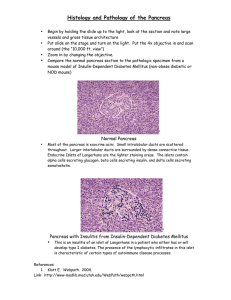
Life or Cell Death: c M o
advertisement

Life or Cell Death: Deciphering c-Myc Regulated Gene Networks In Two Distinct Tissues Sam Robson MOAC DTC, Coventry House, University of Warwick, Gibbet Hill Road, Coventry, CV4 7AL Mco A Outline 1. Introduction to c-Myc 2. Transgenic in vivo models – skin versus pancreas 3. Methods 4. Results 5. Generalised linear models Project Aims • Using two distinct switchable in vivo cMyc models, we aim to: – Analyse differences in gene-expression – Identify c-Myc regulated genes in cell replication and cell death – Improve understanding of complex c-Myc activity in diseases such as cancer – To understand how and why c-Myc can regulate vastly different paradoxical phenotypes in vivo 1: Introduction to c-Myc – Transcription factor involved in wide range of cellular functions – “Dual function” – May regulate up to 15% of all genes – Deregulated in majority of human cancers – Therapeutic target? – Exact mechanisms not well understood – we know WHAT c-Myc does, but we want to know WHY it does it – In vitro studies miss complex interactions of surrounding environment on cell fate c-Myc Regulated Processes Growth External Signals c-Myc Proliferation (eg. mitogens, survival factors) Apoptosis Loss of Differentiation Cell-Cycle Progression Gene Activation MYC MAX Ub CCND2 CDK4 Cyclin D2 CDK4 CACGTG CUL1 CKS p27KIP1 Proteosome p27KIP1 E-Box sequence in promoter sequence of target gene p27KIP1 Cyclin E CDK2 CAK Inactive Cyclin E CDK2 Active P MIZ-1 MYC MAX p15Ink4b (CDKN2B) p27 (not known if Miz-1 is required) Sp1/Sp3 MYC p15Ink4b (CDKN2B) p21Waf1 (CDKN1A) Apoptosis – Cell Death FAS Ligand FAS “Death Receptor” Death Induced Signalling Complex (DISC) FADD BID BCL-2 Apoptosome Procaspase 8 Caspase Cascade Effector ARF caspases FLIP BAX/ BAK tBID APAF-1 MOMP c-Myc Procaspase 9 Cytochrome c Smac DIABLO Mitochondrion ATP IAPs BIM IAPs PUMA Apoptosis p53 AIF NOXA Endo G Cellular targets Omi/ Htra2 Effector caspases 2: Transgenic in vivo models – Controlled activation of c-Myc functions in target cells – Can analyse immediate effects of c-Myc activation – Targetted to pancreatic islet β-cells (insulin promoter) and skin supra-basal keratinocytes (involucrin promoter) – Activation of c-Myc can lead to drastically different phenotypes – Replication in skin, apoptosis in pancreas Inactive Active MycERTAM HAT TRRAP Transgenic Model – c-MycERTAM RNA Polymerase Legend Myc Box I Myc Box II Basic Helix-Loop-Helix Leucine Zipper Estrogen Receptor Myc-Max complex Transformationbinds E-box TRRAP recruits a histone Transcription domain 4-Hydroxytamoxifen Myc sequence ofProtein acetyltransferase (HAT). This Associated Max Max binds Myc target gene acetylates (TRRAP) binds to nucleosomal atMBII leucine histones resulting in with help from helix-loop-helix chromatin remodelling, MBI 4-OHT binds zipper region allowing access by RNA Bound Heat estrogen receptor HSP90 TAM Polymerase for gene ER ShockupProtein CACGTG opening bHLHz90 transcription domain. TAM c-MycER Activation Inactive Skin Active Suprabasal layer Pancreas Pelengaris et al. (1999), Molecular Cell, Vol. 3(5), 565-577 Pelengaris et al. (2002), Cell, Vol. 109(3), 321-334 Suprabasal layer TAM c-MycER Activation • Skin Unchecked proliferation, no apoptosis - Replication • Pancreas Synchronous cell cycle entry and apoptosis – Death • Myc activation regulates two opposing phenotypes Pelengaris et al. (1999), Molecular Cell, Vol. 3(5), 565-577 Pelengaris et al. (2002), Cell, Vol. 109(3), 321-334 3: Methods – Microarrays – High throughput technique – “Transcriptomics” – Analysis at mRNA level – LCM to ensure RNA homogeneity – mRNA very delicate! Degradation by RNAses – Huge amount of work to develop robust protocol for extraction of RNA of suitable quality and yield from LCM – Many technical problems to overcome Workflow 1: Treatment of Transgenics 2: Extraction of Tissue Controlled activation of c-Myc in two diverse tissues 6: Microarray Hybridisation Hybridise cRNA to microarrays 3: Laser Capture Microdissection Excision of target tissue QC 5: 2-Cycle IVT Preparation of cRNA for microarray hybridisation Isolation of homogenous tissue QC 4: mRNA Extraction Isolate mRNA from target cells QC 7: Microarray Data Analysis 8: Validation Studies 9: Functional Validation Analysis of microarray data Validation studies to confirm results Linking results to the biology of the system Experimental Setup Untreated with 4-OHT x3 4 8 16 32 4 x3 8 16 32 x3 Time course Gene Expression Gene Expression Time course Pancreas Tissue x3 Time course Gene Expression Gene Expression Time course Skin Tissue Treated with 4-OHT 4 8 16 32 4 8 16 32 Laser Capture Microdissection • Heterogeneity of tissue may cause problem with in vivo studies • β-cells make up only ~2% of pancreas • LCM allows isolation of homogenous cell populations • Optimisation of protocol for LCM of islets – No other protocols available • LCM of skin not possible – too tough Laser Capture Microdissection 1: Find Islet 2: Cut Islet 3: Lift Islet 4: Extracted Islet Laser Capture Microdissection 1: Find Islet 2: Cut Islet 3: Lift Islet 4: Extracted Islet Laser Capture Microdissection 1: Find Islet 2: Cut Islet 3: Lift Islet 4: Extracted Islet Technical problems • mRNA very unstable – Great care taken to prevent degradation • Pancreas is notorious for being full of RNAses! • Standard LCM protocols very long – Optimisation of suitable protocol for islets • Small mRNA yield from LCM • Logistics of 84 samples – Lots of preparation! • Batching of samples – Randomisation to prevent systematic errors and batching effects • ~1 year for LCM optimisation ~9 months from tissue to microarray results! RNA Integrity Poor quality: Okay quality: Majority of peaks at lower levels 18S and 28S peaks more prominent, but many peaks at lower levels Good quality: Excellent quality: Fewer peaks at lower levels 18S and 28S peaks clear with almost no peaks at lower levels Effect of RNA Quality on Yield RNA Yield vs Quality 100 90 Starting cRNA ug 80 70 60 50 40 30 20 10 0 3.0 4.0 5.0 6.0 7.0 8.0 9.0 10.0 RIN • General trend between RNA quality (RIN) and yield (Starting cRNA) • Only 1 low starting cRNA samples below RIN=5 cutoff • Implies RIN may not be a great estimator of overall RNA yield Effect of RNA Quality on Yield RNA Yield vs Quality Skin 100 90 Starting cRNA ug 80 Pancreas 70 60 50 40 30 20 10 0 3.0 4.0 5.0 6.0 7.0 8.0 9.0 10.0 RIN • In general, skin samples have higher RNA quality and yield than pancreas samples • Many differences between skin and pancreas – Greater number of ribonucleases in pancreas – Homeostasis maintained in skin – More intense processing for pancreas tissue RNA compared to skin Microarray Analysis • Each feature measures one 25mer nucleotide sequence. • 25-mer sequence specifically binds biotin labelled cRNA. • Hundreds of identical 25mers per feature. • Fluorescence readings give relative mRNA concentration - gene expression • 11-20 features per gene. • Very, very expensive! Courtesy of Affymetrix - www.affymetrix.com 4: Results – Quality control of microarray data – Several outliers but generally good quality data – Outliers increase variance – Remove for differential analysis – Outliers spread nicely amongst conditions – importance of randomisation! – Analysis of early time points – Direct c-Myc targets Skin vs Pancreas Skin Pancreas • Clustering – Group similar samples together • Branching tree like structure – samples on the same branch most similar • Data cluster nicely on tissue (some outliers) • Given the protocol, the data looks great! Gene Expression Analysis Pancreas Skin • Differential Expression Look for genes with changing expression across conditions • Statistics Compare distributions between conditions to look for significant changes • Error Biological error, technical error, random error • Functional Analysis Similar expression profile implies related biological mechanisms Tissue-Specific Differentiation Markers Insulin ~4-fold down in pancreas Involucrin ~2-fold down in skin Cell-Cycle Progression Cyclin D2 ~2-fold up E Cyclin in skin ~4-fold up in pancreas • Ccnd2 and CDK4 upregulated in skin – Indicates G1/S cell cycle progression • No change in pancreas – Odd • CDK inhibitor p27 downregulated in both • Cyclin E upregulated in pancreas and not skin – Again, very odd CDK4 ~4-fold up in skin p27KIP1 ~2-fold down in pancreas ~4-fold down in skin Apoptosis p19ARF ~2-fold up in pancreas • Increase in p19 – Oncogenic stress (p53 dependent pathway) • No change in p53 at transcriptional level – Changes may occur at protein level • Massive increase in Fas receptor expression – Extrinsic pathway • Myc seems to drive apotosis through extrinsic and intrinsic pathways Fas Receptor ~6-fold up in pancreas p53 No change 5: Generalised Linear Models – Most microarray studies focus on one or two main parameters – Multi-factorial approach poses problems with significance analysis – Use of generalised linear models – Widely applicable particularly for clinical studies – Collaboration with Agilent – Implementation in Genespring GX Generalised Linear Models • Unsupervised linear regressive technique. • Model gene expression data as a linear combination of parameter variables: y b1 x1 b2 x2 ... b p x p y = (y1,…,yn)T is the response variable (gene expression) for each sample xi = (x1,…,xn)T are the explanatory variables (1 ≤ i ≤ p) for each sample bi is the model coefficient for explanatory variable xi n is the number of samples, p is the number of parameters ε is some error term Generalised Linear Models • Can be used in the following ways: 1. To check how much of an effect other parameters have on gene expression (eg batching effects) 2. To find genes that change based on particular parameters while taking other parameters and interactions into account (eg clinical data) • Makes fewer assumptions of data distribution • Works with unbalanced experiment designs – useful for clinical data. Generalised Linear Models • Program written in statistical programming language R • Written as part of the Bioconductor project • Implemented in GeneSpring GX (Agilent) – Aim to translate into JAVA for complete integration • Close collaboration with Agilent • Currently testing the program on a number of diverse data sets • MOAC (Shameless plug) – First crop of interdisciplinary scientists almost ready Further Work • Analysis of microarray data – Cluster analysis, differential analysis, network analysis, etc. • Use of GLM algorithm and comparison of results with standard methods (ANOVA) • Validation of results – Immunohistology, quantitative real time PCR, etc. • Functional validation – siRNA, ChIP-on-chip, etc. • Translation of GLM program to JAVA for implementation in GeneSpring GX version 8 Conclusion • c-Myc regulates replication and cell death • Web of pathways to decipher – Tissue context in vivo • Seems to initiate apoptosis through combination of extrinsic and intrinsic pathways • Want to find the ‘suicide note’ for the pancreas – why choose death? Acknowledgements Project Supervisors: Michael Khan David Epstein Stella Pelengaris Advisory Committee: Robert Old Manu Vatish James Lynn Special thanks: Helen Bird Lesley Ward Sue Davis Heather Turner Ewan Hunter Sponsors: EPSRC, BBSRC, AICR, Eli Lilly and Amylin Pharmaceuticals Inc. Mco A Acknowledgements Luxian David Stella Mike Vicky Sevi Sylvie