A Novel System for Scoring of Hormone Receptors in Breast... Histopathology Slides

A Novel System for Scoring of Hormone Receptors in Breast Cancer
Histopathology Slides
Adnan M. Khan
† , ‡ , ∗
, Aisha F. Mohammed
† , ∗
, Shama A. Al-Hajri
†
, Hajer M. Al Shamari
†
,
Uvais Qidwai
†
, Imaad Mujeeb
§
and Nasir M. Rajpoot
† , ‡
, Senior Member, IEEE,
Abstract — Grading of breast cancer is often done by an expert pathologist based on their analysis of micro-level structural features of the cancerous tissue specimen as well as the level of presence of certain protein molecules in the specimen.
The process of assessment of the level of presence of estrogen and progesterone receptors molecules is subjective by its very nature and therefore, causes large inter-expert and sometimes even intra-expert variability, potentially adding noise to the process of selecting the treatment regime for the patient.
Quantification of immunohistochemical stains is critical for an objective assessment of breast cancer histopathology specimens.
We present a fast, compact and inexpensive system for scoring the Estrogen and Progesterone hormone receptors in breast cancer histopathology slides using image analysis algorithm.
We describe hardware and software issues in the construction of the system, and present a comparison of scores produced by our system to those produced by many expert pathologists.
I. INTRODUCTION
According to the World Health Organization (WHO), breast cancer is considered the most common cancer among women and the second-leading cause of cancer death after lung cancer
1
. It is generally diagnosed after a visual microscopic examination of a biopsy sample, obtained from the breast tissue of a patient by an expert pathologist. Since tissues are colorless, staining is generally used to obtain information regarding the biochemical makeup of a tissue specimen. This involves analysis of micro-level structural features of the cancerous tissue specimen as well as the level of presence of certain protein molecules in the specimen.
Immunohistochemical (IHC) stains are generally used to detect the level of expression of protein molecules, such as estrogen receptors (ER) and progesterone receptors (PR), in a tissue section.
The hormone receptor status is one of the factors that provide useful information for treatment planning [1]. Examination of the level of abundance of ER and PR hormones can be used as an evidence for whether or not one or both of those hormones stimulate the tumor growth. The receptors are specific proteins that are found in the cell or its surface.
The function of these receptors is to pick up the hormone signals influencing the cells to grow. A result of ER positive
(ER + ) implies that the hormone abundance promotes the growth of tumor. The same is applied if the cancer is PR
*Joint first authors.
†
College of Engineering, Qatar University, Doha, Qatar.
‡
Department of Computer Science, University of Warwick, UK.
§
Department of Pathology, Hamad General Hospital, Doha, Qatar.
Corresponding Author: nasir.rajpoot@ieee.org
1 http://www.who.int/cancer/detection/breastcancer/en/ positive (PR + ), in which case the tumor will respond to hormone therapy. If ER and PR are not present then the cancer is hormone-receptor-negative and other treatments are applied [2].
In the histopathology laboratory at the Hamad General
Hospital (HGH), the ER and PR stained slides are routinely interpreted by the pathologists. A pathologist may face different kinds of problems while he/she examines these tissue slides. One of the problems is that the visual examination of tissue slides can be quite time consuming: usually 7 10 minutes per slide but sometimes it may take more than 20 minutes per slide. Besides, this process of manual examination can be tedious and can potentially lead to erroneous diagnosis, in the worst case. The impact of a wrong diagnosis, potentially leading to a wrong treatment of cancer, can be mitigated by introducing objectivity to the diagnostic process. Advances in image analysis can be leveraged to negotiate both accuracy and efficiency issues.
This paper presents a novel system for objective assessment of breast cancer histopathology slides which incorporates a fast, high resolution camera to acquire a histopathology image, analyze it in near real-time using an indigenously developed image analysis module, and produce scoring results that are objective and reproducible. The rest of the paper is organized as follows: In section II, we present the overall system architecture and the hardware components used in our system. In section III, we present a brief description of our software module which involves input/output subsystems along with our image processing and analysis module that automatically performs scoring of hormone receptors. In section IV and V, we briefly present experimental results and some future directions of this work respectively.
II. SYSTEM ARCHITECTURE
In order to realize efficient machine vision systems, such as those used for medical imaging, adequate architectures and algorithms, which will ultimately lead to industrially acceptable and flexible solutions, must be carefully considered.
These considerations prompt us to develop a system which is capable of adaptive and dynamic processing of visual information in near real time. Figure 1 shows the overall architecture of the proposed system which consists of four major components: ( 1 ) microcontroller based subsystem to dynamically control the stage of the microscope; ( 2 ) image acquisition; ( 3 ) the host PC and ( 4 ) an output unit (printer).
Due to lack of space, we cannot go into the detailed design, however interested reader is welcome to read the product
Input Unit Host PC Output Unit
HPF
Image
Stain Separation
Stain
Quantification
ER/PR
Score specific documentation for detail. Briefly, the functionality associated with each of these components is outlined as follows:
B. Image Acquisition
Camera Interface
Printer Interface
Microcontroller
Interface
Fig. 1.
System Architecture
A. Stage Controller for Microscope
It is a small hardware module that controls the stage of microscope so that the user adjust the viewing field of the microscope automatically. In our design, two stepper motors along with an Arduino board
2 are used to control the adjustment knobs, which dynamically adjusts the stage of the microscope. This is an important modules as scoring of each histopathology tissue require analysis of multiple visual fields of the tissue specimen and this module may be iteratively used to automatically select a different visual field in each iteration to come up with an average score.
Camera
3
, attached to the microscope, is capable of capturing high resolution images of histopathology specimen. The acquired image can be transferred to the host PC through various interfaces. In our implementation, we choose Universal Serial Bus (USB) interface due to its high performance.
However, the system can be customized to use any suitable high performance interface as well.
C. The Host PC
It is the central part of our system. It controls all the activities of the system which essentially involve: ( 1 ) maintaining the system flow; ( 2 ) communication with variety of devices
(possibly using variety of interfaces) to exchange commands and data; ( 3 ) image acquisition, processing and analysis to score a tissue specimen; ( 4 ) printing a user friendly report for patient.
2
We decided to choose Arduino UNO microcontroller
(http://www.arduino.cc/) because it supports the required features (such as I/O ports and USB communication) for the field position controller.
However, any other alternative may also be used.
3
We used Canon EOS D 1100 with amscope adapter in our system.
Fig. 2.
Overview of the ER/PR Scoring Algorithm
D. Printer
Printer is directly connected to the host PC and is used to print patient specific scoring information. In our implementation, we choose USB interface due to its high performance. However, the system can be customized to use any appropriate interface as well.
III. T HE S CORING A LGORITHM
System software consists of three modules: input, image processing and analysis, and output. Here we briefly outline the details of each.
A. Input & Output
Input module is responsible for performing two main tasks: ( 1 ) control the position of the visual field of the tissue specimen (using Arduino board with two servo motors);
( 2 ) live-streaming of camera and image capture to perform digital image processing and analysis. Similarly, the output module is responsible for printing the calculated score as a text document using the attached printer.
B. Image Processing and Analysis
After acquiring the histopathology image from image acquisition module, a series of morphological operations are applied on images to score a slide for hormone receptors.
Following is the brief description of our image processing pipeline:
1) Stain Separation: In 2001 , Ruifork and Johnston [3] proposed a color deconvolution (CD) framework with potential application in histopathology image analysis. Later, it has been used in a variety of applications [4], [5]. The CD framework transforms RGB color space Ψ to a new color space
ˆ defined by the stains used for staining the tissue section.
If image I = ( C , Ψ) is defined as a 2 D set of pixels c ∈ C with associated color space function Ψ assigning red, green and blue intensities to each pixel. The relationship between color spaces Ψ and
ˆ is defined by Lambert-Beers law as follows,
Ψ = exp ( − S
ˆ
(1) where S is the stain matrix that defines the stain vectors
(absorption factors) associated with each stain used on the tissue. More formally, it can be defined as follows.
S =
s r, 1
s r, 2 s r, 3 s g, 1 s g, 2 s g, 3 s b, 1
s b, 2 s b, 3
(2) where s r, 1
, s g, 1 and s b, 1 denote the pre-defined, normalized red, green and blue values respectively, for s
1 channel. Similarly, the second and third rows of S are defined for s
2 and
( a ) ( b )
( c ) ( d )
Fig. 3.
Demonstration of the Stain Quantification Algorithm on an HPF image: ( a ) Original RGB image; ( b ) Overlay of weak, medium and strong signals in ( a ) with yellow, green and red colors respectively; ( c ) Hematoxylin and ( d ) DAB channels obtained by procedure in section III-B.1. Overall score for this image obtained by our automated system is 0 .
56 .
s
3 channels respectively. Ruifork & Johnston [3] provided a closed form solution to equation 1. They demonstrated that the intensity of a pixel c in the new color space
ˆ is defined as
ˆ c ) = D Φ( c )
D = S
− 1
Φ( c ) = − log(Ψ( c ))
(3)
(4)
(5)
Here,
ˆ c ) represents the extent of each stain ( s
1
, s
2 and s
3
) corresponding to the pixel c .
D is the so-called color deconvolution matrix obtained by calculating the inverse of the stain matrix S , Φ is the so-called optical density (OD) space where a linear combination of stains results in a linear combination of OD values.
ˆ c ) , Ψ( c ) and Φ( c ) are 3 × 1 column vectors [3].
2) Stain Quantification: Once the stain channels are separated into and s
3 s
1
, s
2 and s
3
(where s
1
= Hematoxylin, s
2
= Dab
= background), we quantify the amount of chromatin content (nuclei) and the amount of protein content (ER/PR), as these two quantities play critical role in defining the
ER/PR score of tissue specimen. For this purpose, we make use of first two channels ( s
1 which contains nuclei and s
2 which contains protein content), and invert the grayscale values of these channels to get ˆ
1 and ˆ
2
.
Segmentation is then applied on inverted gray scale images using Otsu automatic threshold selection framework [6]. The segmented images are postprocessed using simple morphological operations to clean the segmentation results. If N is the segmentation results of ˆ
1 and P is the segmentation results of ˆ
2
, then the nuclei content image ( K ) is obtained by removing all the common pixels (between N and P ) from
N as follows:
K = N − ( N ∩ P ) (6)
3) ER/PR Scoring: Next, we divide the pixel intensities in s
1
( K ) into 4 groups (non-signal, weak-signal, medium-signal and strong-signal) using empirically estimated thresholds ( τ
1
,
τ
2 and τ
3
), and count the quantity of pixels in each group for overall scoring.
If N ns
, N ws
, N ms and N ss are the percentages of nonsignal, weak-signal, medium-signal and strong-signal pixels in all the ˆ
1
( K ) images respectively, then following relationship is used to estimate the score ( H ):
H = w
1
N ns
+ w
2
N ws
+ w
3
N ms
+ w
4
N ss
100 × N V F
(7) where w
1
, w
2
, w
3
, w
4 are empirically estimated weights associated with each group of pixels in defining the score and N V F is the total number of visual fields used to define the score of tissue specimen.
IV. RESULTS
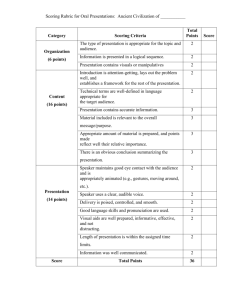
Our dataset consists of histopathology slides from 14 patients. Pathologists’ score (Path-H column in table I) for all the slides is provided by our collaborator at HGH. The score is in the form of x % weak signal, y % medium signal and z % strong signal, we quantify the subjective score using equation 7.
For stain separation, standard stain matrices as recommended in [3] are used. Empirically estimated parameters used in our scoring system are as follows: τ
1
= 0 .
3 , τ
2
=
TABLE I
C OMPARISON OF COMPUTER GENERATED SCORES (H) AGAINST
PATHOLOGISTS ’ SCORES (P ATH -H). L AST COLUMN PRESENTS THE
PERCENTAGE SCORING ACCURACY COMPARED TO P ATH -H.
Fig. 4.
Screenshot of the ER/PR Scoring Software.
Patient Marker NVF
1
2
3
4
5
6
7
8
9
10
ER
PR
ER
PR
ER
PR
ER
PR
ER
PR
ER
PR
ER
PR
ER
PR
ER
PR
ER
PR
Average ER Accuracy
Average PR Accuracy
2
2
2
2
4
3
3
2
3
3
3
3
2
3
3
2
3
3
3
3
H
2.16
0.71
1.63
0.03
1.98
1.43
1.96
2.34
0.00
0.00
2.09
2.03
2.40
0.42
1.59
1.82
1.29
2.03
2.29
0.83
Path-H
2.25
1.50
2.00
0.02
2.75
1.50
2.70
2.70
0.00
0.00
2.85
2.85
2.85
0.90
2.70
2.70
1.80
2.70
3.00
0.60
Accuracy
97.00
73.67
87.67
99.67
86.84
97.67
75.34
88.00
100.00
100.00
74.67
72.67
85.00
84.00
63.00
70.67
83.00
77.67
76.34
92.34
82.89
± 9 .
82
85.64
± 11 .
01
0 .
35 , τ
3
= 0 .
5 , w
1
= 0 , w
2
= 1 , w
3
= 2 , w
4
= 3 . The estimated time needed by the system to process a single image is ≈ 20 seconds. On average, we evaluate 2 4 images per slide to score the slide. Therefore, it approximately takes
1 1 .
5 minutes to score a slide as compared to pathologists’ manual scoring which may take up to tens of minutes.
We evaluate system’s performance by calculating the scoring error , which is defined as the difference between the pathologists’ score and our system’s result. From table
I, it can be seen that the average scoring error for ER hormone receptors is 17 .
11% and for PR hormone receptors is 14 .
36% . Note that these are the results for 10 patients only.
Results for all 14 patients are not presented to save space.
However, the average scoring error approximately remains same even if we include scoring results for all 14 patients.
According to [7], agreement (in terms of kappa score) between the two observers in scoring the hormone receptor status was 0 .
856 and 0 .
711 for ER and PR respectively. Our system produces ER scoring results that are quite comparable to [7] and PR scoring results that are relatively superior, especially considering the fact that the results produced by our system are agreement scores between our system and many pathologists that work at HGH.
V. CONCLUSIONS
The proposed system has been designed to automate the scoring of ER and PR hormone receptors in breast cancer tumor tissues using image processing techniques. The system is capable of scoring the ER/PR prevalence in a histopathology slide in an accurate and efficient manner.
Auxiliary hardware specially developed to help pathologists navigate freely throughout the slide in a more convenient manner. Finally, our system also records all the automated scoring results in an organized database, which makes the documentation of the result easy to analyze for further use.
In future, we intend to extend this system to work for other hormone receptors (e.g. HER 2 ) as well. We have also plans to test the system on even large scale dataset.
ACKNOWLEDGMENTS
This work is partly supported by QNRF grant no NPRP-
5-1345-1-228.
R EFERENCES
[1] M. E. H. Hammond, D. F. Hayes, M. Dowsett, D. C. Allred, K. L.
Hagerty, S. Badve, P. L. Fitzgibbons, G. Francis, N. S. Goldstein,
M. Hayes et al.
, “American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer
(unabridged version),” Archives of pathology & laboratory medicine , vol. 134, no. 7, pp. e48–e72, 2010.
[2] E. Rexhepaj, D. J. Brennan, P. Holloway, E. W. Kay, A. H. McCann,
G. Landberg, M. J. Duffy, K. Jirstrom, and W. M. Gallagher, “Novel image analysis approach for quantifying expression of nuclear proteins assessed by immunohistochemistry: application to measurement of oestrogen and progesterone receptor levels in breast cancer,” Breast
Cancer Research , vol. 10, no. 5, p. R89, 2008.
[3] A. Ruifrok and D. Johnston, “Quantification of histochemical staining by color deconvolution,” Analytical & Quantitative Cytology & Histology , vol. 23, pp. 291–299, 2001.
[4] M. Krajewska, L. H. Smith, J. Rong, X. Huang, M. L. Hyer, N. Zeps,
B. Iacopetta, S. P. Linke, A. H. Olson, J. C. Reed et al.
, “Image analysis algorithms for immunohistochemical assessment of cell death events and fibrosis in tissue sections,” Journal of Histochemistry &
Cytochemistry , vol. 57, no. 7, pp. 649–663, 2009.
[5] C. Taylor and R. Levenson, “Quantification of immunohistochemistryissues concerning methods, utility and semiquantitative assessment II,”
Histopathology , vol. 49, no. 4, pp. 411–424, 2006.
[6] N. Otsu, “A threshold selection method from gray-level histograms,”
IEEE Transactions on Systems, Man and Cybernetics , vol. 9, pp. 62–
66, 1979.
[7] L. Mudduwa, T. Liyanage et al.
, “Immunohistochemical assessment of hormone receptor status of breast carcinoma: Interobserver variation of the quick score,” Indian Journal of Medical Sciences , vol. 63, no. 1, p. 21, 2009.