What do row and column marginals reveal about your dataset?
advertisement

What do row and column marginals reveal about
your dataset?
Behzad Golshan
Boston University
behzad@cs.bu.edu
John W. Byers
Boston University
byers@cs.bu.edu
Evimaria Terzi
Boston University
evimaria@cs.bu.edu
Abstract
Numerous datasets ranging from group memberships within social networks to
purchase histories on e-commerce sites are represented by binary matrices. While
this data is often either proprietary or sensitive, aggregated data, notably row and
column marginals, is often viewed as much less sensitive, and may be furnished
for analysis. Here, we investigate how these data can be exploited to make inferences about the underlying matrix H. Instead of assuming a generative model for
H, we view the input marginals as constraints on the dataspace of possible realizations of H and compute the probability density function of particular entries
H(i, j) of interest. We do this for all the cells of H simultaneously, without generating realizations, but rather via implicitly sampling the datasets that satisfy the
input marginals. The end result is an efficient algorithm with asymptotic running
time the same as that required by standard sampling techniques to generate a single dataset from the same dataspace. Our experimental evaluation demonstrates
the efficiency and the efficacy of our framework in multiple settings.
1
Introduction
Online marketplaces such as Walmart, Netflix, and Amazon store information about their customers
and the products they purchase in binary matrices. Likewise, information about the groups that social
network users participate in, the “Likes” they make, and the other users they “follow” can also be
represented using large binary matrices. In all these domains, the underlying data (i.e., the binary
matrix itself) is often viewed as proprietary or as sensitive information. However, the data owners
may view certain aggregates as much less sensitive. Examples include revealing the popularity of a
set of products by reporting total purchases, revealing the popularity of a group by reporting the size
of its membership, or specifying the in- and out-degree distributions across all users.
Here, we tackle the following question: “Given the row and column marginals of a hidden binary
matrix H, what can one infer about H?”.
Optimization-based methods for addressing this question, e.g., least squares or maximum likelihood,
b of H. However, this
assume a generative model for the hidden matrix and output an estimate H
b may
estimate gives little guidance as to the structure of the feasible solution space; for example, H
be one of many solutions that achieve the same value of the objective function. Moreover, these
methods provide little insight about the estimates of particular entries H(i, j).
In this paper, we do not make any assumptions about the generative process of H. Rather, we
approach the above question by viewing the row and column marginals as constraints that induce
a dataspace X , defined by the set of all matrices satisfying the input constraints. Then, we explore
this dataspace not by estimating H at large, but rather by computing the entry-wise PDF P(i, j), for
every entry (i, j), where we define P(i, j) to be the probability that cell (i, j) takes on value 1 in the
datasets in X . From the application point of view, the value of P(i, j) can provide the data analyst
1
with valuable insight: for example, values close to 1 (respectively 0) give high confidence to the
analyst that H(i, j) = 1 (respectively H(i, j) = 0).
A natural way to compute entry-wise PDFs is by sampling datasets from the induced dataspace
X . However, this dataspace can be vast, and existing techniques for sampling binary tables with
fixed marginals [6, 9] fail to scale. In this paper, we propose a new efficient algorithm for computing
entry-wise PDFs by implicitly sampling the dataspace X . Our technique can compute the entry-wise
PDFs of all entries in running time the same as that required for state-of-the art sampling techniques
to generate just a single sample from X . Our experimental evaluation demonstrates the efficiency
and the efficacy of our technique for both synthetic and real-world datasets.
Related work: To the best of our knowledge, we are the first to introduce the notion of entrywise PDFs for binary matrices and to develop implicit sampling techniques for computing them
efficiently. However, our work is related to the problem of sampling from the space of binary
matrices with fixed marginals, studied extensively in many domains [2, 6, 7, 9, 21], primarily due to
its applications in statistical significance testing [14, 17, 20]. Existing sampling techniques all rely
on explicitly sampling the underlying dataspaces (either using MCMC or importance sampling) and
while these methods can be used to compute entry-wise PDFs, they are inefficient for large datasets.
Other related studies focus on identifying interesting patterns in binary data given itemset frequencies or other statistics [3, 15]. These works either assume a generative model for the data or build
the maximum entropy distribution that approximates the observed statistics; whereas our approach
makes no such assumptions and focuses only on exact solutions. Finally, considerable work has
focused on counting binary matrices with fixed marginals [1, 8, 10, 23]. One can compute the
entry-wise PDFs using these results, albeit in exponential time.
2
Dataspace Exploration
Throughout the rest of the discussion we will assume an n × m 0–1 matrix H, which is hidden. The
input to our problem consists of the dimensionality of H and its row and column marginals provided
as a pair of vectors hr, ci. That is, r and c are n-dimensional and m-dimensional integer vectors
respectively; entry r(i) stores the number of 1s in the ith row of H, and similarly for c(i). In this
paper we address the following high-level problem:
Problem 1. Given hr, ci, what can we infer about H? More specifically, can we reason about
entries H(i, j) without access to H but only its row and column marginals?
Clearly, there are many possible ways of formalizing the above problem into a concrete problem
definition. In Section 2.1 we describe some mainstream formulations and discuss their drawbacks.
In Section 2.2 we introduce our dataspace exploration framework that overcomes these drawbacks.
2.1
Optimization–based approaches
Standard optimization-based approaches for Problem 1 usually assume a generative model for H,
b the best estimate of H optimizing a specific objective function
and estimate it by computing H,
(e.g., likelihood, squared error). Instantiations of these methods for our setting are described next.
Maximum-likelihood (ML): The ML approach assumes that the hidden matrix H is generated by
a model that only depends on the observed marginals. Then, the goal is to find the model parameters
b of H while maximizing the likelihood of the observed row and column
that provide an estimate H
marginals. A natural choice of such a model for our setting is the Rasch model [4, 19], where the
probability of entry H(i, j) taking on value 1 is given by:
eαi −βj
Pr [H(i, j) = 1] =
.
1 + eαi −βj
The maximum-likelihood estimates of the (n + m) parameters αi and βj of this model can be
computed in polynomial time [4, 19]. For the rest of the discussion, we will use the term Rasch to
refer to the experimental method that computes an estimate of H using this ML technique.
Least-squares (LS): One can view the task of estimating H(i, j) from the input aggregates as
solving a linear system defined by equations: r = H × ~1 and c = H T × ~1 Unfortunately, such
2
a system of equations is typically highly under-determined and standard LS methods approach it
b of H to minimize F (H)
b = ||(H
b × ~1) −
as a regression problem that asks for an estimate H
T
b
b
~
r||F + ||(H × 1) − c||F , where || ||F is the Frobenius norm [13]. Even when the entries of H
are restricted to be in [0, 1], it is not guaranteed that the above regression-based formulation will
output a reasonable estimate of H. For example, all tables with row and column marginals r and
c are 0-error solutions; yet there may be exponentially many such matrices. Alternatively, one can
b that minimizes F (H)
b + J(H).
b For the
incorporate a “regularization” factor J() and search for H
2
b
b
rest of this paper, we consider this latter approach with J(H) = (H(i, j) − h) , where h is the
b We refer to this approach as the LS method.
average value over all entries of H.
Although one can solve (any of) the above estimation problems via standard optimization techniques, the output of such methods is a holistic estimate of H that gives no insight on how many
b with the same value of the objective function exist or the confidence in the value of
solutions H
every cell. Moreover, these techniques are based on assumptions about the generative model of the
hidden data. While these assumptions may be plausible, they may not be valid in real data.
2.2
The dataspace exploration framework
To overcome the drawbacks of the optimization-based methods, we now introduce our dataspace
exploration framework, which does not make any structural assumptions about H and considers the
set of all possible datasets that are consistent with input row and column marginals hr, ci. We call
the set of such datasets the hr, ci-dataspace, denoted by Xhr,ci , or X for short.
We translate the high-level Problem 1 into the following question: Given hr, ci, what is the probability that the entry H(i, j) of the hidden dataset takes on value 1? That is, for each entry H(i, j)
we are interested in computing the quantity:
X
P(i, j) =
Pr(H 0 )Pr [H 0 (i, j) = 1] .
(1)
H 0 ∈X
Here, Pr(H 0 ) encodes the prior probability distribution over all hidden matrices in X . For a uniform
prior, P(i, j) encodes the fraction of matrices in X that have 1 in position (i, j). Clearly, for binary
matrices, P(i, j) determines the PDF of the values that appear in cell (i, j). Thus, we call P(i, j) the
entry-wise PDF of entry (i, j), and P the PDF matrix. If P(i, j) is very close to 1 (or 0), then over
all possible instantiations of H, the entry (i, j) is, with high confidence, 1 (or 0). On the other hand,
P(i, j) ' 0.5 signals that in the absence of additional information, a high-confidence prediction of
entry H(i, j) cannot be made.
Next, we discuss algorithms for estimating entry-wise PDFs efficiently. Throughout the rest of the
discussion we will adopt Matlab notation for matrices: for any matrix M , we will use M (i, :) to
refer to the i-th row, and M (:, j) to refer to the j-th column of M .
3
Basic Techniques
First, we review some basic facts and observations about hr, ci and the dataspace Xhr,ci .
Validity of marginals: Given hr, ci we can decide in polynomial time whether |Xhr,ci | > 0 either
by verifying the Gale-Ryser condition [5] or by constructing a binary matrix with the input row and
column marginals, as proposed by Erdös, Gallai, and Hakimi [18, 11]. The second option has the
comparative advantage that if |Xhr,ci | > 0, then it also outputs a binary matrix from Xhr,ci .
Nested matrices: Building upon existing results [18, 11, 16, 24], we have the following:
Lemma 1. Given the row and column marginals of a binary matrix as hr, ci we can decide in
polynomial time whether |Xhr,ci | = 1 and if so, completely recover the hidden matrix H.
The binary matrices that can be uniquely recovered are called nested matrices and have the property
that in their representation as bipartite graphs they do not have any switch boxes [16]: a pair of edges
(u, v) and (u0 , v 0 ) for which neither (u, v 0 ) nor (u0 , v) exist.
3
Explicit sampling: One way of approximating P(i, j) for large dataspaces is to first obtain a uniform
sample of N binary matrices from X : X1 , . . . , XN and for each (i, j), compute P(i, j) as the
fraction of samples for which X` (i, j) = 1.
We can obtain random (near-uniform) samples from X using either the Markov chain Monte
Carlo (MCMC) method proposed by Gionis et al. [9] or the Sequential Importance Sampling
(Sequential) algorithm proposed by Chen et al. [6]. MCMC guarantees uniformity of the samples, but it does not converge in polynomial time. Sequential produces near-uniform samples in
polynomial time, but it requires O(n3 m) time per sample and thus using this algorithm to produce
N samples (N >> n) is beyond practical consideration. To recap, explicit sampling methods are
impractical for large datasets; moreover, their accuracy depends on the number of samples N and
the size of the dataspace X , which itself is hard to estimate.
4
4.1
Computing entry-wise PDFs
Warmup: The SimpleIS algorithm
With the aim to provide some intuition and insight, we start by presenting a simplified version of our algorithm called SimpleIS, also shown in Algorithm 1.
SimpleIS computes the P matrix one column at a
time, in arbitrary order. Each such computation conAlgorithm 1 The SimpleIS algorithm.
sists of two steps: (a) propose and (b) adjust. The
Input: hr, ci
Propose step associates with every row i, weight
Output: Estimate of the PDF matrix P
w(i) that is proportional to the row marginal of row
1:
w = Propose(r)
i. A naive way of assigning these weights is by setr(i)
2: for j = 1 . . . m do
ting w(i) = m . We refer to these weights w as the
x = c(j)
raw probabilities. The Adjust step takes as input 3:
4:
px = Adjust(w, x)
the column sum x = c(j) of the jth column and the
5:
P(:, j) = px
raw probabilities w and adjusts these probabilities
into the final P
probabilities px such that for column j
we have that i px (i) = x. This adjustment is not a simple normalization, but it computes the final
values of px (i) by implicitly considering all possible realizations of the jth column with column
sum x and computing the probability that the ith cell of that column is equal to 1.
Formally, if we use x to denote the binary vector that represents one realization of the j-th column
of the hidden matrix, then px (i) is computed as:
"
#
n
X
px (i) := Pr x(i) = 1 |
x(i) = x .
(2)
i=1
It can be shown [6] that Equation (2) can be evaluated in polynomial time as follows: for any vector
x, let N = {1, . . . , n}, be the set of all possible positions of 1s within x, and let R(x, N ) be the
probability that exactly x elements of N are set to 1, i.e.,
"
#
X
R(x, N ) := Pr
x(i) = x .
i∈N
Using this definition, px (i) is then derived as follows:
px (i) =
w(i)R(x − 1, N \ {i})
.
R(x, N )
(3)
The evaluation of all of the necessary terms R( , ) can be accomplished by the following dynamicprogramming recursion: for all a ∈ {1, . . . x}, and for all B and i such that |B| > a and i ∈ B ⊆ N :
R(a, B) = (1 − w(i))R(a, B \ {i}) + w(i)R(a − 1, B \ {i}).
Running time: The Propose step is linear in the number of the rows and the Adjust evaluates
Equation (3) and thus needs at most O(n2 ) time. Thus, SimpleIS runs in time O(mn2 ).
Discussion: A natural question to consider is: why could the estimates of P produced by SimpleIS
be inaccurate?
4
0
4
To answer this question, consider a hidden 5 × 5 binary table
0
4
with r = (4, 4, 2, 2, 2) and c = (2, 3, 1, 4, 4) and assume that
0
2
SimpleIS starts by computing the entry-wise PDFs of the first
1
2
column. While evaluating Equation (2), SimpleIS generates all
1
2
possible columns of matrices with row marginals r and a column
2 3 1 4 4
with column sum 2 – ignoring the values of the rest of the column
marginals. Thus, the realization of the first column shown in the matrix on the right is taken into
consideration by SimpleIS, despite the fact that it cannot lead to a matrix that respects r and c.
This is because four more 1s need to be placed in the empty cells of the first two rows which in turn
would lead to a violation of the column marginal of the third column. This situation occurs exactly
because SimpleIS never considers the constraints imposed by the rest of the column marginals
when aggregating the possible realizations of column j.
Ultimately, the SimpleIS algorithm results in estimates px (i) that reflect an entry i in a column
being equal to 1 conditioned over all matrices with row marginals r and a single column with column
sum x [6]. But this dataspace is not our target dataspace.
4.2
The IS algorithm
In the IS algorithm, we remedy this weakness of SimpleIS by taking into account the constraints
imposed by all the column marginals when aggregating the realization of a particular column j.
Referring again to the previous example, the input vectors r and c impose the following constraints
on any matrix in Xhr,ci : column 1 must have least one 1 in the first two rows and (exactly) two 1s in
the first five rows. These types of constraints, known as knots, are formally defined as follows.
Definition 1. A knot is a subvector of a column characterized by three integer values h[s, e] | bi,
where s and e are the starting and ending indices defining the subvector, and b is a lower bound on
the number of 1s that must be placed in the first e rows of the column.
Interestingly, given hr, ci, the knots of any column j of the hidden matrix can be identified in linear
time using an algorithm that recursively applies the Gale-Ryser condition on realizability of bipartite
graphs. This method, and the notion of knots, were first introduced by Chen et al. [6].
At a high level, IS (Algorithm 2) identifies the knots within each column and uses them
to achieve a better estimation of P. Here, the process of obtaining the final probabilities
is more complicated since it requires: (a) identifying the knots of every column j (line 3),
(b) computing the entry-wise PDFs for the entries in every knot (denoted by qk ) (lines 4-7), Algorithm 2 The IS algorithm.
and (c) creating the jth column of P by putting
Input: hr, ci
the computed entry-wise PDFs back together
Output: Estimate of the PDF matrix P
(line 8). Note that we use wk to refer to the
1: w = Propose(r)
vector of raw probabilities associated with cells
2: for j = 1 . . . m do
in knot k. Also, vector pk,x is used to store the
3:
FindKnots(j, r, c)
adjusted probabilities of cells in knot k given
4:
for each knot k ∈ {1 . . . l} do
that x 1s are placed within the knot.
5:
for x: number of 1s in knot k do
Step (a) is described by Chen et al. [6], and
6:
pk,x = Adjust(wk , x)
step (c) is straightforward, so we focus on (b),
7:
qk = Ex [pk,x ]
which is the main part of IS. This step con8:
P(:, j) = [q1 ; . . . ; ql ]
siders all the knots of the jth column sequentially. Assume that the kth knot of this column
is given by the tuple h[sk , ek ] | bk i. Let x be the number of 1s inside this knot. If we know the value
of x, then we can simply use the Adjust routine to adjust the raw probabilities wk . But since the
value of x may vary over different realizations of column j, we need to compute the probability
distribution of different values of x. For this, we first observe that if we know that y 1s have been
placed prior to knot k, then we can compute lower and upper bounds on x as:
Lk|y = max {0, bk − y} , Uk|y = min {ek − sk + 1, c(j) − y} .
Clearly, the number of 1s in the knot must be an integer value in the interval [Lk|y , Uk|y ]. Lacking
prior knowledge we assume that x takes any value in the interval uniformly at random. Thus, the
5
0
10
Error
Error
−2
10
LS
MCMC
Sequential
Rasch
SimpleIS
IS
1
10
Running Time (secs)
LS
MCMC
Sequential
Rasch
SimpleIS
IS
0
10
−1
10
−4
10
−2
10
−6
10
0
−3
20
40
60
80
100
10
2
10
0
10
−2
0
20
Matrix Size (n)
(a) Blocked matrices
LS
MCMC
Sequential
Rasch
SimpleIS
IS
4
10
40
60
80
100
Matrix Size (n)
(b) Matrices with knots
10
0
20
40
60
80
100
Matrix Size (n)
(c) Relative running times
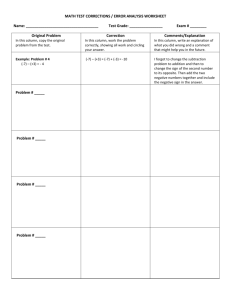
Figure 1: Panels (a) and (b) depict Error (log scale) for six different algorithms, on two classes of
matrices. Panel (c) depicts algorithmic running times.
probability of x 1s occurring inside the knot, given the value of y (i.e., 1s prior to the knot) is:
1
Pk (x|y) =
Uk|y − Lk|y + 1
(4)
Based on this conditional probability we can write the probability of each value of x as
Pk (x) =
c(j)
X
Qk (y)Pk (x|y),
(5)
y=0
in which Pk (x|y) is computed by Equation (4) and Qk (y) refers to the probability of having y
1s prior to knot k. In order to evaluate Equation (5), we need to compute the values of Qk (y).
We observe that for every knot k and for every value of y, Qk (y) can be computed by dynamic
programming as:
Qk (y) =
y
X
Qk−1 (z)Pk−1 (y − z|z).
(6)
z=0
Running time and speedups: If there is a single knot in every column, SimpleIS and IS are
identical. For a column j with ` knots, IS requires O(`2 c(j) + nc(j)2 ) – or worst-case O(n3 ) time.
Thus, sequential implementation of IS has running time O(n3 m). This is the same as the time
required by Sequential for generating a single sample from Xhr,ci , providing a clear indication
of the computational speedups afforded by IS over sampling. Moreover, IS treats each column
independently, and thus it is parallelizable. Finally, since the entry-wise PDFs for two columns with
the same column marginals are identical, our method needs to only compute the PDFs of columns
with distinct column marginals. Further speedups can be achieved for large datasets by binning
columns with similar marginals into a bin with a representative column sum. When the columns in
the same bin differ by at most t, we call this speedup t-Binning.
Discussion: We point out here that IS is highly motivated by Sequential – the most practical algorithm to date for sampling (almost) uniformly matrices from dataspace Xhr,ci . Although
Sequential was designed for a different purpose, IS uses some of its building blocks (e.g.,
knots). However, the connection is high level and there is no clear quantification of the relationship
between the values of P computed by IS and those produced by repeated sampling from Xhr,ci using
Sequential. While we study this relationship experimentally, we leave the formal investigation
as an open problem.
5
Experiments
b
Accuracy evaluation: We measure the accuracy of different methods by comparing the estimates P
they produce against the known ground-truth P and evaluating the average absolute error as:
P b
P(i,
j)
−
P(i,
j)
i,j
b
Error(P, P) =
(7)
mn
6
1
0.16
0.03
InformativeFix
RandomFix
0.14
0.4
Error(P, H)
0.6
0.1
0.08
0.06
0.02
0.015
0.01
0.04
0.2
0.005
0.02
0
0
InformativeFix
RandomFix
0.025
0.12
Error(P, H)
CDF
0.8
0.2
0.4
0.6
0.8
1
P(i, j) values
(a) D BLP distribution of entries
0
0
10
20
30
40
50
60
Percentage of Cells Revealed
(b) D BLP
70
0
0
10
20
30
40
50
60
70
Percentage of Cells Revealed
(c) N OLA
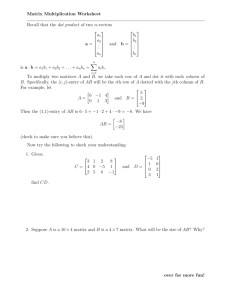
Figure 2: Panel (a) shows the CDF of estimated entry-wise PDFs by IS for the D BLP dataset.
Panels (b) and (c) show Error(P, H) as a function of the percentage of “revealed” cells.
We compare the Error of our methods, i.e., SimpleIS and IS, to the Error of the two optimization methods: Rasch and LS described in Section 2.1 and the two explicit-sampling methods
Sequential and MCMC described in Section 3. For MCMC, we use double the burn-out period (i.e.,
four times the number of ones in the table) suggested by Gionis et al. [9]. For both Sequential
and MCMC, we vary the sample size and use up to 250 samples; for this number of samples these
methods can take up to 2 hours to complete for 100 × 100 matrices. In fact, our experiments were
ultimately restricted by the inability of other methods to handle larger matrices.
Since exhaustive enumeration is not an option, it is very hard to obtain ground-truth values of P for
arbitrary matrices, so we focus on two specific types of matrices: blocked matrices and matrices
with knots.
An n × n blocked matrix has marginals r = (1, n − 1, . . . , n − 1) and c = (1, n − 1, . . . , n − 1). Any
table with these marginals either has a value of 1 in entry (1, 1) or it has two distinct entries with
value 1 in the first row and the first column (excluding cell (1, 1)). Also note that given a realization
of the first row and the column, the rest of the table is fully determined. This implies that there are
exactly (2n − 1) tables with such marginals and the entry-wise PDFs are: P(1, 1) = 1/(2n − 1); for
i 6= 1, P(1, i) = P(i, 1) = (n − 1)/(2n − 1); and for i 6= 1 and j 6= 1, P(i, j) = 2n/(2n − 1).
The matrices with knots are binary matrices that are generated by diagonally concatenating smaller
matrices for which the ground-truth is computed known through exhaustive enumeration. The concatenation is done in such a way such that no new switch boxes are introduced (as defined in Section 3). While the details of the construction are omitted due to lack of space, the key characteristic
of these matrices is that they have a large number of knots.
Figure 1(a) shows the Error (in log scale) of the different methods as a function of the matrix size n;
SimpleIS and IS perform identically in terms of Error and are much better than other methods.
Moreover, they become increasingly accurate as the size of the dataset increases, which means that
our methods remain relatively accurate even for large matrices. In this experiment, Rasch appears
to be the second-best method. However, as our next experiment indicates, the success of Rasch
hinges on the fact that the marginals of this experiment do not introduce many knots.
The results on matrices with many knots are shown in Figure 1(b). Here, the relative performance of
the different algorithms is different: SimpleIS is among the worst-performing algorithms, together
with LS and Rasch, with an average Error of 0.1. On the other hand, IS with Sequential and
MCMC are clearly the best-performing algorithms. This is mainly due to the fact that the matrices
we create for this experiment have a lot of knots, and as SimpleIS, LS and Rasch are all knotoblivious, they produce estimates with large errors. On the other hand, Sequential, MCMC and
IS take knots into account, and therefore they perform much better than the rest.
Looking at the running times (Figure 1(c)), we observe that the running time of our methods is
clearly better than the running time of all the other algorithms for larger values of n. For example,
while both SimpleIS and IS compute P[] within a second, Rasch requires a couple of seconds
and other methods need minutes or even up to hours to complete.
Utilizing entry-wise PDFs: Next, we move on to demonstrate the practical utility of entry-wise
PDFs. For this experiment we use the following real-world datasets as hidden matrices.
7
D BLP: The rows of this hidden matrix correspond to authors and the columns correspond to conferences in DBLP. Entry (i, j) has value 1 if author i has a publication in conference j. This subset of
DBLP, obtained by Hyvönen et al. [12], has size 17, 702 × 19 and density 8.3%.
N OLA: This hidden matrix records the membership of 15, 965 Facebook users from New Orleans
across 92 different groups [22]. The density of this 0–1 matrix is 1.1%1 .
We start with an experiment that addresses the following question: “Can entry-wise PDFs help us
identify the values of the cells of the hidden matrix?” To quantify this, we first look at the distribution
of values of entry-wise PDFs per dataset, shown in Figure 2(a) for the D BLP dataset (the distribution
of entry-wise PDFs is similar for the N OLA dataset). The figure demonstrates that the overwhelming
majority of the P(i, j) entries are small, smaller than 0.1.
We then address the question: “Can entry-wise PDFs guide us towards effectively querying the
hidden matrix H so that its entries are more accurately identified?” For this, we iteratively query
entries of H. At each iteration, we query 10% of unknown cells and we compute the entry-wise
PDFs P after having these entries fixed. Figures 2(b) and 2(c) show the Error(P, H) after each iteration for the D BLP and N OLA datasets; values of Error(P, H) close to 0 imply that our method could
reconstruct H almost exactly. The two lines in the plots correspond to R ANDOM F IX and I NFOR MATIVE F IX strategies for selecting the queries at every step. The former picks 10% of unknown
cells to query uniformly at random at every step. The latter selects 10% of cells with PDF values
closest to 0.5 at every step. The results demonstrate that I NFORMATIVE F IX is able to reconstruct
the table with significantly fewer queries than R ANDOM F IX. Interestingly, using I NFORMATIVE F IX
we can fully recover the hidden matrix of the N OLA dataset by just querying 30% of entries. Thus,
the values of entry-wise PDFs can be used to guide adaptive exploration of the hidden datasets.
Scalability: In a final experiment, we explored the accuracy/speedup tradeoff obtained by tBinning. For the the D BLP and N OLA datasets, we observed that by using t = 1, 2, 4 we reduced
the number of columns (and thus the running time) by a factor of at least 2, 3 and 4 respectively.
For the same datasets, we evaluate the accuracy of the t-Binning results by comparing the values
of Pt computed for t = {1, 2, 4} with the values of P0 (obtained by IS in the original dataset). In
all cases, and for all values of t, we observe that the Error(Pt , P0 ) (defined in Equation (7)) are low
– never exceeding 1.5%. Even the maximum entry-wise difference of P0 and Pt are consistently
about 0.1 – note that such high error values only occur in one out of the millions of entries in P.
Finally, we also experimented with an even larger dataset obtained through the Yahoo! Research
Webscope program. This is a 140, 000 × 4252 matrix of users and their participation in groups. For
this dataset we observe that for 80% reduction to the number of columns of the dataset introduces
an average error of size of only 1.7e−4 (for t = 4).
6
Conclusions
We started with a simple question: “Given the row and column marginals of a hidden binary matrix
H, what can we infer about the matrix itself?” We demonstrated that existing optimization-based
approaches for addressing this question fail to provide a detailed intuition about the possible values
of particular cells of H. Then, we introduced the notion of entry-wise PDFs, which capture the
probability that a particular cell of H is equal to 1. From the technical point of view, we developed
IS, a parallelizable algorithm that efficiently and accurately approximates the values of the entrywise PDFs for all cells simultaneously. The key characteristic of IS is that it computes the entrywise PDFs without generating any of the matrices in the dataspace defined by the input row and
column marginals, and did so by implicitly sampling from the dataspace. Our experiments with
synthetic and real data demonstrated the accuracy of IS on computing entry-wise PDFs as well as
the practical utility of these PDFs towards better understanding of the hidden matrix.
Acknowledgements
This research was partially supported by NSF grants CNS-1017529, III-1218437 and a gift from
Microsoft.
1
The dataset is available at: http://socialnetworks.mpi-sws.org/data-wosn2009.html
8
References
[1] A. Bekessy, P. Bekessy, and J. Komplos. Asymptotic enumeration of regular matrices. Studia Scientiarum
Mathematicarum Hungarica, pages 343–353, 1972.
[2] I. Bezáková, N. Bhatnagar, and E. Vigoda. Sampling binary contingency tables with a greedy start.
Random Struct. Algorithms, 30(1-2):168–205, 2007.
[3] T. D. Bie, K.-N. Kontonasios, and E. Spyropoulou. A framework for mining interesting pattern sets.
SIGKDD Explorations, 12(2):92–100, 2010.
[4] T. Bond and C. Fox. Applying the Rasch Model: Fundamental Measurement in the Human Sciences.
Lawrence Erlbaum, 2007.
[5] R. Brualdi and H. J. Ryser. Combinatorial Matrix Theory. Cambridge University Press, 1991.
[6] Y. Chen, P. Diaconis, S. Holmes, and J. Liu. Sequential monte carlo methods for statistical analysis of
tables. Journal of American Statistical Association, JASA, 100:109–120, 2005.
[7] G. W. Cobb and Y.-P. Chen. An application of Markov chain Monte Carlo to community ecology. Amer.
Math. Month., 110(4):264–288, 2003.
[8] M. Gail and N. Mantel. Counting the number of r × c contigency tables with fixed margins. Journal of
the American Statistical Association, 72(360):859–862, 1977.
[9] A. Gionis, H. Mannila, T. Mielikäinen, and P. Tsaparas. Assessing data mining results via swap randomization. TKDD, 1(3), 2007.
[10] I. J. Good and J. Crook. The enumeration of arrays and a generalization related to contigency tables.
Discrete Mathematics, 19(1):23 – 45, 1977.
[11] S. Hakimi. On realizability of a set of integers as degrees of the vertices of a linear graph. Journal of the
Society for Industrial and Applied Mathematics, 10(3):496–506, 1962.
[12] S. Hyvönen, P. Miettinen, and E. Terzi. Interpretable nonnegative matrix decompositions. In KDD, pages
345–353, 2008.
[13] T. Kariya and H. Kurata. Generalized Least Squares. Wiley, 2004.
[14] N. Kashtan, S. Itzkovitz, R. Milo, and U. Alon. Efficient sampling algorithm for estimating subgraph
concentrations and detecting network motifs. Bioinformatics, 20(11):1746–1758, 2004.
[15] M. Mampaey, J. Vreeken, and N. Tatti. Summarizing data succinctly with the most informative itemsets.
TKDD, 6(4), 2012.
[16] H. Mannila and E. Terzi. Nestedness and segmented nestedness. In KDD, pages 480–489, 2007.
[17] R. Milo, S. Shen-Orr, S. Itzkovirz, N. Kashtan, D. Chklovskii, and U. Alon. Network motifs: Simple
building blocks of complex networks. Science, 298(5594):824–827, 2002.
[18] P. Erdös and T. Gallai. Graphs with prescribed degrees of vertices. Mat. Lapok., 1960.
[19] G. Rasch. Probabilistic models for some intelligence and attainment tests. 1960. Technical Report,
Danish Institute for Educational Research, Copenhagen.
[20] J. Sanderson. Testing ecological patterns. Amer. Sci., 88(4):332–339, 2000.
[21] T. Snijders. Enumeration and simulation methods for 0–1 matrices with given marginals. Psychometrika,
56(3):397–417, 1991.
[22] B. Viswanath, A. Mislove, M. Cha, and K. P. Gummadi. On the evolution of user interaction in Facebook.
In WOSN, pages 37–42, 2009.
[23] B. Wang and F. Zhang. On the precise number of (0,1)-matrices in u(r,s). Discrete Mathematics,
187(13):211–220, 1998.
[24] M. Yannakakis. Computing the minimum fill-in is NP-Complete. SIAM Journal on Algebraic and Discrete Methods, 2(1):77–79, 1981.
9