IV Synthetic genetic switches Reference: Escherichia coli
advertisement

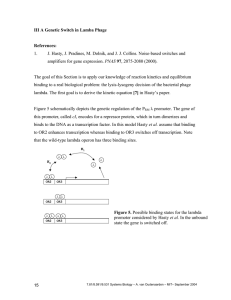
IV Synthetic genetic switches Reference: 1. T. S. Gardner, C. R. Cantor, and J. J. Collins. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339-342 (2000). In this paper Gardner et al. engineered a plasmid containing two repressor genes mutually controlling each others expression. In Box 1 postulate the two equations describing the toggle switch: du α1 = −u dt 1+ v β dv α2 = −v dt 1+ uγ [IV.1] Let’s derive this equation and explore which assumptions were made during the derivation. The fast equilibium reactions for this problem are: K1 P1 + R2β ← → P1R2β K2 P2 + R1γ ← → P2R1γ [IV.2] K3 γR1 ← → R1γ K4 βR 2 ← → R2β The first two reactions model the binding of the repressors to the two promoters inhibiting transcription. We will assume that there is only one binding site for the repressor (in contrast to the lambda phage promoters). However the repressor monomers can form multimers. Repressor 1 multimerizes with γ subunits and repressor 2 with β subunits. Note that K3 and K4 are the effective association constants for multimerization. The units of K3 and K4 are (M)1-γ and (M)1-β respectively. The last two equations assume that intermediate states are not allowed. This is the Hill approximation as we discussed before (see for example [II.22]). Remember that the total number of promoter site is conserved, and the total concentration of both promoters is identical since they are on the same plasmid: [PT ] = [P1T ] = [P1] + [P1R2β ] = [P2T ] = [P2 ] + [P2R1γ ] 21 [IV.3] 7.81/8.591/9.531 Systems Biology – A. van Oudenaarden – MIT– September 2004 Now the rate of synthesis of repressor 1 and 2 can be written as: Rgen1 = k1[PT ] Rgen2 1 k1[PT ] [P1] T = = k [P ] 1 1 + K1[R 2β ] 1 + K1K 4 [R2 ]β [P1] + [P1R2β ] 1 k 2 [PT ] [P2 ] T = k 2 [P T ] = = k [P ] 2 1 + K 2 [R1γ ] 1 + K 2K 3 [R1]γ [P2 ] + [P2R1γ ] [IV.4] The rates k1 and k2 are the effective synthesis rates (including RNA polymerase binding, transcription, translation, and folding) of repressor protein 1 and 2, respectively). Assuming a first order decay process with rate δ, the kinetic equations are: d[R1] k1 [PT ] = − δ[R1 ] dt 1 + K1K 4 [R2 ]β d[R 2 ] k 2 [PT ] = − δ[R 2 ] dt 1 + K 2K 3 [R1]γ [IV.5] ~ When we introduce a dimensionless time t = tδ , equation (50) becomes: d[R1] 1 k1 [PT ] = − [R1 ] ~ δ 1 + K1K 4 [R 2 ]β dt d[R 2 ] 1 k 2 [PT ] = − [R2 ] ~ δ 1 + K 2K 3 [R1]γ dt [IV.6] if we use the following dimensionless concentrations: u = [R1](K 2K 3 )1/γ [IV.7] v = [R 2 ](K1K 4 )1/β Equation [IV.6] becomes: du 1 k1[PT ](K 2K 3 )1/γ −u ~= 1+ v β dt δ dv 1 k 2 [P T ](K1K 4 )1/β −v ~= 1+ uγ dt δ [IV.8] Finally by defining α1 and α2 as: k1[PT ](K 2K 3 )1/γ α1 ≡ δ T k [P ](K1K 4 )1/β α2 ≡ 2 δ [IV.9] we recover the ‘Box’ equation: 22 7.81/8.591/9.531 Systems Biology – A. van Oudenaarden – MIT– September 2004 du α1 −u ~= d t 1+ v β dv α2 −v ~= d t 1 + uγ [IV.10] In steady state both these equations equal zero: α1 1+ v β α2 v= 1+ uγ u= 23 [IV.11] 7.81/8.591/9.531 Systems Biology – A. van Oudenaarden – MIT– September 2004 V Stability analysis Consider the following two coupled differential equations: & = f (x, y ) x & = g ( x, y ) y [V.1] The nullclines are defined as: & = 0 → f (xo , y o ) = 0 x & = 0 → g ( xo , y o ) = 0 y [V.2] in order to solve [V.2] we linearize around the fixed point (xo,yo): ~ x ≡ x−x o ~ y ≡ y − yo [V.3] If f(x,y) and g(x,y) are approximated by a first order Taylor expansion, [V.2] can be written as: ∂f ∂f ≡ a~ x + b~ y +~ y x& ≈ ~ x ∂y ( xo , y o ) ∂x ( xo , y o ) ∂g y& ≈ ~ x ∂x ∂g +~ y ≡ c~ x + d~ y ∂ y ( xo , yo ) ( xo , yo ) [V.4] or in matrix notation: r r X& = AX a b A = c d r x& X& = y& r ~ x X = ~ y [V.5] The matrix A is characterized by its trace and the determinant: τ = trace( A) = a + d [V.6] ∆ = det( A) = ad − bc Let’s try to find a solution of the convenient form: v v v v& = λv = Av 24 [V.7] 7.81/8.591/9.531 Systems Biology – A. van Oudenaarden – MIT– September 2004 This vector is called the eigenvector, λ is the corresponding eigenvalue. [V.7] can be solved by: a − λ det c b =0 d − λ [V.8] leading to: λ1 = λ2 = τ + τ 2 − 4∆ 2 τ − τ 2 − 4∆ [V.9] 2 or ∆ = λ1λ2 τ = λ1 + λ2 [V.10] For a stable fixed point both λ1 and λ2 should be negative. Therefore a stable fixed point is characterized by: ∆>0 τ <0 25 [V.11] 7.81/8.591/9.531 Systems Biology – A. van Oudenaarden – MIT– September 2004

![V Stability analysis [V.1] Consider the following two coupled differential equations: =](http://s2.studylib.net/store/data/013650663_1-c229820fe0c1fe00dd5776745187cd7f-300x300.png)