n
P
pi_k*f_k
Second step: Find optimal starting points
0.10
0.00
0.20
0.10
0.00
10
20
velocity/1000
15
25
The mode tree (Minnotte & Scott, 1993)
h
5
1
0.5
0.1
30
35
AIC (22 modes)
Silverman (3 modes)
BCV (1 mode)
Bandwidth selectors:
is reached.
(Silverman, 1981)
hcrit = inf{h, fˆ(·, h) has at most k modes}
From that bandwidth, climb down the mode tree until the next critical bandwidth
where A = min{σ̂, IQR/1.34}
hopt = 0.9An−1/2,
Calculate Silverman’s optimal bandwidth
Bandwidth selection in 2 steps
−15
−5
0
5
10 15
mass points
used as starting points for the EM algorithm.
0
20
30
EM iterations
10
40
a very accurate estimate for the location of the mass points, which then can be
an estimate for the number of modes, and thus for the number of components
Using hcrit, the mode tree gives
h
5
0.5
0.1
30
20
10
−15
−15
−5
−5
0
0
Climbing down the tree
h
h
5
0.5
0.1
5
0.5
0.1
5
5
10
10
15
15
mass points
mass points
30
20
10
30
20
10
0
0
5
15
20
100
EM iterations
50
30
150
25
EM iterations
10
variations in the bandwidth may change drastically the number of detected modes.
tion for the choice of K. However, this is no reliable automatic routine, as small
Mode trees together with a suitable bandwidth selector give a useful recommenda-
the multimodal structure cannot be seen in the data cloud itself.
Applied more general to the ’residuals’ h−1(Yi) − x0iβ̂ of a GLM, they also work if
The mode tree is a useful instrument to assess visually the number of components.
are so accurate that one hardly needs EM at all!
Starting point selection with mode trees works nicely given K. The starting points
made less sensitive to tuning parameters.
Applying a simple damping procedure, the EM algorithm could be stabilzed and
Summary
www.nuigalway.ie/maths/je/npml.html
..... is being implemented in an R package {npml} (Einbeck, Darnell, & Hinde), see
Variance component models
Random coefficient models
Include explanatory variables
Set an appropriate link function
Replace Gaussian by another exponential family distibution
Everything more general....
SILVERMAN. (1981): Using kernel density estimates to investigate multimodal regression. JRSSB, 43, 97–99.
Components (with discussion) JRSSB, 59, 731–792.
RICHARDSON, S. and GREEN, P. (1997): On Bayesian Analysis of Mixtures with an Unknown Number of
density features. JCGS, 2, 51-68.
MINNOTTE, M. C. and SCOTT, D. W. (1993): The mode tree: A tool for visualization of nonparametric
LAIRD, N. M. (1978): Nonparametric maximum likelhood estimation of a mixing distribution. JASA, 73, 805–811.
mixture. Lecture Notes in Computer Science, 2695, 625–640.
CARREIRA-PERPIÑAN, M. A. and WILLIAMS, C.K.I. (2003): On the number of modes of a Gaussian
ford, UK.
AITKIN, M., FRANCIS, B. and HINDE, J. (2005): Statistical Modelling in GLIM 4 (Second edition). Ox-
and Computing, 6, 251–262.
AITKIN, M. (1996): A general maximum likelihood analysis of overdispersion in generalized linear models. Statistics
References