4 Lecture 4
advertisement

4
Lecture 4
We start with a short revision of the Turing instability.
Consider a 2 variable reaction diffusion model, 1D domain of length L with no-flux boundary
conditions.
@A
@t
@I
@t
=
f1 (A, I) + DA r2 A
=
f2 (A, I) + DI r2 I
f1 , f2 non-linear, stable fixed point for D = 0 , turns unstable for D > 0
We assume perturbations of the form:
✓ ◆ ✓ ◆
A
↵
=
e t cos (kx)
I
k=
n⇡
L
n = 0, 1, 2, . . . is the wavenumber , wavelength = 2⇡
k
✓
◆
a11 a12
J=
a21 a22
✓
◆
a11 Da k 2
a12
JD =
a21
a22 Di k 2
characteristic equation = dispersion relation
2
6
4a11 + a22
|
2
3
7
k 2 (Da + Di )5 + det(JD ) = 0
{z
}
⌧ (JD )<0
det(JD ) = Da Di k 4
(Da a22 + Di a11 ) k 2 + det(J)
| {z }
>0
To obtain an instability we demand that det(JD ) < 0 and obtain the following 3 conditions for a
Turing instability:
I
a11 a22
III Di a11 + Da a22
We define dcrit =
diagonal entries of J opposite signs (I+III)
a11 + a22 < 0
II
⇣
Di
Da
⌘
a12 a21 > 0
p
> 2 Da Di det (J) > 0
crit
k 2 . Since k is a multiple of
off-diagonal entries opposite signs
different diffusion speeds Da 6= Di
and sketch a dispersion plot of det(JD ) versus k 2 and Re( )max verus
⇡
L
we obtain a finite range of wave numbers where the real part of
24
the largest eigenvalue becomes unstable. The growing mode kmax is the one which is closest to the
@
peak of the dispersion curve where @(k
(kmax )
2 ) = 0. max [Re ] = Re
By looking at the sign of the Jacobian we see that there are two principal solutions:
✓
◆
✓
◆
+
+ +
and
+
These can be sketched in the following way: see board
Examples are the Meinhardt model (type 1) and the Bruesselator (type 2).
4.1
The Bruesselator
@u
@t
@v
@t
=
a
=
bu
(b + 1) u + u2 v + Du
u2 v + D v
@2u
@x2
@2v
@x2
a, b, Du , Dv > 0
Please work through Tutorial Problem 2).
4.2
Hydra head regeneration
If cut in half the freshwater polyp Hydra can regenerate two complete animals by reorganising
existing tissue. According to the gradient hypothesis this could be explained by a “head activating
morphogen” which provides positional information. A linear gradient however would require that
cells can accurately detect absolute concentrations. An exponential distribution which could be
achieved by including global degradation (see Lecture 2) would improve things slightly, above all
when combined with an opposing gradient of an inhibitory signal. However, from an engineering
point of view what we would like to have is a more less binary distribution of the activation signal.
A Turing mechanism therefore seems very appropriate.
However, is there any biological evidence that would support an activator/inhibitor mechanism?
Short range head activation
We consider a grafting experiment, whereby head tissue of a labelled Hydra is grafted onto
another animal further down the body column. We observe that the graft stimulates nearby tissue
to transform into a head structure. This suggests that head activation operates on a short range.
Long range inhibition
If we graft tissue from further down the body column onto another animal, the success rate of
growing a head in that region will be very small. This suggests that the presence of the recipient’s
head inhibits the formation of other heads. The inhibition depends on the distance to the recipient’s
head in an almost linear fashion.
Proportion regulation
25
Limiting the production of the activator by including a saturation term for example results in
the length of the activated zone scaling with the length of the system. Activator levels will be
almost constant so that the signal distribution is almost binary.
4.3
The Meinhardt model and a biological interpretation of activator/inhibitor dynamics
You will often encounter the terms “local instability, global stability and lateral inhibition” in the
context of biological models for pattern formation. Another phrase uses for such models is “local
excitation/global inhibition” or LEGI in short.
Let’s consider the following activator-inhibitor model by Meinhardt:
@a
@t
@b
@t
=
⇢a2
b
=
µa + Da r2 a + ⇢0
⇢0 a 2
⌫b + Db r2 b
(17)
(18)
We now want to discuss how these phrases are connected to the dynamics of such a dynamical
system.
1) local instability:
First we turn the equation describing the rate of change for the activator. We radically set all
constants to 1 and assume b to be constant. Also, diffusion is ignored for a moment.
Then eq(17) becomes
da
= a2 a
dt
The steady state is a = 1 . For a > 1 a will grow.
2) now including b and simplifying the second equation we obtain
db
= a2
dt
b
The steady state is b = a2 . Assuming that b equilibrates very rapidly we can replace b in the
activator equation by a2 .
da
a2
a2
=
a⇡ 2 a=1 a
dt
b
a
This means that the presence of b changes the stability of the steady state.
3) Now we add diffusion of b .
At first b can be considered constant as it is fast diffusing. We observe growth of a as described in
1) above (local instability). After a while however b accumulates and cannot be considered constant
anymore. This results in global stabilisation of the pattern (global stabilisation). As inhibitor levels
around an activator peak will be above average the formation of activator peaks nearby an existing
peak will be suppressed (lateral inhibition).
26
4.4
Classification of different mechanisms underlying models for pattern formation
Let’s consider the activator/inhibitor model by Meinhardt:
@a
@t
=
⇢a2
b
|{z}
I
@b
@t
=
µa + Da r2 a + ⇢0
|{z}
(19)
II
⇢0 a 2
|{z}
III
⌫b + Db r2 b
(20)
A) in the model above the inhibitor acts on the activator in a linear way
B) non-linear inhibition would give rise to terms like the following for example
2
I: ⇢a
b2
the inhibitor could for example be a decay product of a , like in
III: ⇢0 a2
For cases A) and B) we can state that the inhibitor slows down activator production.
k
0 m
Generally for terms I: ⇢a
and III: ⇢ ban we obtain globally stable patterns if
bl
lm
>k
n+1
1>0
C) another possibility is that the inhibitor accelerates the activator destruction (enhanced degradation as in the original Turing model)
II:
µba
Often, models of this kind tend to oscillate.
D) Depletion of a substrate or cofactor (see Schnakenberg model for example).
effect: as the activator concentration is limited we obtain broader peaks, which can shift and
split in growing fields
E) Mutual repression: inhibition of inhibition can be equivalent to autocatalysis
F) Mutual activation: leads to very symmetric stripes (Drosophila segmentation models)
5
5.1
Lecture 5
Towards oscillatory behaviour and diffusion coupled oscillators
The instability we discussed so far occurs by det(JD ) changing its sign from positive to negative.
The bifurcation could be either a saddle-node, transcritical, or pitchfork bifurcation. Here, one
of the eigenvalues changes sign from negative to positive. However, biological systems very often
show oscillatory behaviour (for example in the Hodgkin-Huxley model for action potentials in
nerve cells and a simplified caricature, the Fitz-Hugh-Nagumo equations). An important route to
oscillatory behaviour is the appearance of a limit cycle oscillation through a Hopf bifurcation. Here
tr(J) changes its sign from negative to positive. The two conjugate complex eigenvalues cross the
imaginary axis. Coupling of such oscillators by diffusion can give rise to complex dynamic patterns.
27
5.2
Codimension-two Turing-Hopf Bifurcation
See paper by Meixner et al. “Generic spatiotemporal dynamics near codimension-two Turing Hopf
bifurcation”, Physical Review E, 55(6), 6690-6697, 1997.
5.3
Travelling waves in Fisher’s equation
Diffusion problems are often associated with travelling wave solutions. We are looking for solutions
which travel at constant speed and do not change shape.
28
Let’s consider Fisher’s equation
@u
@2u
= ku(1 u) + D
@t
@x
For D = 0 this is the well known logistic growth equation with a carrying capacity of 1.
We non-dimensionalize
by using the following transformations
p
t = k1 ⌧ and x = D/k⇠
so that
@u
@2u
= u(1 u) +
@⌧
@⇠
(21)
We now introduce a wave variable z = ⇠ c⌧ which will allow us to describe the dynamics within
the reference frame of the travelling wave, c being the wave speed.
u(⇠, ⌧ ) = u(⇠
c⌧ ) = u(z)
If we think in terms of populations or concentrations we will demand u(z) > 0 .
Using
@u
@⌧
@u
@⇠
=
c
=
@u
@z
@u
@z
and substituting in eq(21) we obtain
U 00 + cU 0 + U (1
U) = 0
(22)
where the prime denotes differentiation with respect to z .
Eq(22) can be written as a second order system:
U0
=
0
=
V
V
cV
U (1
U)
Steady states (U ⇤ , V ⇤ ) are (0, 0) and (1, 0)
Nullclines are V = 0 and V = U (1c U )
Stability is analysed by linearising around the steady state. The Jacobian is
✓
◆
0
1
J=
2U ⇤ 1
c
For (U ⇤ , V ⇤ ) = (1, 0) we obtain eigenvalues
1,2
=
c±
p
c2 + 4
2
which means that we have one negative and one positive eigenvalue for any value of c and therefore
a saddle node.
For (U ⇤ , V ⇤ ) = (0, 0) we obtain eigenvalues
p
c ± c2 4
=
1,2
2
and thus have a stable node for c > 2 and a stable spiral for 0 < c < 2 .
The latter can be ruled out as it would require U to become negative.
With the boundary conditions
U (+1) = 1 and U ( 1) = 0 the single solution is the heteroclinic connection between the two
fixed points.
The tangent on that trajectory is given by
dV
=
dU
cV
U (1
V
U)
Please try and draw a phase plot in the U,V plane and wave profiles of U(z).
One can show that in case of the Fisher equation the wave speed depends on the initial conditions.
5.4
Summary of biological examples
There is a large number of examples where mathemarical models for spatial pattern formation have
significantly furthered our understanding of biological systems. Some we encountered in the course
of this part of MA265 were:
Head activation and regeneration in Hydra
Segmentation during Drosophila development
Hair follicle spacing in mice
Patterns on animal coats and seashells
Orientation of cells in gradients of an extracellular signal (chemical, mechanical)
Actin waves
Dictyostelium morphogenesis
30
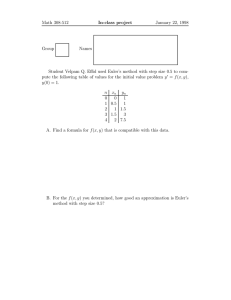
Euler’s method
I
Let’s start with Taylor’s theorem, provided that y (t) 2 C 2 [a, b]:
y (ti+1 ) = y (ti )+(ti+1 ti )y 0 (ti )+
I
We are looking for an approximation to the well-posed initial
value problem
dy
= f (t, y ) , for a t b
dt
I
2
y 00 (⇠i ) , with i = 0, 1, 2, . . . , N 1
⇠i is some number in (ti , ti+1 ) . With h = ti+1
(1)
We discretize the time domain using equally spaced mesh points
We might want to interpolate afterwards to find intermediate
approximations.
ti we can write
h2 00
y (⇠i )
2
Replacing y 0 (ti ) with f (ti , y (ti )) (see the definition of our IVP) we
obtain
y (ti+1 ) = y (ti ) + hy 0 (ti ) +
I
ti = a + ih , with i = 0, 1, 2, . . . , N
I
ti ) 2
(ti+1
h2 00
y (⇠i )
2
By dropping the remainder term we obtain a difference equation
y (ti+1 ) = y (ti ) + hf (ti , y (ti )) +
I
wi+1 = wi + hf (ti , wi ) , with i = 0, 1, 2, . . . , N
1
and starting value w0 = ↵ .
Error bound for Euler’s method
The error bound for the Euler method depends linearly on the step
size h .
If we are able to compute the second derivative y 00 (t) and a constant
M for all t 2 [a, b] exists with |y 00 (t)| M , then the error bound is:
|y (ti )
hM h L(ti
wi |
e
2L
|y (ti ) wi | is the global discretization error.
L is the Lipschitz constant .
a)
i
1
Theorem
Suppose f (t, y ) is defined on a convex set D ⇢ R2 . If a constant
L > 0 exists with
@f
(t, y ) L , for all (t, y ) 2 D
@y
, then f satisfies a Lipschitz condition on D in the variable y with
Lipschitz constant L .
Theorem
Suppose that D = {(t, y ) | a t b, 1 < y < 1} and that f (t, y ) is
continuous on D . If f satisfies a Lipschitz condition on D in the
variable y , then the IVP dy
dt = f (t, y ) , for a t b with initial
conditions y (a) = ↵ has a unique solution y (t) for a t b .
Theorem
Suppose that D = {(t, y ) | a t b, 1 < y < 1} and that f (t, y ) is
continuous on D . If f satisfies a Lipschitz condition on D in the
variable y , then the IVP dy
dt = f (t, y ) , for a t b with initial
conditions y (a) = ↵ is well posed.
Can we decrease the step size infinitely to obtain
better accuracy?
I
I
No, because the previous error bound neglected the round-off
error. When decreasing h , the step size, we have to do more
calculations which increases the round-off error.
Let’s introduce errors in the initial condition ( 0 ) as well as the in
the difference equation (error i at step i):
u0 = ↵ +
I
total error can go to infinity since limh!0
I
0
ui+1 = ui + hf (t, ui ) +
One can show that if h becomes smaller than
hM
2
q
2
M
, then the
+h =1
In practise we rarely set h to so small values that we could be
affected by the round-off error, simply because the speed of the
computation would be too low.
i+1
if | i |
The error bound is then given by
✓
◆h
i
1 hM
|y (ti ) wi |
+
eL(ti a) 1 + | 0 | eL(ti
L
2
h
a)
and no longer depends linearly on h .
The Improved Euler method
I
Euler’s method estimates the derivative only at the left end of the
interval between tn and tn+1 .
I
The improved Euler method uses the average derivative over this
interval.
I
Let’s first approximate the right side of the interval [ti , ti+1 ] using
an Euler step
ŵi+1 = wi + hf (ti , wi )
I
Then we use the mean of the derivative at both ends of the
interval to compute our next approximation.
wi+1
I
1
= wi + h [f (ti , wi ) + f (ti+1 , ŵi+1 )]
2
The truncation error for the improved Euler method is O(h2 ) .
Runge-Kutta methods
Runge-Kutta methods were developed to avoid the computation of
higher order derivatives which higher order Taylor methods may
involve. In place of these derivatives extra values of the given function
f(x,y) are used, in a way which duplicates the accuracy of a Taylor
polynomial
In practice, a good balance between computational cost and
accuracy is achieved by the fourth-order Runge-Kutta method.
I
Runge-Kutta Order Four
w0
=
↵
k1
=
hf (ti , wi )
k2
=
hf (ti + h/2, wi + 1/2k1 )
k3
=
hf (ti + h/2, wi + 1/2k2 )
k4
=
hf (ti+1 , wi + k3 )
wi+1
=
wi + 1/6 (ki + 2k2 + 2k3 + k4 )
for each i = 0, 1, 2, . . . , N
1
Adaptive methods
I
I
I
I
I
The Euler method can be formally classified as Runge-Kutta
method of order 1, the modified Euler as RK of order 2.
The local truncation error is O(h4 ) . Four evaluations per step are
needed. Even higher order methods allow larger timesteps, but
require too many evaluations to obtain the same local truncation
error, so that the trade-off for the Runge-Kutta method of order
four is best.
If wi+1 is the approximation for our method with truncation error
O(hn ) and ŵi+1 the approximation for our method with truncation
error O(hn+1 ) then one can approximate the local truncation
error as
1
⌧i+1 (h) = (ŵi+1 wi+1 )
h
To obtain a new step size qh such that the truncation error is
smaller than " we can use the following formula
q
I
I
✓
"h
|ŵi+1
wi+1 |
◆1/n
The Runge-Kutta-Fehlberg method (ode45() in Matlab) uses a
RK method of order 5 to estimate the local error in a RK method
of order 4. In total only six evaluations per step are needed for
this method.
If the truncation error is greater than " than wi+1 is discarded and
evaluated again using the new step size qh. This step size is
kept for the next iteration. If the truncation error is comparatively
small, then the step size is increased again.
If we have an estimate of the truncation error then we can use an
adaptive step size to cross “easy parts” of a solution in a few big
steps, while the challenging parts are crossed with small
stepsizes. Number and position of our mesh points are varied
such that the truncation error is kept within a specified bound.
|y (ti )
wi | < "
I
The aim is to minimize the number of mesh points needed to
approximate our solution.
I
The idea is to predict the local truncation error by using methods
of differing order. Using the predicted error we can choose a step
size that will keep the local and the global truncation error in
check.
Multivariable Systems
An m ’th order system of first-order initial value problems has the form
du1
(t)
dt
du2
(t)
dt
dum
(t)
dt
=
f1 (t, u1 , u2 , . . . , um )
=
f2 (t, u1 , u2 , . . . , um )
..
.
=
fm (t, u1 , u2 , . . . , um )
for a t b , with initial conditions
u1 (a) = ↵1 , u2 (a) = ↵2 , . . . , um (a) = ↵m
The problem is now to solve for all variables u1 (t) , . . . , um (t).
Deriving finite differences methods for the diffusion
equation
Starting from Fick’s second law:
@c
@2c
=D 2
@t
@x
and using ci,j for the concentration at time j and at position i .
we can first use a Taylor series in the time direction while keeping x
constant:
✓ ◆
✓ 2 ◆
@c
1
@ c
ci,j+1 = ci,j + 4t
+ (4t)2
+ ...
@t i,j 2
@t 2 i,j
which yields:
✓
@c
@t
◆
=
i,j
ci,j+1 ci,j
+ O(4t)
4t
Let’s do the same for the x direction for 4x and 4x , now keeping
the time constant.
✓ ◆
✓ 2 ◆
@c
1
@ c
2
ci+1,j = ci,j + 4x
+ (4x)
+ ...
@x i,j 2
@x 2 i,j
ci
1,j
= ci,j
4x
✓
@c
@x
◆
i,j
1
+ (4x)2
2
✓
@2c
@x 2
◆
...
i,j
Adding both equations gives:
✓ 2 ◆
ci 1,j 2ci,j + ci+1,j
@ c
=
+ O((4x)2 )
@x 2 i,j
(4x)2
Possible numerical instabilities in the explicit scheme
Using both difference equations for
@c
@t i,j
and
⇣
@2c
@x 2
⌘
i,j
following central difference equation for Fick’s 2nd law:
and
ci,j+1 ci,j
ci
=D
4t
ci,j+1 = ci,j + 4t D
2
which is of order O(4t, (4x) )
ci
1,j
1,j
we obtain the
I
2ci,j + ci+1,j
(4x)2
2ci,j + ci+1,j
(4x)2
The previously derived explicit scheme is conditionally stable:
For 1D the following condition must hold:
D ⇤ 4t/(4x)2
I
1
2
Implicit methods have been developed that are unconditionally
stable (eg Crank-Nicolson method).
4th order accurate central differences in 1D
✓
@2c
@x 2
◆
=
i
ci+2 + 16ci+1
30ci + 16ci
12(4x)2
1
cx
2
+ O((4x)4 )
I
Matlab’s standard ODE solver takes only one single column
vector as input for the initial conditions. If we have 2 variables we
must therefore append these before passing them.
I
In the function that evaluates the differential equations it is
sometimes easier to split the column vector first so that the
variables can be indexed easier.
I
No-flux boundary conditions can be easily implemented by
mirroring the boundary values, i.e. if our grid runs from 1:N we
create an index vector for the left neighbour in the form of
xm1=[1,1:N-1]; and for the right neighbour as xp1=[2:N,N];
The Method of Lines
I
The Method of Lines transforms a PDE problem into a ODE
problem which can be approximated using the standard solvers
such as Runge-Kutta.
I
For our case of reaction-diffusion problems each grid point in our
discretized domain becomes an independent variable. The
variables will be coupled according to the particular finite
difference approximation that is used.
I
If we have a 2 variable PDE model with N grid points we will
therefore have to solve a system of 2*N coupled ODEs.
function fisher()
%set up time span
tfinal=100;
%domain length
L=150;
%number of grid points
X=150;
%space step
P.deltax = L/X;
%initial conditions
a0=zeros(X,1);
a0(1)=0.5;
[t,y]=ode45(@dydt_fisher,0:tfinal,[a0],[],P);
%output
colormap(jet(256));
figure(1)
imagesc([0:L],[0:tfinal],y)
figure(2)
plot([0:L-1],y(25,:));
figure(3)
c=0;%2
for t=20:30
plot([1:L]-c*t,y(t,:));
hold on
end
hold off
end
function dydt=dydt_fisher(t,dydt,P)
X=length(dydt);
dydt = dydt.*(1-dydt) + 1/P.deltax^2*(dydt([1,1:X-1])2*dydt+dydt([2:X,X]));
end
!
!
function bruesselator()
%time span
tfinal=100;
tspan=[0:2:tfinal];
%domain length
L=2*14.5;%2*14.5
%number of grid points
X=50;
%space step
P.deltax = L/X;
%model parameters
P.a=1.5;
P.b=2.5 %critical=2.34
%diffusion constants
P.Da = 2.8;
P.Db = 22.4% 22.4;
%initial conditions
a0=P.a + 0.01*(-1+2*rand(X,1) );
b0=P.b/P.a*ones(X,1);
[t,y]=ode45(@dydt_brusselator,tspan,[a0;b0],[],P);
colormap(jet(256));
figure(1)
imagesc([0:L],[0:tfinal],y(:,1:X))
figure(2)
imagesc([0:L],[0:tfinal],y(:,X+1:2*X))
end
function dydt=dydt_brusselator(t,dydt,P)
X=length(dydt)/2;
a = dydt(1:X);
b = dydt(X+1:2*X);
x=1:X;
xm1=[1,1:X-1];
xp1=[2:X,X];
dydt(1:X) = P.a-(P.b+1)*a+a.^2.*b + P.Da/P.deltax^2*(a(xm1)2*a(x)+a(xp1));
dydt(X+1:2*X) = P.b*a-a.^2.*b + P.Db/P.deltax^2*(b(xm1)2*b(x)+b(xp1));
end!




![[t,y]=ode45(‘ ',[0 tlim], [4 4]); dydt= (t,y)](http://s2.studylib.net/store/data/013524672_1-a672587e3d402c460771d7b588302558-300x300.png)



