Applied Topology Henry Adams July 18, 2013 Stanford University MathemaAcs Camp
advertisement

Applied
Topology
Henry
Adams
July
18,
2013
Stanford
University
MathemaAcs
Camp
Filtration parameter
t
Datasets
have
shapes
What
shape
is
this?
0.191
Datasets
have
shapes
Example:
Diabetes
study
145
points
in
5‐dimensional
space
G. M. Reaven and R. G. Miller: The Nature of Chemical Diabetes
19
Fig. 1. Artist's rendition of data as
seen in three dimensions. View is
approximately along 45~line as seen
through Prim 9 program on the computer; coordinate axes are in the
9background
An
A$empt
to
Define
the
Nature
of
Chemical
Diabetes
Using
a
Mul;dimensional
Table 1. Classification of the 145 subjects into three groups on the basis of the oral glucose tolerance test
Analysis
by
G.
M.
Reaven
and
R.
G.
Miller,
1979.
Metabolic characteristics (mean + SD)
Group
Number
Rel.wt.
Glucose area
Insulin area
SSPG
Datasets
have
shapes
Example:
Cyclo‐Octane
(C8H16)
data
1,031,644
points
in
72‐dimensional
space
Non‐Manifold
Surface
Reconstruc;on
from
High
Dimensional
Point
Cloud
Data
by
Shawn
MarAn
and
Jean‐Paul
Watson,
2010.
Datasets
have
shapes
Example:
Cyclo‐Octane
(C8H16)
data
1,031,644
points
in
72‐dimensional
space
Figure 7: Conformation Space of Cyclo-Octane. Here we show how the set of conforma
tions of Non‐Manifold
Surface
Reconstruc;on
from
High
Dimensional
Point
Cloud
Data
by
cyclo-octane can be represented as a surface in a high dimensional space. On the
Shawn
MarAn
and
Jean‐Paul
Watson,
2010.
left, we show
various conformations of cyclo-octane as drawn by PyMol (www.pymol.org)
In the center, these conformations are represented by the 3D coordinates of their atoms
Datasets
have
shapes
Example:
Cyclo‐Octane
(C8H16)
data
1,031,644
points
in
72‐dimensional
space
Figure 7: Conformation Space of Cyclo-Octane. Here we show how the set of conforma
tions of Non‐Manifold
Surface
Reconstruc;on
from
High
Dimensional
Point
Cloud
Data
by
cyclo-octane can be represented as a surface in a high dimensional space. On the
Shawn
MarAn
and
Jean‐Paul
Watson,
2010.
left, we show
various conformations of cyclo-octane as drawn by PyMol (www.pymol.org)
In the center, these conformations are represented by the 3D coordinates of their atoms
Datasets
have
shapes
Example:
Cyclo‐Octane
(C8H16)
data
1,031,644
points
in
72‐dimensional
space
Figure 8: Triangulation of Cyclo-Octane Conformation Space. Here we show the triangulation obtained by our surface reconstruction algorithm on the cyclo-octane conformation
space. The triangulation was carried out in 24 dimensions, but is shown using the reduced
dimensional representation provided by Isomap. Self-intersections are shown in black.
Non‐Manifold
Surface
Reconstruc;on
from
High
Dimensional
Point
Cloud
Data
by
Shawn
MarAn
and
Jean‐Paul
Watson,
2010.
Topology
studies
shapes
A
donut
and
coffee
mug
are
“homotopy
equivalent.”
~
Topology
studies
shapes
A
donut
and
coffee
mug
are
“homotopy
equivalent.”
~
8):11, 1–18
Singh et al.
Topology
studies
shapes
Topological
Analysis
of
Popula;on
Ac;vity
in
Visual
Cortex
by
Singh,
Memoli,
Ishkhanov,
Sapiro,
Carlsson,
and
Ringach,
2008.
Topology
studies
shapes
Torus
Topology
studies
shapes
Topology
studies
shapes
Klein
boale
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
Homology
groups
H0,
H1,
H2,
H3,
…
Hk
“counts
the
number
of
k‐dimensional
holes”.
Homotopy
equivalent
shapes
have
the
same
homology
groups.
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
H0
has
rank
1.
H1
has
rank
1.
H2
has
rank
0.
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
H0
has
rank
1.
H1
has
rank
3.
H2
has
rank
0.
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
H0
has
rank
1.
H1
has
rank
0.
H2
has
rank
1.
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
H0
has
rank
3.
H1
has
rank
4.
H2
has
rank
1.
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
H0
has
rank
1.
H1
has
rank
2.
H2
has
rank
1.
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
H0
has
rank
1.
H1
has
rank
2.
H2
has
rank
1.
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
0‐simplex
1‐simplex
2‐simplex
3‐simplex
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
0‐simplex
1‐simplex
2‐simplex
Simplicial
complexes
3‐simplex
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
0‐simplex
1‐simplex
2‐simplex
3‐simplex
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
0‐simplex
1‐simplex
2‐simplex
3‐simplex
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
0‐simplices
1‐simplices
2‐simplices
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
0‐simplices
1‐simplices
2‐simplices
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
0‐cycle
1‐cycle
A
cycle
has
no
boundary.
2‐cycle
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
0‐cycle
1‐cycle
A
cycle
has
no
boundary.
2‐cycle
+ Bp with c ∈ Z
.
More
formally,
this
collection
is
ca
p
R2
ycles in the same coset are said to be homologous, whic
Topology
studies
shapes
see Figure IV.5. We may take c as the representative
Homology
(
)
Z/2Z
A torus
with three closed curves, each a 1-cycle. Only one
Two
cycles
are
equivalent
if
they
differ
by
a
boundary.
mologous
to the sum of the other two. The sum of the t
Hk
measures
equivalence
classes
of
k‐cycles.
1-boundary, namely of the pair of pants between them.
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
H0
has
rank
1.
H1
has
rank
1.
H2
has
rank
0.
Two
cycles
are
equivalent
if
they
differ
by
a
boundary.
Hk
measures
equivalence
classes
of
k‐cycles.
R2
Topology
studies
shapes
Homology
(
)
Z/2Z
Homology
groups
H0,
H1,
H2,
H3,
…
Hk
“counts
the
number
of
k‐dimensional
holes”.
Homotopy
equivalent
shapes
have
the
same
homology
groups.
“Topology!
The
stratosphere
of
human
thought!
In
the
twenty‐fourth
century
it
might
possibly
be
of
use
to
someone
…” ‐Aleksandr
Solzhenitsyn,
The
First
Circle
Appearance
draw balls
Topology
applied
to
data
analysis
draw complex
Filtration parameter
What
shape
is
this?
t
0.191
Appearance
draw balls
Topology
applied
to
data
analysis
draw complex
Filtration parameter
What
shape
is
this?
t
0.191
Appearance
draw balls
Topology
applied
to
data
analysis
draw complex
Filtration parameter
What
shape
is
this?
Cech
complex
t
0.191
Appearance
draw balls
Topology
applied
to
data
analysis
draw complex
Filtration parameter
What
shape
is
this?
Cech
complex
t
0.191
Appearance
draw balls
Topology
applied
to
data
analysis
draw complex
Filtration parameter
What
shape
is
this?
Cech
complex
t
0.191
Topology
applied
to
data
analysis
What
shape
is
this?
Cech
complex
Topology
applied
to
data
analysis
What
shape
is
this?
Cech
complex
Topology
applied
to
data
analysis
Significant
features
persist.
Image
patch
example
Study
3x3
high‐contrast
patches
from
images
Points
in
9‐dimensional
space
On
the
Local
Behavior
of
Spaces
of
Natural
Images
by
Gunnar
Carlsson,
Tigran
Ishkhanov,
Vin
de
Silva,
and
Afra
Zomorodian,
2008.
Image
patch
example
1st
densest
group
of
patches
Image
patch
example
1st
densest
group
of
patches
"#%
"#&
"#'
"#(
"
!"#(
!"#'
!"#&
!"#%
!!
!"#$
"
"#$
!
InterpretaAon:
nature
prefers
linearity
Image
patch
example
2nd
densest
group
of
patches
Image
patch
example
2nd
densest
group
of
patches
"#%
"#&
"#'
"#(
"
!"#(
!"#'
!"#&
!"#%
!!
!"#$
"
"#$
!
InterpretaAon:
nature
prefers
horizontal
and
verAcal
direcAons
Image
patch
example
3rd
densest
group
of
patches
7
Image
patch
example
3rd
densest
group
of
patches
Image
patch
example
3rd
densest
group
of
patches
Image
patch
example
3rd
densest
group
of
patches
Image
patch
example
3rd
densest
group
of
patches
Image
patch
example
3rd
densest
group
of
patches
Image
patch
example
3rd
densest
group
of
patches
Image
patch
example
3rd
densest
group
of
patches
InterpretaAon:
nature
prefers
linear
and
quadraAc
patches
at
all
angles
Datasets
have
shapes
Example:
Cyclo‐Octane
(C8H16)
data
1,031,644
points
in
72‐dimensional
space
Figure 8: Triangulation of Cyclo-Octane Conformation Space. Here we show the triangulation obtained by our surface reconstruction algorithm on the cyclo-octane conformation
space. The triangulation was carried out in 24 dimensions, but is shown using the reduced
dimensional representation provided by Isomap. Self-intersections are shown in black.
Non‐Manifold
Surface
Reconstruc;on
from
High
Dimensional
Point
Cloud
Data
by
Shawn
MarAn
and
Jean‐Paul
Watson,
2010.
4
a.nb
In[422]:=
Evasion paths in mobile sensor
networks
12
Rips!data1, t, 0, 1"
Rips simplicial complex
Appearance
draw complex
draw balls
Filtration parameter
In[423]:=
In[429]:=
Rips!data3n, t, 0, 1"
In[430]:=
a.nb
Rips!data4n, t, 0, 1"
5
In[431]:=
a.nb
Rips!data5n, t, 0, 1"
11
In[427]:=
10
Rips!data6, t, 0, 1"
a.nb
In[428]:=
a.nb
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
Filtration parameter
t0.358
Out[423]=
Out[429]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[430]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[431]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Printed by Mathematica for Students
a.nb
Filtration parameter
t0.358
Out[427]=
13
Rips data7, t, 0, 1
Rips simplicial complex
Filtration parameter
t
Out[422]=
Rips data2, t, 0, 1
a.nb
t0.358
0.358
Out[428]=
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
9
Coverage problem
• Sensors in a domain D ⊂ Rn .
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Coverage problem
• Sensors in a domain D ⊂ Rn .
• Measure only local connectivity.
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Coverage problem
• Sensors in a domain D ⊂ Rn .
• Measure only local connectivity.
• Čech complex.
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Coverage problem
• Sensors in a domain D ⊂ Rn .
• Measure only local connectivity.
• Čech complex.
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Sensors move in a domain D ⊂ Rn over time interval I .
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Sensors move in a domain D ⊂ Rn over time interval I .
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Sensors move in a domain D ⊂ Rn over time interval I .
• Measure only local connectivity.
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Sensors move in a domain D ⊂ Rn over time interval I .
• Measure only local connectivity.
• Čech complex.
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Sensors move in a domain D ⊂ Rn over time interval I .
• Measure only local connectivity.
• Čech complex.
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Let X ⊂ D × I be the covered region.
D
I
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Let X ⊂ D × I be the covered region.
• Using coordinate-free input, can we determine if an evasion
path exists?
D
I
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Let X ⊂ D × I be the covered region.
• Using coordinate-free input, can we determine if an evasion
path exists?
D
I
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
4
• Let X ⊂ D × I be the covered region.
• Using coordinate-free input, can we determine if an evasion
path exists?
a.nb
In[422]:=
6
Rips data1, t, 0, 1
In[423]:=
In[424]:=
a.nb 8 5 a.nb
Rips!data3y, t, 0, 1"
In[425]:=
Rips!data4y, t, 0, 1"
In[426]:=
a.nb 107
Rips!data5y, t, 0, 1"
In[427]:=
Rips!data6, t, 0, 1"
a.nb
In[428]:=
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
Filtration parameter
t
Filtration parameter
t0.358
Out[423]=
Filtration parameter
t0.358
Out[424]=
Filtration parameter
t0.358
Out[425]=
Filtration parameter
t0.358
Out[426]=
Filtration parameter
t0.358
Out[427]=
a.nb
Rips data7, t, 0, 1
Rips simplicial complex
Filtration parameter
Out[422]=
Rips data2, t, 0, 1
a.nb
t0.358
0.358
Out[428]=
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
9
Evasion problem
• Stacked complex SC ! X encodes all Čech complexes.
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Stacked complex SC ! X encodes all Čech complexes.
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Stacked complex SC ! X encodes all Čech complexes.
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Theorem 7 (reformulated)
If there is an α ∈ Hn (SC, ∂D × I) with
0 != δα ∈ Hn−1 (∂D × I) , then no evasion path exists.
D
α
I
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Theorem 7 (reformulated)
If there is an α ∈ Hn (SC, ∂D × I) with
0 != δα ∈ Hn−1 (∂D × I) , then no evasion path exists.
D
I
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Theorem 7 (reformulated)
If there is an α ∈ Hn (SC, ∂D × I) with
0 != δα ∈ Hn−1 (∂D × I) , then no evasion path exists.
D
I
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Theorem 7 (reformulated)
If there is an α ∈ Hn (SC, ∂D × I) with
0 != δα ∈ Hn−1 (∂D × I) , then no evasion path exists.
• Coordinate-free.
• Not sharp. Can it be sharpened?
D
I
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Evasion problem
• Theorem 7 (reformulated)
If there is an α ∈ Hn (SC, ∂D × I) with
0 != δα ∈ Hn−1 (∂D × I) , then no evasion path exists.
• Coordinate-free.
• Not sharp. Can it be sharpened?
D
I
Coordinate-free Coverage in Sensor Networks with Controlled
Boundaries via Homology by V. de Silva and R. Ghrist
Zigzag persistence
• Form ZSC and take Hn−1 (ZSC) ∼
= Hn−1 (ZX).
Zigzag Persistence by G. Carlsson and V. de Silva
Zigzag persistence
!→
!→
!→
Zigzag Persistence by G. Carlsson and V. de Silva
←!
←!
←!
←!
!→
• Form ZSC and take Hn−1 (ZSC) ∼
= Hn−1 (ZX).
Zigzag persistence
• Form ZSC and take Hn−1 (ZSC) ∼
= Hn−1 (ZX).
Zigzag Persistence by G. Carlsson and V. de Silva
Zigzag persistence
• Form ZSC and take Hn−1 (ZSC) ∼
= Hn−1 (ZX).
• Hypothesis: there is an evasion path ⇔ there is a long bar.
Zigzag Persistence by G. Carlsson and V. de Silva
Zigzag persistence
• Form ZSC and take Hn−1 (ZSC) ∼
= Hn−1 (ZX).
• Hypothesis: there is an evasion path ⇔ there is a long bar.
• ⇒ is true, but ⇐ is false.
Zigzag Persistence by G. Carlsson and V. de Silva
Dependence on embedding X !→ D × I
• SC alone does not determine if an evasion path exists.
• Two networks with the same SC . Top contains evasion path
while bottom does not.
Dependence on embedding X !→ D × I
4
a.nb
In[422]:=
• SC alone does not determine if an evasion path exists.
• Two networks with the same SC . Top contains evasion path
while bottom does not.
6
Rips data1, t, 0, 1
In[423]:=
In[424]:=
a.nb 8 5 a.nb
Rips!data3y, t, 0, 1"
In[425]:=
Rips!data4y, t, 0, 1"
In[426]:=
a.nb 107
Rips!data5y, t, 0, 1"
In[427]:=
Rips!data6, t, 0, 1"
a.nb
In[428]:=
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
Filtration parameter
t
Filtration parameter
t0.358
Out[423]=
Out[424]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[425]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[426]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[427]=
a.nb
Rips data7, t, 0, 1
Rips simplicial complex
Filtration parameter
Out[422]=
Rips data2, t, 0, 1
a.nb
t0.358
0.358
Out[428]=
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
9
Dependence on embedding X !→ D × I
4
a.nb
In[422]:=
• SC alone does not determine if an evasion path exists.
• Two networks with the same SC . Top contains evasion path
while bottom does not.
6
Rips data1, t, 0, 1
In[423]:=
Rips!data3y, t, 0, 1"
In[425]:=
Rips!data4y, t, 0, 1"
In[426]:=
a.nb 107
Rips!data5y, t, 0, 1"
In[427]:=
Rips!data6, t, 0, 1"
a.nb
In[428]:=
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
Filtration parameter
4Out[422]=
a.nb
In[423]:=
Rips data2, t, 0, 1
Filtration parameter
t0.358
6Out[424]=
a.nb
Out[423]=
Rips data1, t, 0, 1
Filtration parameter
t0.358
In[424]:=
In[425]:=
Filtration parameter
t0.358
a.nb 8Out[426]=
5 a.nb
Out[425]=
Rips!data3y, t, 0, 1"
Filtration parameter
t0.358
Rips!data4y, t, 0, 1"
In[426]:=
Rips!data5y, t, 0, 1"
In[427]:=
t0.358
Rips!data6, t, 0, 1"
In[428]:=
Rips data7, t, 0, 1
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
Filtration parameter
t
Filtration parameter
t0.358
Out[423]=
t0.358
Out[424]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[425]=
Printed by Mathematica for Students
Filtration parameter
Out[426]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[427]=
Printed by Mathematica for Students
draw balls
Filtration parameter
t0.358
t0.358
0.358
Out[428]=
Printed by Mathematica for Students
a.nb
9
0.358
a.nb 10
7 a.nb
Out[428]=
Rips simplicial complex
Filtration parameter
9
Filtration parameter
t0.358
Out[427]=
a.nb
Rips data7, t, 0, 1
Appearance
t
Out[422]=
In[424]:=
a.nb 8 5 a.nb
Rips simplicial complex
Filtration parameter
In[422]:=
Rips data2, t, 0, 1
a.nb
Printed by Mathematica for Students
Printed by Mathematica for Students
Dependence on embedding X !→ D × I
4
a.nb
In[422]:=
• SC alone does not determine if an evasion path exists.
• Two networks with the same SC . Top contains evasion path
while bottom does not.
6
Rips data1, t, 0, 1
In[423]:=
Rips!data3y, t, 0, 1"
In[425]:=
Rips!data4y, t, 0, 1"
In[426]:=
a.nb 107
Rips!data5y, t, 0, 1"
In[427]:=
Rips!data6, t, 0, 1"
a.nb
In[428]:=
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
Filtration parameter
4Out[422]=
a.nb
Out[423]=
Rips!data1, t, 0, 1"
Filtration parameter
t0.358
In[423]:=
In[429]:=
Rips!data3n, t, 0, 1"
Filtration parameter
t0.358
12
a.nb
Out[425]=
Out[424]=
Rips data2, t, 0, 1
Filtration parameter
t0.358
In[430]:=
Filtration parameter
t0.358
a.nb Out[426]=
5
Rips!data4n, t, 0, 1"
In[431]:=
Rips!data5n, t, 0, 1"
11
Out[427]=
In[427]:=
t0.358
10
a.nb
Out[428]=
Rips!data6, t, 0, 1"
In[428]:=
a.nb
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
t
Filtration parameter
t0.358
Out[423]=
t0.358
Out[429]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[430]=
Printed by Mathematica for Students
Filtration parameter
Out[431]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[427]=
Printed by Mathematica for Students
draw balls
Filtration parameter
t0.358
13
Rips data7, t, 0, 1
Appearance
Filtration parameter
t0.358
0.358
Out[428]=
Printed by Mathematica for Students
a.nb
9
0.358
Rips simplicial complex
Filtration parameter
9
Filtration parameter
t0.358
a.nb
a.nb
Rips data7, t, 0, 1
Appearance
t
Out[422]=
In[424]:=
a.nb 8 5 a.nb
Rips simplicial complex
Filtration parameter
In[422]:=
Rips data2, t, 0, 1
a.nb
Printed by Mathematica for Students
Printed by Mathematica for Students
Dependence on embedding X !→ D × I
4
a.nb
In[422]:=
6
Rips data1, t, 0, 1
In[423]:=
Rips!data3y, t, 0, 1"
In[425]:=
Rips!data4y, t, 0, 1"
In[426]:=
a.nb 107
Rips!data5y, t, 0, 1"
In[427]:=
Rips!data6, t, 0, 1"
a.nb
In[428]:=
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
Filtration parameter
4Out[422]=
a.nb
Out[423]=
Rips!data1, t, 0, 1"
Filtration parameter
t0.358
In[423]:=
In[429]:=
Rips!data3n, t, 0, 1"
Filtration parameter
t0.358
12
a.nb
Out[425]=
Out[424]=
Rips data2, t, 0, 1
Filtration parameter
t0.358
In[430]:=
Filtration parameter
t0.358
a.nb Out[426]=
5
Rips!data4n, t, 0, 1"
In[431]:=
Rips!data5n, t, 0, 1"
11
Out[427]=
In[427]:=
t0.358
10
a.nb
Out[428]=
Rips!data6, t, 0, 1"
In[428]:=
a.nb
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
t
Filtration parameter
t0.358
Out[423]=
t0.358
Out[429]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[430]=
Printed by Mathematica for Students
Filtration parameter
Out[431]=
Printed by Mathematica for Students
Filtration parameter
t0.358
Out[427]=
Printed by Mathematica for Students
draw balls
Filtration parameter
t0.358
13
Rips data7, t, 0, 1
Appearance
Filtration parameter
t0.358
0.358
Out[428]=
Printed by Mathematica for Students
a.nb
9
0.358
Rips simplicial complex
Filtration parameter
9
Filtration parameter
t0.358
a.nb
a.nb
Rips data7, t, 0, 1
Appearance
t
Out[422]=
In[424]:=
a.nb 8 5 a.nb
Rips simplicial complex
Filtration parameter
In[422]:=
Rips data2, t, 0, 1
a.nb
Printed by Mathematica for Students
Printed by Mathematica for Students
Dependence on embedding X !→ D × I
• The two covered regions are fibrewise homotopy equivalent but
their complements are not.
Zigzag persistence
Fat graphs
• What minimal sensing capabilities might we add?
2
• Let D ⊂ R . A fat graph structure specifies the cyclic
ordering of edges adjacent to each vertex.
• Equivalent to a set of boundary cycles.
Fat graphs
A Delaunay triangulation, with four
• The alpha complex and
fat graphofstructure
ofcentered
a connected
intersection
the !-ball
at p and the Voron
sensor network determine
if anofevasion
path exists.
the union
!-balls centered
at points in P. The alph
the set {B̌! (p) | p ∈ P}. The underlying space |α! (P)| i
immediately implies that the alpha shape is homotop
Edelsbrunner [26] for a self-contained proof.
If the point set P is in general position, the alp
intersection of the Delaunay triangulation of P and th
points in P define a simplex in the alpha complex if an
at most ! that contains no other point in P.
Aleksandrov-Čech complexes and unions of balls for two differe
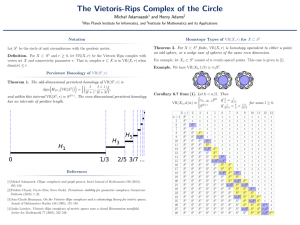
15.1.2 Vietoris-Rips Complexes: Flags and Shadow
The proximity graph N! (P) is the geometric graph who
all pairs of points at distance at most 2!; in other words,
complex. The Vietoris-Rips complex VR! (P) is the fla
An alpha complex and a decomposed union of balls.
graph N! (P).
A set of k + 1 points in P defines a k-simpl
Out[24]=
Fat graphs
4
FatGraphHard_edit.nb
FatGraphHard_edit.nb
3
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Cech simplicial complex
Cech simplicial complex
Appearance
Appearance
draw one simplices
draw one simplices
draw TWEAKED one simplices
draw TWEAKED one simplices
draw Cech complex
draw Cech complex
Filtration parameter
Filtration parameter
t
t
0.249
Out[24]=
0.249
Fat graphs
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Fat graphs
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Fat graphs
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Fat graphs
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Fat graphs
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Fat graphs
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Fat graphs
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Fat graphs
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Fat graphs
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
Filtration parameter
Filtration parameter
t
0.364
Filtration parameter
t
0.364
t
Fat graphs
Filtration parameter
0.364
t
0.364
draw balls
Filtration parameter
Filtration parameter
t
Filtration parameter
t
0.364
0.364
t
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
Printed by Mathematica for Students
2
Printed by Mathematica for Students
4
bNo.nb
bYes.nb
bNo.nb
Printed by Mathematica for Students
6
3
Printed by Mathematica for Students
bYes.nb
bYes.nb
Printed by Mathematica for Students
5
8
Printed by Mathematica for Students
bNo.nb
bNo.nb
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
Filtration parameter
t
0.364
draw balls
Filtration parameter
Filtration parameter
t
4
0.364
bNo.nb
0.364
6
0.364
bNo.nb
Filtration parameter
t
Printed by Mathematica for Students
bNo.nb
draw balls
Filtration parameter
t
Printed by Mathematica for Students
3
draw balls
Filtration parameter
t
Printed by Mathematica for Students
bNo.nb
draw balls
Filtration parameter
t
0.364
Printed by Mathematica for Students
bNo.nb
0.364
t
Printed by Mathematica for Students
5
8
bNo.nb
bNo.nb
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
t
Filtration parameter
0.364
Printed by Mathematica for Students
t
Filtration parameter
0.364
Printed by Mathematica for Students
t
Filtration parameter
0.364
Printed by Mathematica for Students
t
Filtration parameter
0.364
Printed by Mathematica for Students
t
Filtration parameter
0.364
Printed by Mathematica for Students
t
0.364
Printed by Mathematica for Students
Rips simplicial complex
Filtration parameter
7
Pair A: Top row contains evasion path; bottom does not.
draw balls
draw balls
Printed by Mathematica for Students
2
Printed by Mathematica for Students
0.364
7
draw balls
Filtration parameter
0.364
t
Printed by Mathematica for Students
0.364
Printed by Mathematica for Students
Pair B: Top row contains evasion path; bottom does not.
hat minimal sensor capabilites might one add to distinguish these examples? Each covered region
Out[24]=
Fat graphs
4
FatGraphHard_edit.nb
FatGraphHard_edit.nb
3
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
• Are the Čech complex and fat graph structure sufficient?
Cech simplicial complex
Cech simplicial complex
Appearance
Appearance
draw one simplices
draw one simplices
draw TWEAKED one simplices
draw TWEAKED one simplices
draw Cech complex
draw Cech complex
Filtration parameter
Filtration parameter
t
t
0.249
Out[24]=
0.249
Fat graphs
• The alpha complex and fat graph structure of a connected
sensor network determine if an evasion path exists.
• Are the Čech complex and fat graph structure sufficient?
$
&
"
#
#
&
%
%
!
$
"
!
4
a.nb
In[422]:=
12
Rips!data1, t, 0, 1"
In[423]:=
In[429]:=
Rips!data3n, t, 0, 1"
In[430]:=
a.nb
Rips!data4n, t, 0, 1"
5
In[431]:=
a.nb
Rips!data5n, t, 0, 1"
11
In[427]:=
10
Rips!data6, t, 0, 1"
a.nb
In[428]:=
a.nb
a.nb
• Final
thought:
ideas
from
pure
mathemaAcs
are
oien
helpful
in
tackling
problems
arising
from
more
applied
sejngs.
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Rips simplicial complex
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
Appearance
draw complex
draw complex
draw complex
draw complex
draw complex
draw complex
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
draw balls
13
Rips data7, t, 0, 1
Rips simplicial complex
draw complex
Filtration parameter
Filtration parameter
t
Out[422]=
Rips data2, t, 0, 1
a.nb
Filtration parameter
t0.358
Out[423]=
Filtration parameter
t0.358
Filtration parameter
t0.358
Out[429]=
Out[430]=
Filtration parameter
t0.358
Out[431]=
Filtration parameter
t0.358
Out[427]=
t0.358
0.358
Out[428]=
Thank you!
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
Printed by Mathematica for Students
9