Genome-Wide Association and Fine Mapping Brassica oleracea
advertisement

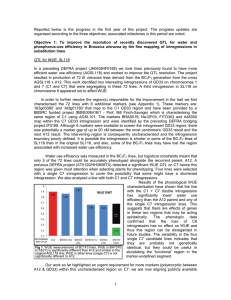
Genome-Wide Association and Fine Mapping of Genetic Loci Predisposing to Water Use Efficiency in Brassica oleracea (L.) Sajjad Awan1, Miriam Gifford1, John Hammond2 and Andrew Thompson3 1. University of Warwick, University of Warwick; 2. University of Western Australia; 3. Cranfield University Introduction Rapid increase in human population has put extreme pressure on agricultural resources especially irrigation water and threatens the food security Brassicas are important vegetable crops grown in the UK for edible oil/vegetables and face periodic water shortages in the UK especially during winter The Current project focuses on locating genes responsible for water use efficiency (WUE) in these plants without the use of transgenic technology. It was achieved as follow; The double haploid A12 (Chinese kale) and GD33 (broccoli) cross pollinated to obtain a mapping population in the ‘C’ genome (Brassica oleracea) The QTL for WUE was found on Chr-7; evidence from substitution line 118 (BC1F1) shown in Figure.1 The BC1F3 (selfing BC1F2)population showed reduction in WUE (Figure.3) due to introgression of GD33 on A12 on Chr-1 & 7. Fine mapping was carried out by using SNP markers (KASPar). The RFLP and SSR markers were used to genotype the introgressed regions (Fig.1) sora93 BRAS023 A48350 FITO302 Na12F03 BRAS019 Ca72 LG C7 Marker Position 0.6 1.1 1.6 2.8 2.8 2.8 3.3 3.5 2.2 Marker M1 M2 M3 M4 M5 M6 M7 M8 M9 11 13 14 14 14 14 17 17 17 17 18 19 24 27 29 30 Marker score BAT070-02 BC1F3 T:T C:C T:T G:G T:T A:A A:A C:C G:G A:A C:C A:A A:A T:T G:G A:A G:G C:C A:A T:T T:T C:C C:C T:T G:G C:C T:T BAT090-04 BC1F4 T:T C:C T:T G:G T:T - A:A C:C - A:A C:C A:A A:A - G:G A:A A:A A:A G:G C:C A:A T:T T:T C:C G:G C:C T:T BAT075-06 BC1F3 T:T C:C T:T G:G T:T A:A A:A C:C C:C G:G T:T G:G G:G C:C A:A G:G - A:A G:G C:C A:A T:T T:T C:C G:G C:C T:T G G A A A A A A A BAT011 SL118-11_H9 A A A/H G H H G/H H H H BAT047 BAT074-05 BC1F3 A:A C:T C:C T:T C:C G:G A:A C:C G:G A:A C:C A:A A:A T:T G:G A:A G:G C:C - T:T T:T C:C C:C T:T G:G C:C T:T SL118-11_C7 A A A/H H G G G/H H A A BAT048 BAT089-06 BC1F4 A:A C:T C:C T:T C:C G:G C:C C:T C:C A:A C:C A:A A:A T:T G:G A:A G:G C:C A:A T:T T:T C:C C:C T:T G:G C:C T:T SL118-11_G3 A A A/H A G G G/H G G G BAT056 BAT074-09 BC1F3 A:A C:T C:C T:T C:C G:G A:C C:T - A:G C:T A:G A:G T:C G:G A:A G:G C:C A:A T:T T:T C:C C:C T:T G:G C:C T:T SL118-8_B1 A A A/H A H H G/H H H G BAT057 BAT075-03 BC1F3 A:A C:T C:C T:T C:C G:G C:C C:T C:C G:G T:T G:G G:G C:C A:A G:G A:A A:A G:G C:C A:A C:C C:C T:T G:G C:C T:T SL118-8_C11 A A/H H T:T G:G T:T A:A A:A C:C G:G A:A C:C A:A A:A T:T G:G A:A G:G C:C A:A T:T T:T C:C C:C T:T G:G C:C T:T G G BAT058 BC1F1 C:C G/H H T:T A H AGSL118 A A12 Recurrent Parent A:A C:T C:C T:T C:C G:G C:C C:T C:C G:G T:T - G:G C:C A:A G:G A:A A:A G:G C:C - T:T T:T C:C G:G C:C T:T 1 1 1 1 1 1 2 2 3 4 4 4 4 4 5 5 6 6 6 6 6 7 7 7 SL118-10_C1 A A A/H A H G G/H G G G BAT059 A A A/H A G G G/H G G H BAT060 SL118-8_C1 A A A/H A G G G/H G H H BAT061 SL118-9_D6 A A A/H A H H G/H H H A BAT068 SL118-11_E8 A A A/H A H H G/H H H A BAT069 SL118-8_B3 G G G A G G G/H G G G BAT70 SL118-10_G6 G SL118-11_C6 G G G A/H G A A H H H H G/H G/H H H H H H H BAT071 BAT072 BINS Fig 1. Genotyping results matrix for BC1F2 lines (left Panel) and BC1F3/BC1F4 lines (upper panel) derived from AGSL118 x A12DHd. Dark green cell colour denotes a GD33 allele; Yellow cell colour denotes an A12 allele; H denotes both parent alleles detected i.e. heterozygous result. A/H or G/H indicates that the marker result is partially ambiguous; - denotes a missing value. The 6 SSR markers were used on Linkage group-7 to select BC1F3 lines. The BC1F3/BC1F4 lines originated from selfing of BC1F2 lines preselected by using SSR/SNP markers. Upper panel shows the genotyping results from KASPar markers in BC1F3/BC1F4 lines. Introgression of GD33 on Chr-7 significantly increased the gs (Figure-2) and reduced WUE (Figure-3) Selected lines were self pollinated to achieve homozygous BC1F3/BC1F4 population The Selected lines were genotyped using KASPar markers on LG-07 only (Fig.1, lower panel) Statistical analysis of BINS in Fig.1 revealed significant association of BIN-3 and BIN-7 with WUEp Genes present in the selected locus will be identified and knockout mutant will be produced to confirm the ‘selected’ genes function. Fig ure.2. Glasshouse characterisation for plant stomatal conductance in selected BC1F3 lines. BAT70 reveals a significantly higher stomatal conductance compared to recurrent parent (A12). Fig ure.3. Glasshouse characterisation for water use efficiency in selected BC1F3 lines. BAT70 reveals a significantly lower WUE compared to recurrent parent. Genome Wide Association Mapping 96 ecotypes from Arabidopsis were used in the analysis SNPs with relatively high ‘diff’ value and OR low ‘P-value’ (<0.0005) were plotted to find the clustering hits (Figure.4) Using ‘R’ and ‘Virtual Plant’ genes were selected associated with the SNPs (Circled in Fig.4). Figure.4. Selection of putative SNPs involved in controlling water use efficiency in Arabidopsis. Figure.5. Polymorphism in Beta Glucosidase Gene. Green Nucleotides represent confirmed SNPs, red ‘rejected SNPs’ 32 M10 M11 M12 M13 M14 M15 M16 M17 M18 M19 M20 M21 M22 M23 M24 M25 M26 M27 Genotype Pedigree Backcross Progeny 14 SL118-8_A6 G SL118-8_E6 Three genotypes were selected to analyse the effect of introgression of GD33 on plant stomatal conductance (gs) and WUE At3g01180 Sample Name Ni4B10 Fine mapping the introgressed regions in A12DHd x SL118 progeny At3g03380 LG LG C1 C6 The PCR products from the selected genes were sequenced for polymorphism (Fig.5) Conclusion Use of recent technologies such as SNP based assays (KASPar) has made it possible for efficient genotyping on large scale. This technology can be used to find QTL responsible for nitrogen and phosphorus use efficiency in relatively short period of time. The orthologues controlling the WUE in arabidopsis can be identified with the availability of well annotated whole genome sequence of Brassica oleracea . The approaches implemented in the current research project can also be used in other important crops such as oil seed rapes, wheat , soybean and maize. Warwick Crop Centre www.warwick.ac.uk/go/wcc