Learning genetic networks from gene expression data
advertisement

Learning genetic networks from gene
expression data
Dirk Husmeier
Biomathematics & Statistics Scotland (BioSS)
JCMB, The King’s Buildings, Edinburgh EH9 3JZ
United Kingdom
http://www.bioss.ac.uk/∼dirk
Paradigm Shift in Molecular Biology
Paradigm Shift in Molecular Biology
Pre-Genomic
• Reductionist (DNA or RNA or protein)
• Generally qualitative, non-numeric
• Hypothesis driven
Paradigm Shift in Molecular Biology
Pre-Genomic
• Reductionist (DNA or RNA or protein)
• Generally qualitative, non-numeric
• Hypothesis driven
Post-Genomic
• Holistic, systems approach: DNA and RNA and protein
• Quantitative, highly numeric
• Data driven
Paradigm Shift in Molecular Biology
Pre-Genomic
• Reductionist (DNA or RNA or protein)
• Generally qualitative, non-numeric
• Hypothesis driven
Post-Genomic
• Holistic, systems approach: DNA and RNA and protein
• Quantitative, highly numeric
• Data driven
=⇒ Need for machine learning and statistics
Inferring genetic networks
from
microarray gene expression data
From http://www.csu.edu.au/faculty/health/biomed/subjects/molbol/images
.
.
Transcription
Translation
.
DNA
.
mRNA
Protein
.
+
q
+
+
+
hysteretic
oscillator
ligand
binding
F
B
A
eq
e
E
-
f
b
a
ab
+
f2
g
-
G
switch
c2
c
+
-
H
h
-
C
cascades
-
d2
d
D
-
g2
external
ligand
J
j
.
.
Transcription
Translation
.
DNA
mRNA
Protein
Microarrays
.
.
From http://www.nhgri.nih.gov/DIR/Microarray/NEJM Supplement/
Reverse engineering
Learn the network structure from
postgenomic data.
Problem: Noise, sparse data
Bayesian networks
Probabilistic framework for
robust inference of interactions
in the presence of noise
Nir Friedman et al. (2000)
Journal of Computational Biology 7: 601-620
Outline of the talk
• Recapitulation: Bayesian networks
• Reverse engineering:
Learning networks from data
• Estimating the accuracy of inference
Outline of the talk
• Recapitulation: Bayesian networks
• Reverse engineering:
Learning networks from data
• Estimating the accuracy of inference
Revision: Bayes’ Rule
G: A certain gene is over-expressed
C: A patient is suffering from cancer
Ω
G
G^C
P (G, C)
P (G|C) =
P (C)
C
P (G, C)
P (C|G) =
P (G)
Revision: Bayes’ Rule
G: A certain gene is over-expressed
C: A patient is suffering from cancer
Ω
G
G^C
P (G, C)
P (G|C) =
P (C)
P (G, C) = P (G|C)P (C)
C
P (G, C)
P (C|G) =
P (G)
P (G, C) = P (C|G)P (G)
Revision: Bayes’ Rule
G: A certain gene is over-expressed
C: A patient is suffering from cancer
Ω
G
G^C
P (G|C) =
P (G, C)
P (C)
P (G, C) = P (G|C)P (C)
C
P (C|G) =
P (G, C)
P (G)
P (G, C) = P (C|G)P (G)
P (G|C)P (C)
P (C|G) =
P (G)
A
C
B
D
E
Nodes
A
C
B
D
E
Edges
A
C
B
D
E
Edges = directed
A
C
B
D
E
No directed cycles !
A
C
B
D
E
P (A, B, C, D, E) =
Y
i
P (nodei|parentsi)
A
C
B
D
E
P (A, B, C, D, E) =
P (A)
A
C
B
D
E
P (A, B, C, D, E) =
P (A)P (B|A)
A
C
B
D
E
P (A, B, C, D, E) =
P (A)P (B|A)P (C|A)
A
C
B
D
E
P (A, B, C, D, E) =
P (A)P (B|A)P (C|A)P (D|B, C)
A
C
B
D
E
P (A, B, C, D, E) =
P (A)P (B|A)P (C|A)P (D|B, C)P (E|D)
A
B
C
A
B
A
C
B
C
When are A and B conditionally independent given C?
P (A, B|C) = P (A|C)P (B|C)
A
B
C
A
B
C
A
B
C
When are A and B marginally independent?
P (A, B) = P (A)P (B)
A
B
C
A
B
C
P (A, B, C) = P (A|C)P (B|C)P (C)
A
B
C
P (A, B, C) = P (A|C)P (B|C)P (C)
P (A, B, C)
P (A, B|C) =
= P (A|C)P (B|C)
P (C)
A
B
C
P (A, B, C) = P (A|C)P (B|C)P (C)
P (A, B, C)
P (A, B|C) =
= P (A|C)P (B|C)
P (C)
But: P (A, B) 6= P (A)P (B)
Babies
Storks
Babies
Storks
Environment
A
B
C
A
B
C
P (A, B, C) = P (B|C)P (C|A)P (A)
A
B
C
P (A, B, C) = P (B|C)P (C|A)P (A)
P (A, B|C) =
P (A,B,C)
P (C)
=
P (C|A)P (A)
P (B|C) P (C)
A
B
C
P (A, B, C) = P (B|C)P (C|A)P (A)
P (A, B|C) =
P (A,B,C)
P (C)
=
P (C|A)P (A)
P (B|C) P (C)
P (A, B|C) = P (B|C)P (A|C)
A
B
C
P (A, B, C) = P (B|C)P (C|A)P (A)
P (A, B|C) =
P (A,B,C)
P (C)
(A)
= P (B|C) P (C|A)P
P (C)
P (A, B|C) = P (B|C)P (A|C)
But: P (A, B) 6= P (A)P (B)
Cloudy
Rain
Grass wet
Cloudy
Rain
Grass wet
A
B
C
A
B
C
P (A, B, C) = P (C|A, B)P (A)P (B)
A
B
C
P (A, B, C) = P (C|A, B)P (A)P (B)
X
P (A, B) =
P (A, B, C) = P (A)P (B)
C
A
B
C
P (A, B, C) = P (C|A, B)P (A)P (B)
X
P (A, B) =
P (A, B, C) = P (A)P (B)
C
But: P (A, B|C) 6= P (A|C)P (B|C)
Battery
Petrol
Engine
Battery
Petrol
Engine
A
B
A
C
A
B
A
B
C
C
C
A
B
C
A
B
B
A
B
C
A
B
C
A
B
A
B
?
A
B
A
B
No
Unobserved node
Observed node
Blocked path
Open path
• Two variables A, B are d-separated if every path from A to B is blocked.
• A⊥B iff A and B are d-separated.
A
C
B
D
E
P (A, B, C, D, E) =
Y
P (nodei|parentsi)
i
Bayesian network = DAG + distribution family + parameters
Example: multinomial CPD
P(true) P(false)
0.5
0.5
Cloudy
Cloudy
true
false
P(true) P(false)
0.3
0.7
0.8
0.2
Cloudy
Rain
Sprinkler
true
false
Wet grass
Sprinkler
Rain
P(true)
true
true
true
false
1.0
0.8
0.0
0.2
false
true
1.0
0.0
false
false
0.0
1.0
P(false)
P(true) P(false)
0.8
0.2
0.0
1.0
Linear Gaussian CPD
P (A|P1, . . . , Pn) = N
w0 +
n
X
i=1
wiPi, σ
2
!
Learning network structure from data
Find the best network structure M :
M ∗ = argmax{P (M |D)}
Find the best model M , that is, the best network
P (M |D) ∝ P (D|M )P (M )
R
P (D|M ) = P (D|θ, M )P (θ|M )dθ
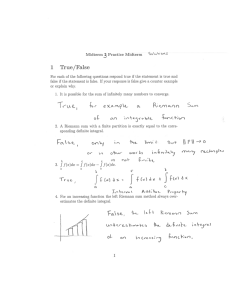
When is the integral analytically tractable?
Find the best model M , that is, the best network
P (M |D) ∝ P (D|M )P (M )
R
P (D|M ) = P (D|θ, M )P (θ|M )dθ
When is the integral analytically tractable?
• Complete observation: No missing values.
• P (D|θ, M ) and P (θ|M ) must satisfy certain regularity
conditions.
• Examples: Multinomial with a Dirichlet prior, linear
Gaussian with a normal-gamma prior.
A
C
B
D
E
P (A, B, C, D, E) =
Y
P (nodei|parentsi)
i
Bayesian network = DAG + distribution family + parameters
Biological example
Yeast cell cycle
Clustering
Spellman et al. 1998
Molecular Biology of the Cell 9 (12) :3273-97
G
e
n
e
s
.
Experiments
G
e
n
e
s
Experiments
From Spellman et al., http://cellcycle-www.stanford.edu/
Advantage of clustering
Fast, computationally cheap
Shortcoming of clustering
It is NOT reverse engineering
SLT2 clusters with low-osmolarity response genes
Biological example
Yeast cell cycle
Nir Friedman et al. (2000)
Journal of Computational Biology 7: 601-620
Low osmolarity response genes
...
SLT2
Rlm1p
MAP kinase
Transcription factors
Swi4/6
SLT2
Low osmolarity
response genes
Low osmolarity response genes
...
SLT2
Rlm1p
MAP kinase
Transcription factors
Swi4/6
SLT2
Low osmolarity
response genes
Low osmolarity response genes
...
SLT2
Rlm1p
MAP kinase
Transcription factors
Swi4/6
SLT2
Low osmolarity
response genes
Low osmolarity response genes
...
SLT2
Rlm1p
MAP kinase
Transcription factors
Swi4/6
SLT2
Low osmolarity
response genes
Low osmolarity response genes
...
SLT2
Rlm1p
MAP kinase
Transcription factors
Swi4/6
SLT2
Low osmolarity
response genes
Can we learn causal relationships
from conditional dependencies ?
Can we learn causal relationships
from conditional dependencies ?
A
B
C
Bayesian network:
A node is independent of its nondescendants, given its parents.
Causal network:
A node is independent of its earlier causes, given its immediate causes.
Causal
Network
Bayesian
Network
Causal
Network
Bayesian
Network
Causal
Network
Bayesian
Network
Problem 1
Hidden variables
Bayesian networks versus
causal networks
True
causal
graph
A
B
C
Node A
unknown
A
B
C
Problem 1
Hidden variables
Problem 2
Equivalence classes
and
PDAGs
A
B
C
A
B
C
A
B
C
A
B
C
A
B
C
A
B
A
B
C
C
A
B
C
P(A,B,C) =
P(B|C) P(C|A) P(A)
P(A|C) P(B|C) P(C)
P(A|C) P(C|B) P(B)
P(C|A,B) P(A) P(B)
A
B
C
A
B
A
B
C
C
A
B
C
P(A,B,C) =
P(B|C) P(C|A) P(A)
P(A|C) P(C)
P(A|C) P(B|C) P(C)
P(A|C) P(C|B) P(B)
P(B|C) P(C)
P(C|A,B) P(A) P(B)
A
B
A
C
B
A
B
A
C
C
B
C
P(A,B,C) =
P(B|C) P(C|A) P(A)
P(A|C) P(B|C) P(C)
P(A|C) P(C|B) P(B)
P(A|C) P(C)
A
B
C
P(C|A,B) P(A) P(B)
P(B|C) P(C)
A
B
C
A
B
C
A
B
C
A
B
A
C
B
B
C
C
A
A
A
B
C
B
C
• Two DAGs are equivalent iff they have the same skeleton (= the underlying
undirected graph) and the same v-structure.
• v-structure: Converging directed edges into the same node without an edge
between the parents.
An equivalence class of DAGs can be represented by a PDAG
(partially directed acyclic graph).
A
C
B
D
E
We can only learn PDAGs from the data!
• Observation: A passive measurement of the
domain of interest.
• Intervention: Setting the values of some
variables using forces outside the causal model,
e.g., gene knockout or over-expression
• Observation: A passive measurement of the
domain of interest.
• Intervention: Setting the values of some
variables using forces outside the causal model,
e.g., gene knockout or over-expression
• Interventions can destroy the symmetry within
an equivalent class.
A
B
A
B
A
B
A
B
A
B
A
B
Interventional data
A and B are correlated
A
B
inhibition of A
A
A
B
down-regulation of B
B
A
B
no effect on B
n
n
i =1
i =1
−{i}
P ( D | M ) = ∏ P ( X i = Di | pa[ X i ] = D pa[ X i ] ) → ∏ P ( X i = Di−{i} | pa[ X i ] = D pa
[ Xi ] )
Learning with interventions
No intervention:
Q
P (D|M ) = i P (Xi|P a(Xi))
Two models M, M0 with the same score are
structure equivalent.
Int: Set of interventions −→ Modified score:
Y
P (D|M ) =
P (Xi|P a(Xi))
i,Xi∈Int
/
This score is no longer structure equivalent.
A
B
C
D
E
.
.
A
B
C
D
E
A
.
B
A
B
A
B
A
B
A
B
C
C
C
C
C
D
D
D
D
D
E
E
E
E
E
.
A
B
C
D
E
A
.
B
A
B
A
B
A
B
A
B
C
C
C
C
C
D
D
D
D
D
E
E
E
E
E
.
A
B
C
D
E
A
.
B
A
B
A
B
A
B
A
B
C
C
C
C
C
D
D
D
D
D
E
E
E
E
E
.
A
B
C
D
E
A
.
B
A
B
A
B
A
B
A
B
C
C
C
C
C
D
D
D
D
D
E
E
E
E
E
.
A
B
C
D
E
A
.
B
A
B
A
B
A
B
A
B
C
C
C
C
C
D
D
D
D
D
E
E
E
E
E
.
A
B
C
D
E
A
.
B
A
B
A
B
A
B
A
B
C
C
C
C
C
D
D
D
D
D
E
E
E
E
E
.
A
B
C
D
E
A
.
B
A
B
A
B
A
B
A
B
C
C
C
C
C
D
D
D
D
D
E
E
E
E
E
.
A
B
C
D
E
A
.
B
A
B
A
B
A
B
A
B
C
C
C
C
C
D
D
D
D
D
E
E
E
E
E
.
A
B
C
D
E
A
.
B
A
B
A
B
A
B
A
B
C
C
C
C
C
D
D
D
D
D
E
E
E
E
E
.
Active learning
Based on preliminary inference:
Predict the intervention that maximizes the
information content of the expected response.
.
.
Experiment
.
Modelling
.
.
.
Experiment
Measurements
.
Modelling
.
.
.
Experiment
Measurements
.
Modelling
Inference
.
.
.
Experiment
Intervention
.
Modelling
Inference
.
.
.
Experiment
Intervention
.
Modelling
Inference
.
Dynamic Bayesian Networks
Symmetry breaking by the direction of time:
Cause precedes its effect
Modelling recurrent structures and
feedback loops
t=1
t=2
t=3
t=4
Outline of the talk
• Recapitulation: Bayesian networks
• Reverse engineering:
Learning networks from data
• Estimating the accuracy of inference
Learning the network from data
Find the best network structure M :
M ∗ = argmax{P (M |D)}
Find the best parameters θ
∗
θ∗ = argmax{P (θ|D, M ∗)}
Find the best model M , that is, the best network
P (M |D) ∝ P (D|M )P (M )
Find the best model M , that is, the best network
P (M |D) ∝ P (D|M )P (M )
R
P (D|M ) = P (D|θ, M )P (θ|M )dθ
When is the integral analytically tractable?
Find the best model M , that is, the best network
P (M |D) ∝ P (D|M )P (M )
R
P (D|M ) = P (D|θ, M )P (θ|M )dθ
When is the integral analytically tractable?
• Complete observation: No missing values.
• P (D|θ, M ) and P (θ|M ) must satisfy certain regularity
conditions.
• Examples: Multinomial with a Dirichlet prior, linear
Gaussian with a normal-gamma prior.
BDe:
Multinomial with a Dirichlet prior
Heckerman, Geiger, Chickering (1995)
Learning Bayesian Networks:
The Combination of Knowledge and Statistical Data
Machine learning 20, 245-274
BGe:
Linear Gaussian with a normal-gamma prior
Geiger and Heckerman (1994)
Learning Gaussian networks
Proceedings of the Tenth Conference on
Uncertainty in Artificial Intelligence
Morgan Kaufmann publisher, San Francisco, 235-243
Naive approach
• Compute P (M |D) for all possible network structures M .
• Select network structure M ∗ that maximizes P (M |D)
Naive approach
• Compute P (M |D) for all possible network structures M .
• Select network structure M ∗ that maximizes P (M |D)
Problem 1:
Number of different network structures increases super-exponentially
with the number of nodes.
N of nodes
2 4
6
8
10
N of structures 3 543 3.7 × 106 7.8 × 1011 4.2 × 1018
−→ Optimization problem intractable for large N of nodes
Naive approach
• Compute P (M |D) for all possible network structures M .
• Select network structure M ∗ that maximizes P (M |D)
Problem 2:
Data are sparse → Intrinsic uncertainty of inference
P(M|D)
P(M|D)
M
M*
Large data set D:
Best network structure M* well defined
M
M*
Small data set D:
Intrinsic uncertainty about M*
Objective: Sample from the posterior distribution
P (D|Mk )P (Mk )
P (Mk |D) = P
i P (D|Mi)P (Mi)
P
Direct approach intractable due to i P (D|Mi)P (Mi)
Markov chain Monte Carlo (MCMC):
• Proposal move: Given network Mold, propose a new
network Mnew with probability Q(Mnew |Mold).
• Acceptance/Rejection:
Accept this new network
with
n
o
Q(Mold|Mnew )
new |D)
probability min 1, PP(M
×
(M |D)
Q(Mnew |M )
old
old
Markov chain Monte Carlo (MCMC)
Reject
Propose
Propose
Accept
Propose
(
P (Mnew )
Q(Mold |Mnew )
new )
Acceptance probability: min 1, PP(D|M
×
×
(D|Mold )
P (Mold )
Q(Mnew |Mold )
)
Marginal likelihood
P (D|Mnew )
P (D|Mold)
×
P (Mnew )
P (Mold)
×
Q(Mold|Mnew )
Q(Mnew |Mold)
Find the best model M , that is, the best network
P (M |D) ∝ P (D|M )P (M )
R
P (D|M ) = P (D|θ, M )P (θ|M )dθ
When is the integral analytically tractable?
• Complete observation: No missing values.
• P (D|θ, M ) and P (θ|M ) must satisfy certain regularity
conditions.
• Examples: Multinomial with a Dirichlet prior, linear
Gaussian with a normal-gamma prior.
Example: multinomial CPD
P(true) P(false)
0.5
0.5
Cloudy
Cloudy
true
false
P(true) P(false)
0.3
0.7
0.8
0.2
Cloudy
Rain
Sprinkler
true
false
Wet grass
Sprinkler
Rain
P(true)
true
true
true
false
1.0
0.8
0.0
0.2
false
true
1.0
0.0
false
false
0.0
1.0
P(false)
P(true) P(false)
0.8
0.2
0.0
1.0
Multinomial CPD
Disadvantage
• Requires discretization −→ Loss of information.
• Number of free parameters exponential in the number
of parents.
Advantage
• Can model nonlinear interactions.
Linear Gaussian CPD
!
n
X
P (A|P1, . . . , Pn) = N w0 +
wiPi, σ 2
i=1
Advantage
• Number of free parameters linear in the number of
parents.
• No discretization −→ No loss of information.
Disadvantage
• Can only model linear interactions.
A
B
A
B
C
C
C= w1 A + w2 B
C= w1 A + w2 B
B
A
A
B
C
C
C= w1 A + w2 B
C= w1 A + w2 B
Alternatives to the BDe and BGe scores:
Nonlinear and continuous relations, possibly including
hidden variables
• Imoto et al. (2003): heteroscedastic regression, Laplace
approximation
• Beal (2003): BNets with hidden variables, VBEM
• Pournara (2005): nonlinear regression with Gaussian
processes, RJMCMC
Prior probability
P (D|Mnew )
P (D|Mold)
×
P (Mnew )
P (Mold)
×
Q(Mold|Mnew )
Q(Mnew |Mold)
.
.
Fan-out unrestricted
Fan-in restricted
not permissible
.
.
Proposal probability
P (D|Mnew )
P (D|Mold)
×
P (Mnew )
P (Mold)
×
Q(Mold|Mnew )
Q(Mnew |Mold)
MCMC moves
Delete
edge
Reverse
edge
Create
edge
Proposal probability = ?
Proposal probability = ?
Proposal probability = 1/5
Proposal probability = 1/6
Neighbourhood
Neighbourhood
Delete
Reverse
Add
Delete
Add
Delete
Reverse
Add
Reverse
Add
Add
Add
Problem: Statistical significance of the networks
• Complex models: Transcript levels of hundreds of genes.
• Sparse data: Typically a few dozen samples.
P(M|D)
P(M|D)
M
M*
Large data set D:
Best network structure M* well defined
M
M*
Small data set D:
Intrinsic uncertainty about M*
• Posterior probability P (M |D) diffuse: Global network inference is meaningless.
Solution: Focus on features and subnetworks
Feature: Indicator variable for a property of interest,
e.g.: Are X and Y close neighbours in the network?
1 if M satisfies the feature
f (M ) =
0 otherwise
Solution: Focus on features and subnetworks
Feature: Indicator variable for a property of interest,
e.g.: Are X and Y close neighbours in the network?
1 if M satisfies the feature
f (M ) =
0 otherwise
X
Posterior probability of features: P (f |D) =
f (M )P (M |D)
M
T
1X
Approximate this sum with MCMC: P (f |D) =
f (Mi)
T i=1
where {Mi} is a sample from the posterior obtained with MCMC.
Model network, data set size: N = 50
6
4
14
9
16
5
3
10
13
8
15
2
1
11
12
17
7
18
32
31
34
19
39
35
30
21
29
33
22
28
20
36
40
38
37
26
23
24
27
25
Model network, data set size: N = 50
6
4
14
9
16
5
3
10
13
8
15
2
1
11
12
17
7
18
32
31
34
19
39
35
30
21
29
33
22
28
20
36
40
38
37
26
23
24
27
25
Predicted connectivity spectrum
10
9
8
Connectivity
7
6
5
4
3
2
1
0
0
10
20
30
Node
40
50
Outline of the talk
• Recapitulation: Bayesian networks
• Reverse engineering:
Learning networks from data
• Estimating the accuracy of
inference