vii i ii
vii
1
2
TABLE OF CONTENTS
TITLE CHAPTER
THESIS TITLE
DECLARATION
DEDICATION
ABSTRACT
ABSTRAK
ACKNOWLEDGEMENTS
TABLE OF CONTENTS
LIST OF TABLES
LIST OF FIGURES
LIST OF ABBREVIATIONS
LIST OF APPENDICES
INTRODUCTION
1.1
1.2
Background Study
Scope and Objectives of study
LITERATURE REVIEW
2.1 Introduction
2.2 Dyes and Pigments
2.2.1 History of Dyes
2.2.2 The basic of Colour in Dyes
2.2.3 Classification of Dyes
2.2.3.1 Azo dyes
2.2.3.2 Anthraquinone Dyes
2.3 Colour Removal
PAGE
v
vi
vii
xii
i
ii iii
iv
xiii
xv xviii
1
1
3
6
7
7
9
10
4
4
5
5
3
2.3.1 Measurement of Colour Removal
2.3.2 Mechanisms of Colour Removal
2.3.3 Factors Affecting Colour Removal
2.3.3.1 Oxygen
2.3.3.2 Temperature
2.3.3.3 pH Value
2.3.3.4 Dye concentration
2.3.3.5 Dye structure
2.3.3.6 Electron donor
2.3.3.7 Redox potential
2.3.3.8 Redox mediator
2.4 Wastewater Treatment Containing Dye
2.4.1 Bacterial Decolourization and Biodegradation
2.4.1.1 Anaerobic System
2.4.1.2 Aerobic System
2.4.1.3 Combined Anaerobic-Aerobic
Biodegradation
2.5 Azoreductase as Dye degrading enzyme
2.4.2 Others microorganism in biodegradation
2.6 Concluding Remarks
GENERAL MATERIALS AND METHODS
3.1 Growth Medium Preparation
3.1.1 Nutrient agar
3.1.2 Nutrient Broth
3.2 Synthetic Wastewater Medium: Chemical Defined
Medium
3.3 Culture Preparation
3.3.1 Growing of Microorganisms
3.3.2 Single Culture Preparation of Bacteria
3.3.3 Preparation of Inoculum and Maintenance
3.4 Growth Curve of Bacteria strain A
3.5 Decolourization of Azo Dyes viii
10
10
13
13
14
14
11
12
12
13
15
15
16
21
22
23
24
25
25
27
27
27
27
28
28
28
29
30
30
30
4
3.6 Chemicals and DNA Kits
3.7 General Outline of Research Methodology
SCREENING AND CHARACTERIZATION OF DYE
DEGRADER BACTERIA FROM TEXTILE
WASTEWATER
4.1 Introduction
4.2 Materials and Methods
4.2.1 Microorganisms
4.2.2 Media and Reagents Preparation
4.2.3 Inoculum Preparation and Culture
Maintenance
4.2.4 Screening of Potential Azo Dye-Discoloring
Bacteria
4.2.5 Optimization of Azo Dye SF BLACK EXA
Decolourization
4.2.6 Relationship of Growth and Decolourization
4.2.7 Analytical Methods
4.2.7.1 Determination of Azo Dye
Decolourization
4.3
4.2.7.2 Determination of Bacterial Growth
4.2.8 Identification of Selected Bacteria using
16S rDNA
4.2.8.1 DNA Extraction
4.2.8.2 Agarosegal Electrophoresis
4.2.8.3 Polymerase Chain Reaction (PCR)
Procedure
4.2.8.4 Purification of PCR Product
4.2.8.5 Sequencing of the 16S rRNA Gene
4.2.8.6 Analysis of 16S rRNA Gene and
Phylogenetic Tree Construction
Result and Discussion
4.3.1 Screening of Bacteria for Azo Dye
Decolourization ix
31
31
36
37
37
35
35
33
33
34
34
34
37
38
39
41
41
38
38
38
42
42
42
5 x
4.3.2 Relationship of Growth and Decolourization
4.3.3 Optimization of SF BLACK EXA
4.8
Decolourization 48
4.3.4 Identification of Selected Strain of Bacterium A 55
4.3.4.1Sequencing of the 16S rRNA Gene
4.3.4.2 Sequencing Analysis of Gene
55
Conclusion
Encoding for the 16S rRNA
LOCALIZATION OF AZOREDUCTASE AND
56
59
DEGRADATION ANALYSIS OF REMAZOL BLACK B
(RBB) BY Bacillus cereus STRAIN A
5.1 Introduction
5.2 Materials and Methods
5.2.1 Localization and Detection of Enzymatic
Activities
5.2.1.1 Preparation of Bacterial Cell Fraction
5.2.1.2 Azoreductase Assay
5.2.1.3 Effects of pH value and Temperature on theAzoreductase Activity and
60
60
62
62
62
63
Stability
5.2.2 Preparation of Inoculums and Biodegradation of Remazol Black B
5.2.2.1 Determination of CDW, ORP, pH,
COD and Colour Removal
5.2.3 Product Detection and Determination using HPLC
5.2.3.1 Sample Preparation
5.2.3.2 Chromatographic Apparatus
5.3 Result and Discussion
5.3.1 Localization of Azoreductase Produced by
Bacterium A
5.3.2 Optimization of Azoreductase Assay
5.3.2.1 Effect of pH
46
63
64
64
65
65
65
66
66
67
67
6 xi
5.3.2.2 Effect of Temperature
5.3.2.3 Effect of Incubation Time
5.3.2.4 Effect of NADH
5.3.2.5 Effect of FAD
5.3.2.6 Effect of Dye Concentration
5.3.3 Decolourization and Degradation of
Remazol Black B by Bacillus cereus Strain A
5.3.3.1 Time Course of RBB Degradation in Batch Culture
5.3.3.2 Kinetic Study on RBB Decolourisation by Bacillus cereus Strain A
5.3.3.3 Correlation between Specific
Decolourization Rate and Cell
Dry Weight
5.3.3.4 Correlation between Colour Removal,
COD and pH value
5.3.4 Product Detection and Determination using
HPLC under Facultative Anaerobic-Aerobic
Condition
5.3.4.1 Product Degradation Detection Using
5.4 Conclusion
Reverse phase-HPLC
68
69
70
72
73
72
74
75
77
78
79
79
83
CONCLUSION AND SUGGESTIONS
6.1 Conclusion
6.2 Suggestion for Future Work
REFERENCES
APPENDICES
84
84
86
87
107
xii
LIST OF TABLES
TITLE TABLE NO.
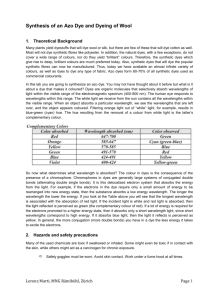
2.1 Region of the electromagnetic spectrum and relationship between wavelength and color
2.2
3.1
4.1
4.2
4.3
4.4
4.5
4.6
B iodegradation of dyes using single bacterial strain
Component of Chemically Defined Medium
Component mixes for electrophoresis
2X PCR Master Mix compositions
The universal primers used for the amplification of
16S rRNA
Components for PCR reaction
Thermal Profile for PCR Reaction
Decolourisation rate of azo dye under three different conditions: aerobic with shaking, partial aerobic (without
PAGE
6
17
28
39
39
40
40
41 shaking) and facultative anaerobic. The effect of azo dye decolourisation by all bacteria was tested using four
5.1 different types of azo dyes: SF RED 3BS, SFN BLUE
150%, SF BLACK EXA and SF YELLOW EXF.
Azoreductase activity determined from different fractions of cells assayed under anaerobic and aerobic condition
43
66
xiii
FIGURE NO.
2.1
2.2
LIST OF FIGURES
TITLE
Example of dye-auxochromes and dye-chromophores of azo dyes
PAGE
9
General overview of the fate of azo dyes and aromatic amines during anaerobic–aerobic treatment 23
3.1
4.2
A general outline of research methodology
Time course of the bacterial growth and decolourisation under (a) facultative anaerobic and (b) aerobic conditions in the medium adjusted
32
4.3
4.4 to pH 7, incubated at 37 ºC.
Decolourization of SF BLACK EXA by bacterium
47
A in CDM at 37 °C under facultative anaerobic condition at different pH values.
Decolourization of SF BLACK EXA by bacterium
49
A in CDM at 37°C after 7 days incubation under
4.5
4.6 facultative anaerobic condition at different
SF BLACK EXA concentration.
Decolourization of SF BLACK EXA by bacterium
51
A in CDM at 37°C after 14 days incubation under facultative anaerobic condition using different nitrogen sources at varying concentrations. 52
Decolourization of SF BLACK EXA by bacterium
A in CDM at 37 °C after 7 days incubation under facultative anaerobic condition using different carbon sources at varying concentrations. 53
5.1
5.2
5.3
5.4
5.5
5.6
5.7
5.8
4.7
4.8
4.9
5.9
5.10
5.11
5.12 xiv
Decolourization of SF BLACK EXA by bacterium
A in CDM at 37 °C after 7 days incubation under facultative anaerobic condition using different inoculum sizes.
The 1.5 kb of PCR product of 16S rRNA fragment obtained via PCR amplification.
Phylogram show phylogenetic relationships of
54
55 bacterium A based on partial 16S rRNA sequences retrieved from raw textile wastewater.
Effect of pH value on azoreductase activity.
Effect of temperature on azoreductase activity.
Effect of incubation time on azoreductase activity.
57
68
69
70
Effect of NADH concentration on azoreductase activity. 71
Effect of FAD concentration on azoreductase activity.
Effect of dye concentration on azoreductase activity.
73
74
Profile of decolourization rate, COD removal rate and cell dry weight by Bacillus cereus Strain A against time. 75
Kinetic study on decolourisation of Remazol Black B by Bacillus cereus Strain A in CDM at 35 °C under facultative anaerobic condition. 76
Correlation between decolourization rate and cell dry weight. 77
Time course of RBB biodegradation by Bacillus cereus
Strain A in CDM under facultative anaerobic-aerobic condition at 35 ºC.
The HPLC chromatogram of combined facultative anaerobic-aerobic treatment.
Chromatogram of sulfanilic acid standard in HPLC analysis.
79
81
82
LIST OF ABBREVIATIONS
ADMI
APHA
4-AP
ANS
BC
BOD
C.I
CaCl
2
°C
%
%h
-1
λ
ABS
3-ABS
4-ABS
5-ABS
CDM
CDW
Cl
Cl
-1
CO
2
COD d -1 dATP dCTP dGTP
DNA
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
Degree Celsius
Percentage percentage per hour
Wavelength
Aminobenzene sulphate
3-Aminobenzene sulphate
4-aminobenzene sulphate
5-aminobenzene sulphate
American Dye Manufacturer Institute
American Public Health Association
4-Aminophenol
Aminonaphythyl sulphate
Before Century
Biological Oxygen Demand
Colour Index calcium chloride
Chemically Defined Medium cell dry weight chloride ion chloride
Carbon dioxide chemical oxygen demand per day deoxyadenosine 5’-triphosphate deoxycytosine 5’ triphosphate deoxyguanosine 5’ triphosphate deoxyribonucleic acid xv
dNTP dTTP
K
2
HPO
4 kb
KH
2
PO
4 kg mg mgh
-1 min mol mgL -1
MgCl
2
MgSO
4
.7H
2
O mL mm mM mV
NA
NAD +
NADH
NADP
EDTA
Et Br
FAD
FAD
+
FADH
2
Fe
2+
FMNH
2 g gL -1 h h -1
H
2
O
HPLC
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
- deoxynucleotide triphosphate deoxythymidine 5’-triphosphate ethylene diamine tetra acetic acid
Ethidium bromide flavin adenine dinucleotide(oxidized) ion flavin adenine dinucleotide flavin adenine dinucleotide(reduced) ion ferum flavin adenine mononucleotide (reduced) gram gram per litre hour per hour hydrogen dioxide high performance liquid chromatography dipotassium hydrogen phosphate kilobase potassium dihydrogen phosphate kilogram milligram milligram per hour minute mole milligram per litre magnesium chloride magnesium sulphate heptahydrate milliliter millimeter milimol miliVolt nutrient agar nicotinamide adenine dinucleotide(oxidized) nicotinamide adenine dinucleotide(reduced) nicotinamide adenine dinucleotide phosphate xvi
TSS
U
UV
UV-vis v/v w/v
µgmL
-1
µL ƿM
NADPH
NB
NH
4
Cl
(NH
4
)
2
SO
4 ngmL
-1 nm
NO
3-
OD
600nm
PAAB
PCR pH ppm
Pt-Co
RBB
RM rpm rRNA
SO
4
2-
TAE
Tris
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
-
- xvii nicotinamide adenine dinucleotide phosphate(reduced) nutrient broth ammonium chloride ammonium sulphate nanogram per liter nanometer nitrate optical density at 600nm p-Aminoazobenzene polymerase chain reaction potential ion hydrogen part per million platinum cobalt
Remazol Black B redox mediator rotation per minute ribosomal RNA sulphate tris-acetate buffer
2-hydroxymethyl-2-methyl-1,3-propanediol total suspended solid enzyme unit ultraviolet ultraviolet-visible volume per volume weight per volume microgram per milliliter microliter picomolar
xviii
F
G
C
D
LIST OF APPENDICES
APPENDIX
A
B
TITLE
Full sequences of 16S rDNA for Bacillus cereus
Alignment score of full sequence of bacterium A obtained using BLASTn
Standard Curve of Remazol Black B concentration at
max
= 597nm
Correlation between OD 600nm and cell dry
E weight of Bacillus sp strain A
Standard Methods for the Examination of Water and Wastewater
Reversed phase-HPLC analytical parameters
Ln Cell Dry Weight µ1 and µ2 in Facultative
Anaerobic and Aerobic Conditions
PAGE
108
109
113
114
115
118
119