From: FLAIRS-01 Proceedings. Copyright © 2001, AAAI (www.aaai.org). All rights reserved.
Building a Large KnowledgeBase Semi-Automatically
Udo Hahn
Stefan Schulz
[-~ Text KnowledgeEngineering Lab
Freiburg University
Werthmannplatz 1
D-79085 Freiburg, Germany
hahn@coling,
uni-freiburg,de
Medical Informatics Department
Freiburg University Hospital
Stefan-Meier-Str. 26
D-79104 Freiburg, Germany
s tschulz@uni-freiburg,
de
Abstract
Wedescribe a knowledgeengineeringapproachby which
conceptualknowledge
is extracted froman informal, semantically weakmedicalthesaurus (UMLS)
and automatically convertedinto a formallysounddescriptionlogics
system. Our approachconsists of four steps: concept
definitions are automatically generated fromthe UMLS
source, integrity checkingof taxonomicand partonomic
hierarchiesis performed
by the terminologicalclassifier,
cyclesand inconsistenciesare eliminated,andincremental
refinementof the evolvingknowledge
base is performed
by a domainexpert. Wereport on experimentswith a terminological knowledgebase composedof 164,000 concepts and76,000relations.
Introduction
Over several decades, an enormousbody of medical knowledge, e.g, disease taxonomies, medical procedures, anatomical terms etc., has been assembled in a wide variety of
medical terminologies, thesauri and classification systems.
The conceptual structuring of a domainthey allow is typically restricted to the provision of broader/narrowerterms,
related terms or (quasi-)synonymous terms. This is most
evident in the UMLS,the Unified Medical LanguageSystem (McCray & Nelson 1995), an umbrella system which
covers more than 50 medical thesauri and classifications.
Its metathesaurus componentcontains more than 600,000
concepts which are structured in hierarchies by 134 semantic types and 54 relations between semantic types. Their
semanticsis shallow and intuitive, whichis due to the fact
that their usage is primarily intended for humansengaged
in various forms of clinical knowledgemanagement.
Givenits size, evolutionary diversity and inherent heterogeneity, there is no surprise that the lack of a formal
foundationleads to inconsistencies, circular definitions, etc.
(Cimino1998). This maynot cause utterly severe problems
whenhumansare in the loop and its use is limited to disease
encoding, accountancy or documentretrieval tasks. However, anticipating its use for moreknowledge-intensiveapplications such as natural language understanding of medical narratives (Hahn, Romacker,&Schulz 1999) or medical
°Copyright
(~) 200I, American
Association
for Artificial Intelligence(www.aaai.org).
All rights reserved.
222 FLAIRS-2001
decision support systems (Reggia & Tuhrim1985), those
shortcomingsmight lead to an impasse.
As a consequence, formal models for dealing with medical knowledgehave been proposed, using representation
mechanismsbased on conceptual graphs, semantic networksor description logics (Volot et al. 1994; Mayset al.
1996; Rector et al. 1997). Not surprisingly, however,there
is a price to be paid for more expressiveness and formal
rigor, viz, increasing modelingefforts and, hence, increasing maintenance costs. Therefore, concrete systems making full use of this rigid approach, especially those which
employ high-end knowledgerepresentation languages are
usually restricted to rather small subdomains.
The knowledgebases developed within the frameworkof
the above-mentionedterminological systems have all been
designed from scratch - without makingsystematic use of
the large bodyof knowledgecontained in those medical terminologies. An intriguing approach would be to join the
massive coverage offered by informal medical terminologies with the high level of expressivenesssupported by formal inferencing systems, as developed in the AI knowledge
representation community,in order to develop formally
solid medical knowledgebases on a larger scale. This idea
has already been fostered by Pisanelli, Gangemi,&Steve
(1998) who extracted knowledge from the UMLSsemantic networkas well as from parts of the metathesaurusand
merged them with logic-based top-level ontologies from
various sources. In a similar way, Spackman& Campbell
(1998) describe how SNOMED
(C6t6 1993) evolves
a multi-axial coding systeminto a formally foundedontology. Unfortunately, the efforts madeso far are entirely focused on generalization-based reasoning along is-a hierarchies and lack a reasonable coverage of partonomies.
Part-Whole Reasoning
As far as medical knowledge is concerned, two main
hierarchy-buildingrelationships can be identified, viz. is-a
(taxonomic) and part-whole (partonomic) relations. Unlike
generalization-based reasoning in concept taxonomies, no
fully conclusive mechanismexists up to nowfor reasoning
along part-whole hierarchies in description logic systems.
For medical domains, however,the exclusion of part-whole
reasoning is far from adequate. Anatomical knowledge,
a central portion of medical knowledge,is principally organized along part-whole hierarchies. Hence, any proper
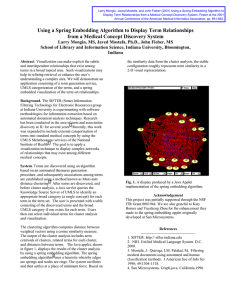
Figure 1:
onomies
SEPTriplets: Partitive Relations within Tax-
medical knowledgerepresentation has to take account of
both hierarchy types (Haimowitz,Patil, &Szolovits 1988).
Various approaches to the reconstruction of partwhole reasoning within the object-centered representation
paradigm are discussed by Artale et al. (1996). In the
description logics communityseveral language extensions
have been proposedbased on special constructors for partwhole reasoning (Rector et al. 1997; Horrocks & Sattler
1999), though at the cost of increasing computationalcomplexity. Motivated by informal approaches sketched by
Schmolze & Mark (1991) we formalized a model of partwhole reasoning (Hahn, Schulz, & Romacker1999) that
does not exceed the expressiveness of the well-understood,
parsimonious concept language .AEC (Schmidt-Schaul3
Smolka 1991))
Our proposal is centered arounda particular data structure, so-called SEPtriplets, especially designed for partwholereasoning(cf. Figure 1). Theydefine a characteristic
pattern of IS-A hierarchies which support the emulation of
inferences typical of transitive PART-OF
relations. In this
formalism, the relation ANATOMICAL-PART-OF
describes
the partitive relation betweenphysical parts of an organism.
A triplet consists, first of all, of a composite’structure’
concept, the so-called S-node (e.g., HAND-STRUCTURE).
Each ’structure’ concept subsumesboth an anatomical entity and each of the anatomicalparts of this entity. Unlike
entities and their parts, ’structures’ have no physical correlate in the real world-- they constitute a representational
artifact required for the formal reconstruction of systematic
patterns of part-whole reasoning. The two direct subsumees
of an S-nodeare the correspondingE-node(’entity’) and
node (’part’), e.g., HAND
and HAND-PART,
respectively.
Unlike an S-node, these nodes refer to specific ontological
objects. The E-node denotes the whole anatomical entity
to be modeled, whereas the P-node is the commonsubsumer of any of the parts of the E-node. Hence, for every P-nodethere exists a correspondingE-nodefor the role
ANATOMICAL-PART-OF.
Some basic anatomical relations
in terms of SEPtriplets are illustrated in Figure 2.
I.A£Callowsfor the constructionof hierarchies of concepts
and relations, whereE denotessubsumption
and -- definitional
equivalence.
Existential(B) anduniversal(V)quantification,negation (-~), disjunction(I-1) andconjunction
(t_l) are supported.
filler constraints(e.g., typingby(7) are linkedto the relationname
R by a dot, BR.(7.
Figure 2: Partonomic Hierarchy of the Concept HAND
The reconstruction of the relation ANATOMICAL-PARTOF by taxonomic reasoning proceeds as follows. Let us
assume that CE and DE denote E-nodes, Cs and Ds denote the S-nodes that subsumeCEand DE, respectively,
and Cp and Dp denote the P-nodes related to CEand DE,
respectively, via the role ANATOMICAL-PART-OF
(cf. Figure 1). Theseconventions can be captured by the following
terminological expressions:
CE E_ Us E Dp E_ Ds
(1)
DE E_ DS
(2)
The P-node is defined as follows (note the disjointness
between D~ and Dp):
Dp - Ds n -~DE r7 Banatomical-part-of .DE
(3)
Since CEis subsumedby Dp(1) we infer that the relation ANATOMICAL-PART-OF
holds between CE and DE :
C E E 3anatomical-part-of.De
(4)
Knowledge Import and Refinement
Our goal is to extract conceptual knowledge from two
highly relevant subdomainsof the UMLS,
viz. anatomyand
pathology, in order to construct a formally sound knowledge base using a terminological knowledgerepresentation
language. This task will be divided into four steps: (I)
the automatedgeneration of terminological expressions, (2)
their submissionto a terminological classifier for consistency checking, (3) the manualrestitution of formal consistency in case of inconsistencies, and, finally, (4) the manual rectification and refinementof the formal representation
structures.
KB SYSTEMS223
~7
I
CHD " C0014261
~7
C0005847
CHD
CHD
C0014261
C0025862
C0005847
CHD
C0026844
~47
C0005847
CHD
GriD
C0005847CHD
C0005847 CHD
~7
CHD
C0005847CHD
C0005847CHD
CSP98
isa
C0026844 partof
C0034052
C0035330 rsa
C0042366
partof
C0042367
pad_of
C0042367
C0042449 IS8
CSP98
SNMI98
MSI-IgQ
MSI-I~
SNM2
MSH99
MSH99
C6P98
MSH99
M81-1~
CSP98
6NMI98
MSH99
MSI-199
MSH99
SNM2
M81-199
Figure 3: SemanticRelations in the UMLS
Metathesaurus
Step 1: Automated
Generationof TerminologicalExpressions. Sourcesfor concepts and relations were the
UMLS
semantic network and the mrrel, mrconand mrsty
tables of the 1999release of the UMLS
metathesaurus.
The mrrel table whichcontains approximately7,5 million
records (cf. Figure3) exhibits the semanticlinks between
two UMLS
CUIs(concept unique identifier), 2 the mrcon
table contains the conceptnamesand mrsty keepsthe semantictype(s) assignedto each CUI.Thesetables, available as ASCIIfiles, wereimportedinto a MicrosoftAccess
relational database and manipulatedusing SQLembedded
in the VBAprogramminglanguage. For each CUIin the
mrrel subset its alphanumeric
codewassubstituted by the
Englishpreferred term foundin mrcon.
After a manualremodelingof the 135 top-level concepts
and 247 relations of the UMLS
semanticnetwork, weextracted, froma total of 85,899concepts, 38,059anatomy
and 50,087 pathology concepts from the metathesaurus.
Thecriterion for the inclusion into oneof these sets was
the assignmentto predefinedsemantictypes. Also, 2,247
conceptswerefoundto be includedinto both sets, anatomy
and pathology. Since wewantedto keep the two subdomainsstrictly disjoint, wemaintainedthese 2,247concepts
duplicated, and prefixed all conceptsby ANAor PAT-accordingto their respective subdomain.This can be justified by the observationthat these hybridconceptsexhibit,
indeed, multiple meanings.For instance, TUMOR
has the
meaningof a malignantdisease on the onehand, and of an
anatomicalstructure on the other hand.
As target structures for the anatomydomainwechose
SEPtriplets. Thesewere expressedin the terminological
language LOOM
which we had previously extended by a
special DEFTRIPLET macro(cf. Table 1 for an example).
OnlyUMLS
part-of, has-part and is-a relation attributes
are consideredfor the constructionof taxonomic
andpartonomichierarchies. Hence,for each anatomyconcept, one
SEPtriplet is created. Theresult is a mixedIS-AandPARTWHOLE
hierarchy.
For the pathologydomain,wetreated CHD(child) and
RN(narrower relation) from the UMLS
as indicators
2As a coding convention in UMLS,any two CUIsmust be connected by at least a shallow
relation(in Figure3, CHilD
relations
in the columnRELare assumedbetween CUIs). These shallow relations maybe refined in the columnRELA,
ifa thesaurus is available which contains more precise information. Some CUIs are
linked either by pan-of or is-a. In any case, the source thesaurus
for the relations and the CUIsinvolved is specified in the columns
X and Y (e.g., MeSH1999, SNOMED
International 1998).
224
FLAIRS-2001
(deftriplet HEART
:is-primitive
HOLLOW-VISCUS
:has-part (:p-and
FIBROUS-SKELETON-OF-HEART
WALL-OF-HEART
CAVITY-OF-HEART
CARDIAC-CHAMBER-NOS
LEFT-SIDE-OF-HEART
RIGHT-SIDE-OF-HEART
AORTIC- VALVE
PULMONARY-VALVE
Table 1: GeneratedTriplets in LOOM
Format
taxonomiclinks. Nopart-wholerelations wereconsidered,
since this categorydoesnot applyto the pathologydomain.
Furthermore,
for all anatomy
conceptscontainedin the definitional statementsof pathologyconceptsthe ’S-node’is
the default conceptto whichthey are linked, thus enabling
the propagation
of roles across the part-wholehierarchy.
In both subdomains,shallowrelations, such as the extremely frequent sibling SIB relation, wereincluded as
comments
into the codeto give someheuristic guidancefor
the manualrefinementphase.
Step 2: Submission to the LOOM
Classifier. The
import of UMLS
anatomyconcepts resulted in 38,059
DEFTRIPLET
expressions for anatomicalconcepts and and
50,087 DEFCONCEPT
expressions for pathological concepts. Each DEFTRIPLET
was expandedinto three DEFCONCEPT(S-, E-, and P-nodes), and two DEFRELATION
(ANATOMICAL-PART-OF-X,INV- ANATOMICAL-PART-OF-
x) expressions, summingup to 114,177concepts. This
yielded (togetherwith the conceptsfromthe semanticnetwork)a total of 240,764definitory LOOM
expressions.
From38,059anatomytriplets, 1219DEFTRIPLET
statementsexhibited a :HAS-PART
clause followedby a list of
a variable numberof triplets, containing morethan one
argumentin 823 cases (average cardinality: 3.3). 4043
DEFTRIPLET
statements containeda :PART-OF
clause, only
in 332 cases followedby morethan one argument(average cardinality: 1.1). Theresulting knowledgebase was
then submittedto the terminologicalclassifier andchecked
for terminologicalcycles and coherence.In the anatomy
subdomain,one terminologicalcycle and 2328incoherent
conceptswerefound, in the pathologysubdomain
355 terminologicalcycles thoughnot a single incoherentconcept
weredetermined(cf. Table2).
Step 3: ManualRestitution of Consistency.The inconsistencies of the anatomypart of the knowledgebase
identified bythe classifier couldall be tracedbackto the
simultaneous
linkageof twotriplets by both is-a andpartof links, an encodingthat raises a conflict dueto the disjointness required for correspondingP- and E-nodes.In
mostof these cases the affectedparentsbelongedto a class
of concepts that obviouslycannot be appropriately modeled as SEPtriplets, e.g., SUBDIVISION-OF-ASCENDINGAORTA, ORGAN-PART. The meaningof each of these concepts almostparaphrasesthat of a P-node,so that in these
Triplets
defconcept
statements
cycles
inconsistencies
II Anatomy
Pathology
38,059
114,177
1
2,328
50,087
355
0
\
Table 2: Classification Results for the ConceptImport
Anltomlr.al-Pad.Of
(
cases the violation of the SEP-internal disjointness condiIsA between P - and S - nodes i
Has-Anatomical-Part
tion was resolved by substituting the involved triplets with
simple LOOM
concepts, by matching them with already exFigure 4: Part-whole ReasoningPatterns with SEPTriplets
isting P-nodes or by disabling IS-A or PART-OF
links.
cept definitions required the inclusion of anatomic or
In the pathologypart of the knowledgebase, we expected
partonomiclinks. Often, necessary taxonomicor partoa large numberof terminological cycles, as a consequence
nomic parents were already available, but not coded
of interpreting the thesaurus-style narrowerterm and child
as UMLSparents or broader concepts. 7 from 100
relations through taxonomic subsumption (IS-A). Bearanatomyconcepts had to be considered as misclassified
ing in mind the size of the knowledgebase, we consider
by the UMLS.
355 cycles a tolerable amount. Those cycles were primarily due to very similar concepts, e.g., ARTERIOSCLERO- ¯ Check of the :has-part arguments assuming ’real
SIS vs. ATHEROSCLEROSIS,
AMAUROSIS
vs. BLINDNESS,
anatomy’. In the UMLS
sources part-of and has-part
and residual categories ("other", "NOS"= not otherwise
relations are considered as symmetric. According to
specified). These were directly inherited from the source
our transformation rules, the attachment of a role HASterminologies and are notoriously difficult to interpret out
ANATOMICAL-PART
tO an E-node BE, with its range
of their definitional context, e.g., OTHER-MALIGNANT- restricted to AEimplies the existence of a concept A
NEOPLASM-OF-SKINvs. MALIGNANT-NEOPLASM-OFfor the definition of a conceptB. Onthe other hand, the
SKIN-NOS.
The cycles were analyzed and a negative list
classification of AEas being subsumedby the P-node
which consisted of 630 concept pairs was manuallyderived.
Be, the latter being defined via the role ANATOMICALIn a subsequentextraction cycle weincorporated this list in
PART-OF
restricted to BE, implies the existence of BE
the automated construction of the LOOM
concept definigiven the existence of AE.These constraints do not altions, and given these new constraints, a fully consistent
ways conformto ’real’ anatomy, i.e., anatomical conknowledgebase was generated.
cepts that mayexhibit pathological modifications. FigStep 4: ManualRectification and Refinement of the
ure 4 (left) sketches a concept A that is necessarily
KnowledgeBase. This step - when performed for the
ANATOMICAL-PART-OF
a concept B, but whose exiswhole knowledge base - is time-consuming and requires
tence is not required for the definition of B. This is typbroad and in-depth medical expertise. An analysis of ranical of the results of surgical interventions, e.g., a large
domsamples from both subdomainsis currently being perintestine without an appendix.
formedby a domainexpert. The preliminary data we supply
Results: The analysis of 15 triplet definitions that exrefer to the analysis of two randomsamples of each onehibit automatically generated :HAS-PART
clauses rehundred anatomy and one-hundred pathology concepts.
vealed
that
34%
of
the
concepts
should
be
eliminated
From the experience we gained so far, the following
from
the
:HAS-PART
list
in
order
not
to
obviate
a coworkflowsteps can be derived:
herent classification of pathologically modifiedanatom¯ Checking the correctness of both the taxonomic and
ical objects) The opposite situation is also common
partitive hierarchies. Taxonomic
and partitive links are
(cf. Figure 4, right): the definition of AEdoes not
manually added or removed. Primitive subsumption
imply that the role ANATOMICAL-PART-OF
be filled by
is substituted by non-primitive subsumptionwhenever
BE, but BEdoes imply that the inverse role be filled
possible. This is a crucial point, because the automatby AE. As an example, a LYMPH-NODE
necessarily
ically generated hierarchies contain only information
contains LYMPH-FOLLICLES,
but there exist LYMPHabout the parent concepts and necessary conditions. As
FOLLICLES
that are not part of a LYMPH-NODE.
an example, the automatically generated definition of
¯ Analysis of the sibling relations and defining concepts
DERMATITIS
includes the information that it is an INas being disjoint. In UMLS,SIB relates concepts that
FLAMMATION,
and that the role HAS-LOCATION
must
share the same parent in a taxonomicor partonomichibe filled by the concept SKIN. An INFLAMMATION
that
erarchy. Pairs of sibling concepts may have common
HAS-LOCATION
SKIN, however, cannot automatically
be classified as DERMATITIS.
3In the example
of Table1, the conceptsprintedin italics, viz.
Results: Taxonomiclinks had to be removed from 8
AORTIC-VALVEand PULMONARY-VALVE
should be eliminated
out of 100 sampled pathology concept definitions, but
fromthe :HAS-PART
list, becausethey maybe missingin certain
from none of the anatomyconcept definitions. On the
cases as a result of congenitalmalformations,inflammatory
procontrary, in 68 cases from this sample, anatomy concessesor surgicalinterventions.
KB SYSTEMS226
descendantsor not. If not, they constitute the root of two
disjoint subtrees. In a taxonomichierarchy, this means
that one conceptimplies the negation of the other (e.g.,
a benignant tumor cannot be a malignant one, et vice
versa). In a partitive hierarchy, this canbe interpreted as
spatial disjointness, viz. one conceptdoes not spatially
overlap with another one. As an example, ESOPHAGUS
and DUODENUM
are spatially disjoint, whereas STOMACHand DUODENUMare not (they share a common
transition structure, called PYLORUS),
such as all neighbor structures that have a surface or region in common.
Spatial disjointness can be modeledsuch that the definition of the S-nodeof the concept A implies the negation
of the S-nodeof the concept B.
Results: The large numberof sibling concepts (on the
average 7.3 siblings per concept in the anatomy,8.8 in
the pathology subdomain) makes the modeling of disjointness a time-consumingtask, as every pair of concepts must be analyzed. At first glance, our data indicate
that conceptualdisjointness holds for at least two-thirds
of the sibling conceptsin both domains,and spatial disjointness for over three quarters in the anatomydomain.
¯ Completion and modification of anatomy-pathologyrelations. Surprisingly, only very few pathology concepts contained an explicit reference to a corresponding
anatomy concept. These relations must, therefore, be
added by a domainexpert. In each case, the decision
must be madewhether the E-node or the S-node has to
be addressed as the target concept for modification. In
the first case, the propagationof roles across part-whole
hierarchies is disabled, in the secondcase it is enabled.
Results: In an analysis of a random sample of 100
pathology concepts, only 17 were found to be linked
with an anatomyconcept. In 15 cases, the default linkage to the S-node was considered to be correct, in one
case the linkage to the E-nodewas preferred. In another
case, the linkage was consideredfalse.
Conclusions
Instead of developing sophisticated medical knowledge
bases from scratch, we here propose a ’conservative’ approach -- reuse existing large-scale resources, but refine
the data from these resources so that advancedrepresentational requirements imposedby more expressive knowledge
representation languagesare met.
The knowledge engineering approach we propose does
exactly this. It provides a formally solid description logics frameworkwith a modeling extension by SEPtriplets
so that both taxonomicand partonomic reasoning are supported equally well. While pure automatic conversion from
semi-formal to formal environmentscauses problemsof adequacyof the emergingrepresentation structures, the refinement methodologywe propose already inherits its power
from the terminological reasoning framework. In our concrete work, we found the implications of using the terminological classifier, the inference engine which computes
subsumptionrelations, of utmost importance and of out226
FLAIRS-2001
standing heuristic value. Hence, the knowledgerefinement
cycles are truly semi-automatic,fed by medicalexpertise on
the side of the humanknowledgeengineer, but also driven
by the reasoning system which makes explicit the consequencesof (im)proper concept definitions.
Acknowledgements.Wewould like to thank our colleagues
in the CLIFgroupfor fruitful discussionsandinstant support.St.
Schulzwas partly supportedby a grant fromDFG(Ha2097/5-2).
References
Artale, A.; Franconi,E.; Guarino,N.; and Pazzi, L. 1996.Partwholerelations in object-centeredsystems:an overview.Data
&KnowledgeEngineering20(3):347-383.
Cimino,J. J. 1998.Auditingthe UnifiedMedicalLanguage
System with semantic methods.Journal of the AmericanMedical
lnformaticsAssociation5(1):41--45.
C6t6, R. 1993. SNOMED
International. Northfield, IL: College
of American
Pathologists.
Hahn, U.; Romacker,M.; and Schuiz, S. 1999. Howknowledge drives understanding:matchingmedical ontologies with
the needsof medicallanguageprocessing.Artificial Intelligence
in Medicine15(1):25-51.
Hahn,U.; Schuiz, S.; and Romacker,M.1999. Part-wholereasoning: a case study in medical ontology engineering. IEEE
Intelligent Systems&their Applications14(5):59-67.
Haimowitz,
I. J.; Patii, R.S.; andSzolovits,P. 1988.Representing medicalknowledge
in a terminologicallanguageis difficult.
In Proceedingsof the 12th AnnualSymposium
on ComputerApplications in MedicalCare- SCAMC’88,
101-105.
Horrocks,I., and Sattler, U. 1999. A description logic with
transitive andinverseroles androle hierarchies.Journalof Logic
and Computation9(3):385-410.
Mays,E.; Weida,R.; Dionne,R.; Laker, M.; White,B.; Liang,
C.; and Oles, E J. 1996.Scalableandexpressivemedicalterminologies. In Proceedingsof the 1996AMIAAnnualFall Symposium (formerly SCAMC)
- AMIA’96, 259-263.
McCray,A. T., and Nelson,S.J. 1995. The representation of
meaningin the UMLS.
Methodsof Information in Medicine
34( 1/2): 193-201.
Pisanelli, D. M.; Gangemi,
A.; and Steve, G. 1998.Anontological analysis of the UMLS
metathesaurus.In Proceedingsof the
1998AMIAAnnualFall Symposium
- AMIA’98, 810--814.
Rector,A. L.; Bechhofer,
S.; Goble,C. A.; Horrocks,I.; Nowlan,
W. A.; and Solomon, W.D. 1997. The GRAILconcept modelling languagefor medicalterminology.Artificial Intelligence
in Medicine9:139-171.
Reggia,J. A., and Tuhrim,S., eds. 1985. Computer-Assisted
MedicalDecisionMaking,volume1 &2. NewYork: Springer.
Schmidt-SchauB,
M., and Smolka,G. 1991. Attributive concept
descriptionswithcomplements.
Artificial Intelligence48:1-26.
Schmolze,J. G., and Mark,W.S. 1991. TheNIKLexperience.
Computational
Intelligence 6:48-69.
Spackman,K. A., and Campbell, K.E. 1998. Compositional
concept representation using SNOMED:
towards further convergenceof clinical terminologies.In Proceedings
of the 1998
AMIAAnnualFall Symposium
- AMIA’98, 740-744.
Volot, E; Zweigenbaum,
P.; Bachimont,B.; Ben Said, M.;
Bouaud,J.; Fieschi, M.; and Boisvieux,J.-F. 1994. Structuration and acquisition of medicalknowledge:using UMLS
in the
ConceptualGraphformalism. In Proceedingsof the 17th Annual Symposiumon ComputerApplications in MedicalCareSCAMC’93,
710-714.