1.89, Environmental Microbiology Prof. Martin Polz Lecture 16 Recap
advertisement

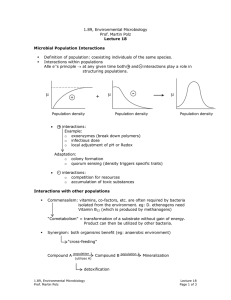
1.89, Environmental Microbiology Prof. Martin Polz Lecture 16 Recap Hybridization Environmental sample extraction Q PCR Quantification DNA PCR Specific genes (rRNA) cloning Phylogenetic relationships Diversity with in samples Compare community Sequences Structure between samples Overall Major phylogenetic lineages have remained uncultured ⇒ existence only known from clone libraries Neutral mutation – in 3rd codon so don’t matter Number of unique sequences found Estimation of Diversity Example: coastal ocean bacterioplankton Statistical tools: chao 1 test S = Sobs + Total number of sequences Number clones sequenced a2 2b a=number sequences found once b=number sequences found twice 1000 1600 rRNA genes 1.89, Environmental Microbiology Prof. Martin Polz Lecture 16 Page 1 of 3 "unstructured tree" average branch length is approximately the same Clusters of closely related sequences (similar clusters observed for different genes sequenced) Functional cluster of organisms; carry out same ecological functions because arose from common organism. Functional cluster of organisms; carry out same ecological functions because arose from common organism Question: How can such structures arise? What does it mean in an ecological context? see here Organisms takes Adaptive mutation can be point or lateral gene transfer or … ove r Adaptive mutation Adaptive mutation Same niche Time Clonal diversification from mutant; have similar function (via neutral mutations) Time selective sweep Same niche We can detect/quantify microbial diversity: Community Fingerprinting: Only works on abundant organisms. Quick way of seeing if two communities contain the same types of organisms (temporal or spacial heterogeneity) Techniques: 1. ARDRA (Restriction digestion of PCR amplified rDNA) 2. T-RF (introduce RE, cut at various places, specific patterns revealed on electropherograms) Fluorescent molecule RE cut Primer rDNA 1.89, Environmental Microbiology Prof. Martin Polz Lecture 16 Page 2 of 3 3. DGGE (Denaturant Gradient Gel Electrophoresis) will not denature. Run on gel to get patterns that reveal ecologically significant patterns. - Increasing denaturant gradient GC Region G C H-bond [ + Melting Get specific patterns Detection/Quantification in Environment Techniques: 1. In situ hybridization Ribosomes (rRNAs) Filters Mix with cells carrying fluorescent labels (oligonucleotides) Fix cells with format dehyde. Must kill cells before live cells impermeable to probes and would digest probes. Wash away unbound probe Binding of probe to rRNA in ribosome Count labeled cells 2. QPCR (Quantitative PCR) see handout 1.89, Environmental Microbiology Prof. Martin Polz Lecture 16 Page 3 of 3