Centre for Computational Science: Biological and Biomedicine research highlights
advertisement

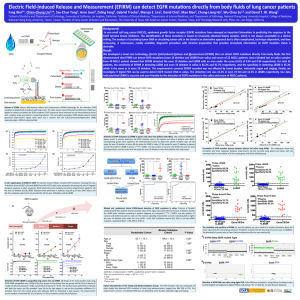
Centre for Computational Science: Biological and Biomedicine research highlights Director: Professor Peter Coveney • Centre established 2003 • Involved in many UK, EU and international projects. • Approximately 20 full time members • Range of problems: theoretical & computational science, computer science, distributed computing, visualisation and advanced networking. • Our different computational techniques span time and lengthscales from the macro-, through the meso- and to the nano- and microscales. • We are committed to studying new approaches and techniques that bridge these scales. • For more information see: http://ccs.chem.ucl.ac.uk Modelling for Clinical Decision Support: HIV Protease Simulation for Surgeons: Cerebral Blood Flow Our tool, HemeLB, simulates the flow in a region of interest and interactively generates images of flow velocity, pressure and vessel wall stress (such as shown to the left). These may eventually help surgeons make more informed decisions as to if and how to intervene. 1 Personal genetic sequence data soon available as we head towards $100 Genome Sequencing 3 cctcaaatcactctttggcaacgaccc atcgtcacaataaagataggagggc aactaagggaagctctattagataca ggagcagatgatacagtattagaaga cataaatttaccaggaagatggaaac caaaaatgatagggggaattggaggt tttgccaaagtaagacagtatgatcag atacccgtagaaatctgtggacataaa gttataggtacagtattagtagg ranking of drug binding affinities The causes of neurovascular pathologies (such as intracranial aneurysms) are not well understood but are likely to include properties of neurovascular blood flow. Simulation Wild-type & mutants LPV 2 A schematic representation of the process is shown above: vessel geometries are reconstructed from medical imaging data (typically CT rotational angiography), which are then input to the fluid flow simulation and results returned to inform decision making. Catalytic ASP dyad Build patient specific models of receptors Rank binding affinity against available drugs Select the one best suited to the individual 4 HPC allows atomic simulation on clinically relevant timescales Include in clinical decision support system Mutations In order to be of diagnostic relevance, the tool we aim to create must be fast and accurate. Our current work is focused on demonstrably improving these aspects. To the right are two images of the velocity field error (compared to high-resolution CFX simulation) in a realistic vessel geometry, before (left) and after (right) preliminary work. Sequence interpretation already routine in HIV treatment. Ideal examplar system to develop clinically relevant models. The same methodology can be applied to the treatment of other infectious diseases and medical conditions such as cancer. I. Stoica, S. K. Sadiq, and P. V. Coveney, JACS, 130, (8), 2639-2648, (2008) Drug Efficacy and Kinase Activation: Epidermal Growth Factor Receptor (EGFR) EGFR is a major target for drugs in treating lung carcinoma since it promotes cell growth and tumour progression. Its mutations promote conformational changes which affect its activation and drug efficacy. we have studied changes in drug binding affinities due to cancer mutations of EGFR using ensemble molecular dynamics simulations, and addressed an enhanced activation mechanism thought to be associated with mutations. Active Inactive state state Translocation of Polynucleotides through Protein Nanopores Polynucleotide (e.g. single stranded DNA) translocation through confined protein-pores is a system with experimental relevance in single channel current recording (SCCR, see right), which is a candidate for next gen genome sequencing. A We can use simulations to gain insight, providing knowledge to improve SCCR to the point of genetic sequencing. Since the system is complex and represents relatively large time scales, we speed up the process in order to allow highly detailed, atomistic, simulations. Binding Free Energy Calculator Binding Affinity Calculator (BAC) is used to determine patient specific response to lung cancer drugs. The workflow is fully automated for execution of each component and transfer of files across distributed HPC resources. Methods of rapid translocation include cv-SMD (left) or ABF (right). Mapping the conformational free energies of wild-type and mutant EGFRs onto a principal component space enables identification of population changes in various conformations upon mutation, EGFR mutation introduces conformational changes in the active and inactive states. These mutation-associated changes alter the drug efficacy by distorting the ATP- and drug-binding cleft, and adjust the activation of the EGFR kinase by affecting the relative stabilities of the active and inactive conformations. S. Wan and P. V. Coveney, J R Soc interface (2011) DOI: 10.1098/rsif.2010.0609 (right) Our simulations reproduce the experimental finding that polyadenine (“a”) translocation more slowly than polydeoxycytidine (“b”) (right) We confirm the observed trend, and provide additional insight: for example, a phosphate-lysine interaction between the DNA and protein, and a contribution of base-stacking