Modern Biology in Two Lectures (Part II) Gil Alterovitz Harvard
advertisement

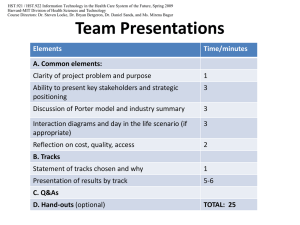
Modern Biology in Two Lectures (Part II) Gil Alterovitz Harvard-MIT Division of Health Science & Technology Course Administration Handouts Open Courseware form- please turn in before leaving class Matlab form- for free copy of Matlab for students in class for use in 6.092/HST.480 course. You can also use server Matlab or lab cluster. Background sheet- complete and turn in by end of class so we can put you on course email list. Homework 1 (Due next Thurs.) See assignments section in course site. Harvard-MIT Division of Health Science & Technology Background Student Department Affiliation Student Subject Area Strengths 50 45 45 R elative Strength- N ormalized to 100 40 40 35 Percent 30 25 20 15 10 5 35 30 25 20 15 10 5 0 6 HST 2 Department 7 0 Bio Comp Sci Engineering Harvard-MIT Division of Health Science & Technology Today Introduction, Part II- Gil Alterovitz Review Part I Splicing Alternative Splicing Post-Translational Modifications Sequence Analysis- Manolis Kellis Harvard-MIT Division of Health Science & Technology Genes to Proteins Transcription Translation DNA: “Lifetime Plan” mRNA: “Task List” 5’ATCTACAGATCAGCTACGACGCGACGAT TTAGCAGCAGCGACGCGACAGCAGCTAGTG ACGATAGCACATAGTTAGCACAGAGCAGAC ACAGACAGCACAGCGACAGCGACGACG-3’ 5’AUCUACAGAUCAGCUACGACGCGACGAU UUAGCAGCAGCGACGCGACAGCAGCUAGUG ACGAUAGCACAUAGUUAGCACAGAGCAGAC ACAGACAGCACAGCGACAGCGACGACG-3’ Protein: Machines MWTRFDSALPRSTPSTAKLVMPOILLLLEE EDTYESAQYKTWLMVCSDETTTE Protein mRNA Ribosome Figure by MIT OCW DNA Sequencing Source: HPCGG Relative Expression Levels Figure by MIT OCW Identification Post translation modification Splicing variants Relative expression levels Harvard-MIT Division of Health Science & Technology Genes Gene 2 Gene 1 Intergenic Sequence Communication analogy: start, message, stop. Source: Ehsan Afkhami Harvard-MIT Division of Health Science & Technology Stereo Rack Analogy Amplifier Harvard-MIT Division of Health Science & Technology Alternative Splicing 5' UTR Protein coding sequence Enhancer regulatory element 3' UTR Stabilizing domain * * AAAAAA A Altered translation Change in protein expression levels AAAAAA A AAAAAA A mRNA instability Altered protein structure and function Change in protein expression levels Change in protein isoform ratios Figure by MIT OCW Harvard-MIT Division of Health Science & Technology Sequence Ordering Coding Strand (Codons) 5' > > > - - - - - - T T C - - - - - - > > > 3' Template Strand (Anti-codons) 3' < < < - - - - - - A A G - - - - - - < < < 5' RNA Message (Codons) 5' > > > - - - - - - U U C - - - - - - > > > 3' Protein Amino Acid DNA Amino > > > Phenylalanine > > > Carboxy Epigenetic factors Feedback loops DNA Transcription RNA Processing mRNA Translation Protein >200 known post-translational modifications (eg, phosphorylation, glycosylation lipid attachment, peptide cleavage) Effects Transcriptional Post-transcriptional Translational and degradation Activity controls control control (eg, alternative splicing, controls (eg, post-translational modifications, alternative polyadenylation, Translational frameshifting degradation, compartmentalisation) RNA editing) Catalytic activity, association, stability, half-life, localisation, activity, etc) Figure by MIT OCW Harvard-MIT Division of Health Science & Technology Post-translational Modifications RESID ID AA0039 Name O4'-phospho-Ltyrosine SequenceSpec L-tyrosine Weight Keyword Fc=243.1 5, Fp=243.0 296, Cc=79.98 Cp=79.96 63, Feature Enzyme PIR:Binding site: phosphate (Tyr) (covalent) PIR:Binding site: phosphate (Tyr) (covalent) (by ...) Phosphoprotein SP:MOD_RES PHOSPHORYL ATION SP:MOD_RES PHOSPHORYL ATION (AUTO-) proteintyrosine kinase (EC 2.7.1.112) Phosphate H 339 modifications in RESID Database H Amino group H H H OH N O Carboxyl group O OH H O P H H Tyrosine side chain Figure by MIT OCW Harvard-MIT Division of Health Science & Technology Bioinformatics: Trends, Tools, and Databases What kind of problems need to be solved? How have previous problems in the field been approached? Databases Needed to Store Growing List of Sequence Data Entrez Human Protein Sequences Entrez Human Protein Sequences Entrez Hum an Nucleotide Sequences Entrez Human Nucleotide Sequences 250000 8000000 7000000 200000 Number of Sequences Number of Sequences 6000000 5000000 4000000 3000000 150000 100000 2000000 50000 1000000 0 1993 1995 1997 1999 Year 2001 2003 0 1993 1995 1997 1999 2001 2003 Year * Alterovitz, G., Afkhami, E. & Ramoni, M. in Focus on Robotics and Intelligent Systems Research, ed. Columbus, F. Nova Science Publishers, Inc., New York, 2005 (In press). Harvard-MIT Division of Health Science & Technology Paradigm Shifts in Bioinformatics Sequencing (1980’s to early 1990’s) DNA/RNA/Protein Sequence Analysis/sequence storage 3-D Protein Structure Prediction (Mid-1980’s-late 1990’s) Databases of Protein structures DNA/RNA Microarray Expression Experiments (Mid1990’s to 2000’s) Databases of expression data Protein interaction experiments (Early 2000’s to Present) Databases with pairwise interactions Mass Spec proteomic pattern experiments (Early 2000’s to Present) Databases with mass spec, protein identifications, proteomic patterns Integration of multiple modalities (Ongoing) Harvard-MIT Division of Health Science & Technology Human Genome Project ~ 99% of human genome has been sequenced (2004). Nature 431: 931-945. Error rate: ~1 event per 100,000 bases Number of protein-coding genes: 20,000-25,000 Number of protein-coding genes in worm: ~18,000 Genes comprise only about 2% of the human genome. The rest consists of non-coding regions: functions may include providing chromosomal structural integrity and gene regulation. Harvard-MIT Division of Health Science & Technology