Harvard-MIT Division of Health Sciences and Technology
advertisement

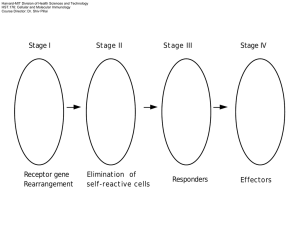
Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai secreted form membrane form 13 aa K K V V K K 26 aa 3aa Hapten Polyvalent antigen monosaccharide polysaccharide antigen receptor NO SIGNAL SIGNAL TRANSDUCTION Igα Igβ YY YY Src-family kinase ITAM ITAM YxxL/IxxxxxxYxxL/I Immune-receptor Tyrosine based Activation Motif Igα YY YY Inactive Syk YY Activated Syk YY Igβ ITAM The T cell receptor α γ ε β ε δ ζζ ITAM CO-RECEPTORS CELL SURFACE PROTEINS THAT BIND TO THE SAME ANTIGEN AS THE ANTIGEN RECEPTOR DISTINGUISHABLE FROM COSTIMULATORS CD4 TCR α γ β ε ε δ ζζ Lck Fyn Zap-70 CO-RECEPTORS • CD4 ON HELPER T CELLS • CD8 ON CYTOTOXIC T CELLS • CD21/CR2 ON B CELLS CD21/CR2 is a co-receptor and a positive regulator of BCR signaling CR2/CD21 YYYY Immune compl e x BCR CD19 β α CD81 Lyn Fyn Syk Vav PI3K Generation of Diversity • 1. V(D)J Recombination • Combinatorial Diversity • Junctional Diversity – N Regions – P nucleotides • 2. Somatic Mutation V D VDJ DNA junctional diversity, bases added and removed mRNA J - C Rearrangement is temporally ordered • B LINEAGE • T LINEAGE • IgH D to J • Then Vto DJ • Then Ig Kappa V to J • TCR D beta to J beta • Then V beta to DJ • Then TCR V alpha to J alpha µ L VDJ Cµ1 Cµ2 Cµ3 Poly A sites µM1 µM2 Cµ4 Choice of first polyadenylation site AAAA AAAA AAAA AAAA Choice of second polyadenylation site µs mRNA µm mRNA V(D)J Recombination • Lymphoid specific • Locus-specific, cell-type specific, stage specific • Accessibility model Heptamer Coding segment 12-bp spacer Nonamer 23-bp spacer Nonamer Heptamer Coding segment RAG-2 RAG-1 N ICK PRECEDES CUT CUT OH P CLEAVE AND MAKE HAIRPINS HAIRPINS SIGNAL JOINT T RIM RIM ADD N REGIONS REGIONS JOIN RAG-1/RAG-2 still required TdT DNA-PK/Ku70/Ku80 XRCC4/DNA ligase CODING JOINT + N-region RAG time • RAG-1 dimers associate with RAG-2 dimers • RAG-1 dimers bind to nonamers, cleave DNA • RAG-2 has a beta-propeller like structure probably brings in other proteins such as accessibility factors • Rag genes evolved from a transposable element? coding segment RSS RSS coding segment Looping out and Deletion Inversion The 12/23 Rule Heptamer Coding segment 12 bp spacer 23 bp spacer Nonamer Nonamer Heptamer Coding segment Cleave at heptamer junction ATTCTTTGAGATAGCTCGA TAAGAAACTCTATCGAGCT heptamer ATTCTTTGAGATAGCTCGA TAAGAAACTCTATCGAGCT Open up hairpin ATTCTTTGAGATAGCTCGATCG TCG TAAGAAACTCTATCGAGCTAGC AGC Add P nucleotides HAIRPINS Resolved by Artemis, an enzyme which works in conjunction with DNA-PKcs Severe Combined Immunodeficiency Rag-1 and Rag-2 deficiencies: SCID No VDJ recombination Artemis deficiency: SCID Defective coding joints Many other causes of SCID V(D)J recombination and human lymphomas • Many human lymphomas involve chromosomal translocations • In some lymphomas (e.g. follicular lymphomas, lymphomas associated with Ataxia Telangiectasia, etc) the machinery involved in V(D)J recombination drives the translocation process Regulation of V(D)J recombination 1. Allelic exclusion - role of pre-antigen receptors 2. Receptor editing VH to DJH rearrangement Ψ pre-BCR Ψ Large pre-B Large pre-B pre-BCR MEDIATED POSITIVE SELECTION (defective in X-linked agammaglobulinemia) pre-BCR C' C' I gD Ψ I gM pre-BCR Y YY Early pro-B Intermediate pro-B Late pro-B Large pre-B Small pre-B Immature B Mature B A B C C' D E F DH to JH rearrangement Ψ pre-BCR Large pre-B C' VL to JL rearrangement Ψ pre-BCR Large pre-B BONE MARROW C' Pre-B receptor µ vPreB λ5 Igα Igβ Syk Blk/Lyn/Fyn ITAM Pre-TCR No known ligand β γ pTα ε ε δ ζζ ITAM Pre-antigen receptors • Select cells that have made in-frame rearrangements • Signals for survival, proliferation and allelic exclusion (rearrangement at second allele is shut off) Central lymphoid organs P e r i p h e r y antigen receptor "The death/ editing wi ndow" Rearrangement of Ig heavy chain or TCR β chain genes Selection by pre-antigen receptor of cells with in- frame Ig heavy chain or TCR β c h a i n r e a r r a n g e m e n t s Elimination or editing of self reactive cells Pro li ferat i on and act i vat i on F i n a l r e p e r t o i r e in responders For more information and examples, see Immunobiology, by Janeway,C., Travers, P., Walport, M. and Capra, J., Garland Publishing, 5th edition, 2001 & Cellular and Molecular Immunology by Abbas, A., Pober, J., and Lichtman, A., W B Saunders; 4th edition.