20.453J / 2.771J / HST.958J Biomedical Information Technology MIT OpenCourseWare .
advertisement

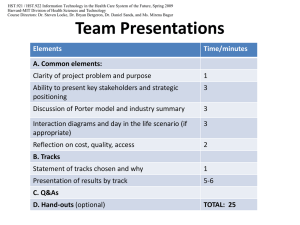
MIT OpenCourseWare http://ocw.mit.edu 20.453J / 2.771J / HST.958J Biomedical Information Technology Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms. 2.771J 20.453J HST.958J SMA5304 Biomedical Information Technology Fall, 2008 Biomedical Information Technology 2.771J 20. 453J HST.958J SMA 5304 Fall 2008 Instructions for the Term Project Each student in the course is required to present a term project that illustrates the use of the course material in a real information technology case in biology or medicine. The actual content of the case can vary depending upon the student’s interests and existing skills. Projects can range from general studies of a class of problems and the recommendation of a solution to detailed implementations in running software. Preliminary Project proposals should be submitted using email by Tuesday, October 18 to the respective faculty on your side of the Pacific. Microsoft Word documents are preferred because they can be easily edited electronically. If that is not possible, use a PDF format. These proposals will be read and approved during the period October 20-30, with any required changes being negotiated using email during that time. Please use email during the week of October 13 if you have any questions regarding suitability of the chosen topic, etc. In the cover sheet of your proposal, please list: • Your name • Your email • The title of your proposal • One to three paragraphs describing the project and your rationale for choosing it You may wish to include more than these elements to illustrate some of the implementation thoughts you have. Diagrams are often useful. The project should be realistically scaled to represent 36-40 hours of work, i.e. about 9-10 hours of work for the four weeks between October 30 and November 20. You will be required to turn in a report on your work and also to present a 30 minute talk to the class on your results on December 2, 4, or 9. Those days we will schedule additional hours to hold all of the presentations. Depending on broadcast availability, you may not be able to present your project simultaneously in Cambridge and Singapore. Here are some examples of topics that might give you a flavor for what is expected: 1. An information object definition for biological data produced in a FACS experiment. (FACS = Fluorescence Activated Cell Sorting). Project would require going to someone doing these experiments and © 2008 C. F. Dewey, Jr., MIT 1 2.771J 20.453J HST.958J SMA5304 Biomedical Information Technology Fall, 2008 compiling a view of the information required to completely characterize the results. This should be compared with the existing standards that have been proposed for FACS. 2. A system for adding genetic data to other experimental data in a clinical trial. The rationale here is that the patient’s test results should be correlated with individual genetic information and that information used to see if genetic factors affect the test results. What data would be useful to compile? What are the queries that one would like to run against the data? Can you build a demonstrator? 3. An extension to the DICOM standard to accommodate the use of microbubbles as a contrast agent in ultrasound. This will require some background work to understand the DICOM standard. 4. A general review of the National Cancer Institute program supported by the Center for Bioinformatics. This program supports a large number of innovative efforts in bioinformatics. See http://ncicb.nci.nih.gov/. This could involve a serious critique of what is done right and what is done wrong, as you have learned it in this course. 5. Design of a system to anticipate the physical results of breast cancer surgery. It was suggested at a recent NIH meeting that a system to help breast cancer patients anticipate the effects of forthcoming surgery would be emotionally and psychologically helpful because it would take away some of the fear of the unknown. How would such a system be designed and what would be its attributes? 6. How would one make a personal human genome viewer? Suppose we have the DNA profile (whatever that is) for a person. How would that person use that profile intelligently to, for example, run their profile against the known risk of contracting specific diseases? 7. Work flow scenarios for accessing stored biological data. Obtain and document real work flow scenarios where biologists use experimental data and data queried from existing genetic and proteomic databases. How does the type of data depend on the query? Do the data drive the query? Do the results of the query (or subsequent manipulation of the returned data) produce a subsequent query (iteration)? Something relating to web services would be very attractive. Instructions for the Presentation and Report The most important deliverable for the Term Paper is the oral presentation that will occur December 2, 4, or 9 depending on when you are scheduled. This presentation should be scheduled for no longer than 20 minutes with 5 minutes of questions and 5 minutes between presentations. Class hours on those days may be extended a bit to allow all of the class to present. We will issue the final schedule by the end of November. In addition to the oral presentation, you should turn in Microsoft Word/Powerpoint documents that contain your slides and a synopsis of your presentation for each slide. To make this easy, you can use the “Lecture Notes” feature of PowerPoint and simply write out your presentation and then it is easy to edit the description © 2008 C. F. Dewey, Jr., MIT 2 2.771J 20.453J HST.958J SMA5304 Biomedical Information Technology Fall, 2008 and the slides in a single stand-alone Powerpoint document. (This can also be very helpful in practicing your presentation.) The documents are due on December 10, the last official day of classes for the Fall term at MIT. © 2008 C. F. Dewey, Jr., MIT 3