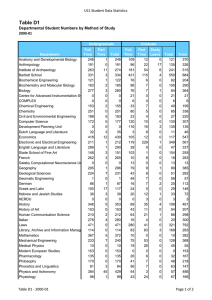
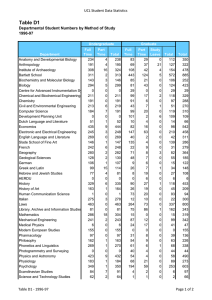
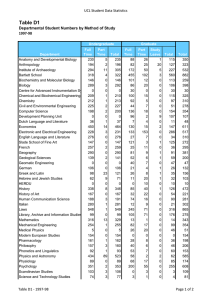
Cell Biology
advertisement

Cell Biology Electron cryomicroscopy and single-particle analysis were used to determine the 30-Å structure of the self-assembling coat protein complex-II cage nanoparticle. Shown are the 2-fold (bottom), 3-fold (middle), and 4-fold (top) fold symmetry axes of the biologically unprecedented cuboctahedron structure responsible for directing cargo selection and membrane curvature during endoplasmic reticulum vesicle budding. Reprinted from Stagg, S.M., Gurkan, C., Fowler, D.M., LaPointe, P., Foss, T.R., Potter, C.S., Carragher, B., Balch, W.E. Structure of the Sec13/31 COPII coat cage. Nature 439:234, 2006. This work is a collaboration between the laboratories of William Balch, Ph.D., Bridget Carragher, Ph.D, and Clinton Potter, B.S., at the National Resource for Automated Molecular Microscopy. Gaudenz Danuser, Ph.D., Associate Professor Dinah Leorke, Ph.D., Research Associate James Lim, Graduate Student Department of Cell Biology CELL BIOLOGY DEPAR TMENT OF CELL BIOLOGY S TA F F Sandra L. Schmid, Ph.D.* Professor and Chairman Francisco Asturias, Ph.D.** Associate Professor William E. Balch, Ph.D.* Professor Kristin Baldwin, Ph.D.*** Assistant Professor Bridget Carragher, Ph.D.** Associate Professor Benjamin Cravatt, Ph.D.**** Professor Director, Helen L. Dorris Child & Adolescent NeuroPsychiatric Disorder Institute 2006 Stephen P. Mayfield, Ph.D.***** Professor Associate Dean of Graduate Studies Mark Mayford, Ph.D.*** Associate Professor Lindsey Miles, Ph.D. Associate Professor Ronald A. Milligan, Ph.D.** Professor Director, Center for Integrative Biosciences Ulrich Müller*** Professor Clinton Potter , B.S.** Associate Professor Philip E. Dawson, Ph.D.***** Associate Professor Lisa Stowers, Ph.D. †† Assistant Professor Velia Fowler, Ph.D.** Professor Heidi Stuhlmann, Ph.D. Associate Professor Shelley Halpain, Ph.D.*** Associate Professor Natasha Kralli, Ph.D. Associate Professor Peter Kuhn, Ph.D.** Associate Professor David Loskutoff, Ph.D. Professor Emeritus Mari Manchester, Ph.D.** Associate Professor Robert Fischer, Ph.D. Alan Bell, B.S.C.S. Xerox Palo Alto Research Center Palo Alto, California Elizabeth Wilson, Ph.D. 19 SENIOR RESEARCH A S S O C I AT E S Richard Bruce, Ph.D. Xerox Palo Alto Research Center Palo Alto, California Douglas Curry, B.S. (E.E.C.S.) Xerox Palo Alto Research Center Palo Alto, California Brian Adair, Ph.D. Barbara Calabrese, Ph.D. Mark Daniels, Ph.D. Jeremiah Joseph, Ph.D. Edward Korzus, Ph.D. ††† University of California Riverside, California Matthew Ritter, Ph.D. Ardem Patapoutian, Ph.D. † Associate Professor James Quigley, Ph.D. Professor Larry R. Gerace, Ph.D.* Professor ADJUNCT APPOINTMENTS Bertil Daneholt, M.D. Karolinska Institutet Stockholm, Sweden Gaudenz Danuser , Ph.D.** Associate Professor Martin Friedlander, M.D., Ph.D. Professor THE SCRIPPS RESEARCH INSTITUTE Ph.D. ††† Kevin F. Sullivan, University of Ireland Galway, Ireland Peter N.T. Unwin, Ph.D.** Professor Clare Waterman-Storer, Ph.D.** Associate Professor Elizabeth Winzeler, Ph.D. † Associate Professor John R. Yates III, Ph.D. Professor Mark J. Yeager, M.D., Ph.D. Professor Scott Elrod, Ph.D. Xerox Palo Alto Research Center Palo Alto, California Mark Ginsberg, M.D. University of California San Diego, California David Goldberg, Ph.D. Xerox Palo Alto Research Center Palo Alto, California Martin Schwander, Ph.D. Gina Story, Ph.D. ††† Washington University St. Louis, Missouri Defne Yarar, Ph.D. Andries Zijlstra, Ph.D. R E S E A R C H A S S O C I AT E S Xiaohua Gong, Ph.D. University of California Berkeley, California Jessica Alexander, Ph.D. Klaus Hahn, Ph.D. University of North Carolina Chapel Hill, North Carolina Veronica Ardi, Ph.D. Eric Peeters, Ph.D. Xerox Palo Alto Research Center Palo Alto, California Andrea Bacconi, Ph.D. Geza Ambrus-Aikelin, Ph.D. Angelique Aschrafi, Ph.D.†††† Hongdong Bai, Ph.D. Kent Baker, Ph.D. S TA F F S C I E N T I S T S Claudia Barros, Ph.D. Michael Bracey, Ph.D. Maria Beligni, Ph.D. Anchi Cheng, Ph.D. Richard Belvindrah, Ph.D. Elena Deryugina, Ph.D. Edward Brignole III, Ph.D. 20 CELL BIOLOGY 2006 THE SCRIPPS RESEARCH INSTITUTE Florence Brunel, Ph.D. Anna Durrans, Ph.D. Paul LaPointe, Ph.D. Jacobus Neels, Ph.D. Anja Bubeck, Ph.D. ††† Gen-Probe, Inc. San Diego, California Samer Eid, Ph.D. ††† Merck Research Laboratories, Neuroscience Drug Discovery West Point, Pennsylvania Nicole Lazarus, Ph.D. Sherry Niessen, Ph.D. Donmienne Leung, Ph.D. ††† Applied Molecular Evolution San Diego, California Silvia Ortega-Gutierrez, Ph.D. Gang Cai, Ph.D. Lesley Page, Ph.D. Gregory Cantin, Ph.D. Eric Carlson, Ph.D. Ph.D. ††† Michael Fitch, Tanabe Research Laboratories U.S.A. San Diego, California Lujian Liao, Ph.D. Aurelia Cassany, Ph.D. Santos Franco, Ph.D. Yuriy Chaban, Ph.D. Margaret Gardel, Ph.D. Pablo Chamero, Ph.D. Emily Chen, Ph.D. Ihsiung Chen, Ph.D. ††† CODA Genomics Laguna Hills, California Yei Hua Chen, Ph.D. Charmian Cher, Ph.D. Smita Chitnis, Ph.D. Esther Choi, Ph.D. Parag Chowdhury, Ph.D. John Lewis, Ph.D. ††† Dalhousie University Halifax, Nova Scotia, Canada Ph.D. ††† Maria Gonzalez, Rincon Pharmaceuticals La Jolla, California Jorg Grandl, Ph.D. Nicolas Grillet, Ph.D. Cemal Gurkan, Ph.D. †††† Johannes Hewel, Ph.D. Michael Hock, Ph.D. Ke Hu, Ph.D. Maria Lillo, Ph.D. ††† University of Salamanca Salamanca, Spain Ana Maria Pasaperi-Limon, Ph.D. Olivier Pertz, Ph.D. ††† University of California San Diego, California Barbie Pornillos, Ph.D. Jennifer Lin, Ph.D. Anita Pottekat, Ph.D. Ryan Littlefield, Ph.D. ††† University of Washington Seattle, Washington Judith Prieto, Ph.D. Dinah Loerke, Ph.D. Natalie Prigozhina, Ph.D. ††† Vala Sciences, Inc. La Jolla, California Darren Logan, Ph.D. Thomas Pucadyil, Ph.D. Bingen Lu, Ph.D. Rajesh Ramachandran, Ph.D. Matthias Machacek, Ph.D. Kalotina Machini, Ph.D. Vandana Ramachandran, Ph.D. Jill Chrencik, Ph.D. Michael Huber, Ph.D. Mark Madsen, Ph.D. Abbas Razvi, Ph.D. Michael Churchill, Ph.D. Darren Hutt, Ph.D. Valentina Marchetti, Ph.D. Leon Reijmers, Ph.D. Francesco Conti, Ph.D. Eric Hwang, Ph.D. Julia Marin-Navarro, Ph.D. Anna Reynolds, Ph.D. Judith Coppinger, Ph.D. Khuloud Jaqaman, Ph.D. Michael Matho, Ph.D. Kaustuv Datta, Ph.D. Anass Jawhari, Ph.D. †††† Naoki Matsuo, Ph.D. Edwin Romijn, Ph.D. ††† Philips Scientific Equipment Division Eindhoven, the Netherlands Leif Dehmelt, Ph.D. Lin Ji, Ph.D. Daniel McClatchy, Ph.D. Ajay Dhaka, Ph.D. Nobutaka Kato, Ph.D. Caroline McKeown, Ph.D. Anouk Dirksen, Ph.D. Claire Kidgell, Ph.D. †††† Marcel Mettlen, Ph.D. Meng-Qui Dong, Ph.D. Katsuhiro Kita, Ph.D. Helena Mira, Ph.D. Michael Dorrell, Ph.D. Kevin Koehntop, Ph.D. Jennifer Mitchell, Ph.D. Alan Saghatelian, Ph.D. ††† Harvard University Cambridge, Massachusetts Kelly A. Dryden, Ph.D. Jenny Kohler, Ph.D. Machiko Muto, Ph.D. Kumar Saikatendu, Ph.D. Jerome Dupuy, Ph.D. Atanas Koulov, Ph.D. Andromeda Nauli, Ph.D. Tomoyo Sakata, Ph.D. Cristian Ruse, Ph.D. Mohsen Sabouri-Ghomi, Ph.D. CELL BIOLOGY 2006 Cleo Salisbury, Ph.D. Ge Yang, Ph.D. Ian Schneider, Ph.D. Christina Schroeder, Ph.D. Masahiro Yasuda, Ph.D. ††† University of Michigan Ann Arbor, Michigan Stephan Sieber, Ph.D. ††† Ludwig-Maximilians University Munich, Germany Rie Yasuda, Ph.D. ††† Osaka University Osaka, Japan Pratik Singh, Ph.D. Zhongmin Zou, Ph.D. ††† Institute of Combined Injury of PLA, Third Military Medical University Chongqing, P.R. China THE SCRIPPS RESEARCH INSTITUTE * Joint appointment in the Department of Molecular Biology ** Joint appointment in the Center for Integrative Molecular Biosciences *** Joint appointment in the Institute for Childhood and Neglected Diseases **** Joint appointments in the Department of Chemistry, the Scott Stagg, Ph.D. Mark Surka, Ph.D. Skaggs Institute for Chemical Biology, and the Helen L. Dorris Child and Adolescent NeuroPsychiatric Disorder Institute ***** Joint appointment in the Skaggs Institute for Chemical Biology † Patricia Szainer, Ph.D. Joint appointments in the Institute for Childhood and S C I E N T I F I C A S S O C I AT E S Neglected Diseases and the Claire Tiraby Nguyen, Ph.D. Tuija Uusitalo, Ph.D. ††† University of Helsinki Helsinki, Finland Genomics Institute of the Novartis Hilda Edith Aguilar de Diaz, M.D. Research Foundation †† Joint appointments in the Helen L. Dorris Child and Adolescent Alexei Brooun, Ph.D. Neuro-Psychiatric Disorder Institute Valerie Uzzell, Ph.D. Claire Delahunty, Ph.D. Mohammed El-Kalay, Ph.D. Josep Villena, Ph.D. Tinglu Guan, Ph.D. Xiaodong Wang, Ph.D. ††† Medical University of Ohio Toledo, Ohio Anand Kolatkar, Ph.D. Eranthie Weerapana, Ph.D. BinQing Wei, Ph.D. Scott Westenberger, Ph.D. Ann Wheeler, Ph.D. Torsten Wittmann, Ph.D. ††† University of San Francisco San Francisco, California James Wohlschlegel, Ph.D. Catherine Wong, Ph.D. Aaron Wright, Ph.D. Lihua Wu, Ph.D. ††† Department of Immunology Scripps Research Appointment completed; new location shown John Venable, Ph.D. Kari Bradtke Weber, Ph.D. ††† †††† Appointment completed 21 22 CELL BIOLOGY 2006 Sandra Schmid, Ph.D. Cell Biology Overview embers of the Department of Cell Biology continue to excel in the rich environment of Scripps Research, and their successes are being recognized by others. Clare Waterman-Storer, who launched her independent career at Scripps Research, received the National Institutes of Health Director’s Pioneer Award this year, along with a 5-year grant to support her research program. Dr. Waterman-Storer was 1 of 13 scientists chosen from more than 800 applicants for this extremely prestigious award; the Director’s Pioneer Award recognizes leading scientists in the United States and invests in their potential to make important advances in biomedical research. Also this year John Yates received the prestigious Christian B. Anfinsen Award from the Protein Society, which recognizes significant technical achievements in the field of protein science. Gaudenz Danuser, Martin Friedlander, Dr. Waterman-Storer, and I have given plenary addresses recognizing notable scientific achievements and leadership in our respective fields. Finally, Dr. Danuser and Natasha Kralli were promoted to associate professor, and Stephen Mayfield, who also serves as associate dean of Graduate Studies, was promoted to full professor—positions befitting their academic and scientific accomplishments. M THE SCRIPPS RESEARCH INSTITUTE A unique attribute of Scripps Research that distinguishes it from academic institutions whose faculty must meet diverse educational obligations is its ability to build research efforts around areas of strength, ensuring critical mass and leadership in important areas of biomedical research. Although the department is proud of the success of its individual scientists, we believe that this success reflects in part the strong synergies that have developed as we have built on areas of strength within cell biology. Synergy is defined as 2 or more groups working together in such a way that the result is greater than the sum of their individual capabilities. Although there are numerous foci of synergy driving innovation and research in the department, I highlight here the 3 areas that represent the strengths on which we will continue to build. An early and unique strength of our department, which synergizes with the structural efforts in the Department of Molecular Biology, is the use of electron cryomicroscopy to provide structural insights into the workings of complex, multisubunit cellular machines. Francisco Asturias, Bridget Carragher, Clint Potter, Ron Milligan, Nigel Unwin, Mark Yeager, and their colleagues have built an internationally preeminent center for electron cryomicroscopy. Together, these groups are developing new methodologies, solving important structures, and training the next generation of electron cryomicroscopy structural biologists, not only among students and fellows at Scripps Research, but through popular intensive summer courses offered to students from around the world. These synergistic interactions have driven unprecedented productivity. Many important structures have been solved in the past year, including (1) the chloroplast ribosome to reveal functionally important differences between it and its bacterial progenitor (Dr. Milligan in collaboration with members of Dr. Mayfield’s laboratory), (2) the intact infectious P22 bacteriophage to reveal the mechanisms of viral DNA transfer into the host cell (Drs. Carragher and Potter in collaboration with Jack Johnson in the Department of Molecular Biology), (3) the coat protein complex II cage to reveal a unique architecture for deforming a membrane and collecting cargo molecules into transport vesicles (Drs. Carragher and Potter in collaboration with members of Bill Balch’s laboratory), (4) the structure of DNA polymerase epsilon, providing new insight into its interaction with its DNA template (Dr. Asturias), (5) the structure of a minus-end directed kinesin to reveal the mechanism of its unusual directionality (Dr. Milligan in collaboration with Ron Vale at the University of Cali- CELL BIOLOGY 2006 fornia, San Francisco), and (6) the structure of an integrin complexed to its substrate revealing the activated form of this cell adhesion molecule (Dr. Yeager). These remarkable accomplishments reveal the accelerated pace of electron cryomicroscopy structural biology made possible through the relatively recent expansion of these efforts within the Center for Integrative Molecular Biosciences, and the resulting innovations in technology and efforts to automate this historically slow and tedious technology. The importance of these efforts, under the leadership of Drs. Carragher and Potter, has been recognized with funding from the National Center for Research Resources and designated as a National Resource for Automated Molecular Microscopy. A second area of synergy, which has been built in association with the Institute for Childhood Diseases, is in cellular and molecular neurobiology. Kristen Baldwin recently joined these efforts from Columbia University after completing her postdoctoral training with Richard Axel, the recipient of the 2004 Nobel Prize in Physiology and Medicine for his work on olfaction. Using the olfactory system as a model, Dr. Baldwin plans to dissect the mechanisms governing neuronal diversity and the establishment of neural connectivities that enable us to process and respond to complex sensory input. She joins a group of investigators at the Institute for Childhood Diseases that includes Shelley Halpain, Mark Mayford, Uli Mueller, Ardem Patapoutian, and Lisa Stowers, who work on diverse but complementary aspects of neuronal development, sensory perception (especially touch, smell, and hearing), the establishment of neuronal circuitry, and higher-order functions of learning and memory. Their combined expertise allows them to tackle the complexities of how the brain is wired for higher-order functioning. The outcomes of their studies have important implications for childhood diseases such as autism, as well as for mental retardation, schizophrenia, and neurodegenerative diseases such as Alzheimer’s and Parkinson’s. A third area of synergy on which we plan to build is in the quantitative spatial and temporal analyses of higherorder cellular processes, such as cell migration, signal transduction, the establishment of polarity, and intracellular trafficking. These efforts, also being carried out within the Center for Molecular Biosciences, are spearheaded by Dr. Danuser, Velia Fowler, and Dr. WatermanStorer. Traditionally, cell biologists have focused their efforts on dissecting a single cellular process or a single piece of the cellular machinery, often in isolation from THE SCRIPPS RESEARCH INSTITUTE 23 the cell. The work of Drs. Danuser and Waterman-Storer is revealing the complex molecular and physical interactions between multiple moving parts of the cell that are required for directed cell locomotion. Dynamic interactions and interdependencies between the actin cytoskeleton and the endocytic machinery are being revealed in collaboration with scientists in my laboratory. Spatial and temporal regulation of signaling events that direct cellular behavior are being analyzed in collaboration with Gary Bokoch in the Department of Immunology. The sophisticated microscopy, image analysis, and mathematical modeling techniques being developed in the Center for Molecular Biosciences, combined with innovations in molecular biology, are enabling cell biologists to manipulate and study the complex behavior of whole cells, rather than the traditional and more limited “divide and conquer” approaches of the past. Although the Department of Cell Biology at Scripps Research is clearly leading in these important areas of research and innovation, in this fast-paced and competitive arena if you’re not moving forward, you’ll quickly slip backward. Therefore, we hope to continue to build on these areas of strength with the recruitment of talented and creative faculty who bring new and complementary expertise to our efforts and who will contribute to and benefit from the synergistic environments we have established. 24 CELL BIOLOGY 2006 INVESTIGATORS’ R EPORTS Structural Characterization of Macromolecular Machines F.J. Asturias, Y. Chaban, J. Brown, E. Brignole, G. Cai e use state-of-the-art electron microscopy and image analysis techniques to determine the 3-dimensional structures of macromolecular complexes involved in a variety of cellular processes, including DNA transcription, DNA replication, chromatin modification and remodeling, and fatty acid synthesis. Macromolecular electron microscopy is an ideal technique for these studies because it requires only a small amount of material and the conditions for preparing samples are physiologically relevant. Images of individual macromolecules are recorded and then computationally combined to obtain structures of low to moderate (25–10 Å) resolution. These structures are often interpreted by docking atomic resolution structures of component subunits in the lower resolution map of an entire complex. Our ultimate goal is to use a combination of biochemical and structural information to reveal the mechanism by which a macromolecular complex carries out its function. In our current research on DNA transcription and its regulation, we are analyzing the basal machinery and assembly of the RNA polymerase II preinitiation complex. We are also studying complexes involved in the regulation of transcription during initiation and in previous steps in which the structure of chromatin is altered to control access to DNA. We are particularly interested in the structure and function of Mediator, a complex that plays a central role in regulating transcription in eukaryotes at the time transcription begins. We have developed a reproducible protocol for purifying Mediator that will enable us to pursue biochemical and structural studies to get to the heart of the mechanism of regulation by Mediator. We are also gearing up to use fluorescence microscopy to validate the in vivo relevance of our in vitro studies. In the past year we also made progress in analyzing the structure and mechanism of DNA polymerase ε (Pol ε). We used electron microscopy and single-particle image analysis to calculate a 16-Å resolution of the polymerase, the first of a multisubunit eukaryotic DNA polymerase. We were able to determine the location of W THE SCRIPPS RESEARCH INSTITUTE 2 of the 4 subunit components of Pol ε and to document and measure changes in the relative orientation of the 2 large domains that constitute the structure. Although no atomic resolution structures of the component subunits were available to help in interpreting the electron microscopy map, we used the structural information to design template elongation assays that resulted in a model for interaction of Pol ε with DNA that explains the intrinsic processivity and capacity of Pol ε to interact with a variety of templates (Fig. 1). F i g . 1 . Structure of Pol ε and a model for its interaction with DNA. Electron microscopy and single-particle image analysis were used to calculate the structure of the polymerase at a resolution of 16 Å. The structure of the catalytic Pol2 subunit was calculated independently, as was the structure of the Pol2–Dpb2 subunit complex. Image analysis also revealed changes in the relative orientation of the Pol2 and Dpb2-Dpb3-Dpb4 domains made possible by a flexible connection. These changes in relative orientation might result in a conformation that would allow access of the DNA to the active-site cleft (top). A return of the tail to its normal conformation after entry of double-stranded DNA into the active-site cleft would result in close interaction of the nucleic acid with the extended tail domain (bottom). This mode of interaction with DNA would explain the intrinsic processivity of Pol ε and the involvement of the Dpb3–Dpb4 subunit complex in double-stranded DNA binding. The processivity dependence of the length of the double-stranded primer region revealed by elongation assays would be explained by the requirement for a minimal length of double-stranded DNA to ensure proper interaction with the full length of the extended tail domain in the Pol ε structure. CELL BIOLOGY 2006 Finally, we continue to investigate the role that conformational changes play in the function of mammalian fatty acid synthase (FAS), the enzyme responsible for the synthesis of long-chain fatty acids. In this true macromolecular assembly line, the different enzymes involved in the synthesis of fatty acids have fused into a single polypeptide chain that includes 6 catalytic and 1 acyl carrier protein domains. Using a novel approach in which FAS point mutants were imaged in the presence of substrates (effectively pausing the enzyme at a given catalytic step), we were able to determine an FAS structure that led to a revised model for FAS organization. That model has now been confirmed by a recently published partial x-ray structure of FAS. As a result of the molecular flexibility that appears to be essential for the function of FAS, 2 of the FAS domains were not observed in the x-ray structure. Further electron microscopy analysis of FAS will reveal the location of all domains and provide information about the variety of conformational states that make possible the multitude of interdomain interactions required for the function of the enzyme. PUBLICATIONS Asturias, F.J., Cheung, I., Sabouri, N., Chilkova, O., Wepplo, D., Johansson, E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat. Struct. Mol. Biol. 13:35, 2006. Takagi, Y., Chadick, J.Z., Davis, J.A., Asturias, F.J. Preponderance of free Mediator in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 280:31200, 2005. Chemical Biology of Conformational Disease and Membrane Traffic W.E. Balch, Y. An, C. Chen, J. Conkright-Johnson, D. Fowler, C. Gurkan, D. Hutt, A. Koulov, P. LaPointe, J. Matteson, A. Nauli, L. Page, H. Plutner, A. Pottekat, A. Razvi, S. Stagg, P. Szajner, I. Yonemoto major challenge is to understand and treat the many protein-misfolding diseases that affect human health, including cystic fibrosis, emphysema, type 2 diabetes, and amyloidosis. These abnormalities are classified as membrane-trafficking conformational diseases because a defect in protein folding at some stage of the eukaryotic secretory pathway results in loss of activity or protein aggregation. A key concern is to determine the underlying defect in A 25 protein folding and how that defect affects the ability of the protein to function normally within the context of the cell’s intracellular transport machinery or in the extracellular environment of the host. Our broad objective is to define the molecular basis for the trafficking of normal and misfolded proteins through the secretory pathway of eukaryotic cells. We use chemical, structural, biological, and bioinformatics approaches. Eukaryotic cells are highly compartmentalized; each compartment of the exocytic and endocytic pathways provides a unique chemical landscape in which protein function and folding may be modulated. Movement between these compartments involves the activity of both anterograde and retrograde transport tubules and vesicles. Many conformational diseases are a consequence of dysfunction at different stages of this transport pathway or outside the cell. Transport through the secretory pathway involves a selective mechanism in which cargo molecules are concentrated into carrier vesicles. Vesicle-mediated transport is regulated by a diverse group of small GTPases belonging to the Ras superfamily. Each of these molecules acts as a “molecular sensor” to regulate different steps in the reversible assembly of vesicle coats and targeting-fusion complexes. During export from the first compartment of the secretory pathway, the endoplasmic reticulum, coat recruitment to budding sites involves activation of the GTPase Sar1. After activation, the cytosolic coat components Sec23/24 and Sec13/31 form the coatomer complex II coat (COPII) that polymerizes to promote budding from the surface of the endoplasmic reticulum. This machinery directs exit from the endoplasmic reticulum of proteins encoded by nearly one third of the genome in eukaryotes. Recently, in collaboration with C. Potter and B. Carragher, Department of Cell Biology, we solved the 2-dimensional electron cryomicroscopy structure of the Sec13/31 cage (Fig. 1). This cage is a self-assembling nanoparticle that collects cargo by assembling into a polymer scaffold that interacts with an adaptor protein complex bound to “exit codes” found on the cytoplasmic domains of cargo and cargo receptors. These exit codes bind to a multivalent adaptor platform found on the surface of Sec24 facing the lipid layer. With J.R. Yates, Department of Cell Biology, we are using state-ofthe-art proteomics (multidimensional protein identification technology or MudPIT) to identify unknown components involved in cargo selection. 26 CELL BIOLOGY 2006 F i g . 1 . Structure of the self-assembling COPII cage. Illustrated are the 3 different symmetry-related views of the Sec13/31 complex that self-assembles to form unprecedented cuboctahedron geometry on the surface of the endoplasmic reticulum to form a molecular scaf- fold (cage) that collects cargo for export. By coordinating cargo concentration with membrane curvature and fission, the cage can generate a transit vesicle that mobilizes cargo to the cell surface. Reprinted from Stagg, S.M., Gurkan, C., Fowler, D.M., LaPointe, P., Foss, T.R., Potter, C.S., Carragher, B., Balch, W.E. Structure of the Sec13/31 COPII coat cage. Nature 439:234, 2006. After budding and fusion of COPII transport vesicles from the endoplasmic reticulum, targeting and fusion of the vesicles to generate the next compartment of the secretory pathway, the Golgi apparatus, require a different class of Ras-like GTPases that belong to the Rab family. Members of the large Rab family (>70 members) act as molecular switches that assemble complexes involved in vesicle tethering and fusion. Using a bioinformatics approach involving hierarchial clustering and mRNA expression profiling (microarray), we found that each Rab GTPase executes targeting and fusion decisions at a distinct step in the exocytic or endocytic pathway. By integrating the interactions of multiple distinct effectors at each step, Rab GTPases act as hubs to define the highly distinctive membrane THE SCRIPPS RESEARCH INSTITUTE architecture of eukaryotic cells found in different tissues. This systems biology approach provides for the first time a global view of membrane traffic from the top down, integrating form with function. Of particular importance is our characterization of the structure of the Rab1 tether p115, done in collaboration with I.A. Wilson, Department of Molecular Biology. The structure reveals a superhelical coiled coil multivalent assembly platform that facilitates Rab-dependent maturation of tethering-fusion complexes. In addition, Rab proteins are recycled for use in multiple rounds of tether assembly. We recently showed the surprising importance of the Hsp90 chaperone system in Rab recycling after vesicle fusion. Many mutation disrupt cargo traffic from the endoplasmic reticulum by preventing proper protein folding during synthesis, resulting in loss of recognition by the COPII selection machinery. Other protein conformational diseases have mutations that disrupt function at later steps of the secretory pathway and outside the cell in new chemical environments that can alter the protein fold. In collaboration with J. Kelly, Department of Chemistry, we are studying the link between trafficking defects and the protein-folding energetics of a number of conformational diseases, including cystic fibrosis, hereditary childhood emphysema, Gaucher disease, familial amyloidosis of Finnish type, Parkinson’s disease, and transthyretin amyloidosis. These analyses have led to a new understanding of the function of the endoplasmic reticulum in normal physiology, suggesting that this compartment functions as a capacitor for protein folding and human evolution. Our analysis of cystic fibrosis has revealed that system-wide modification of the chaperone folding pathways (the chaperone) can alter the steady-state energetic pools of unfolded and folded macrostates (conformational populations) that allow for rescue of the trafficking defect and restore the function of chloride channels at the cell surface. Through a multidisciplinary approach that combines the tools of chemistry, biology, systems biology, bioinformatics, and structure, we hope to gain critical insight into the fundamental principles of cargo trafficking and the basis for a variety of inherited transport diseases. Knowledge of the function of these cargo selection pathways will enable the development of small-molecule chemical chaperones to encourage export and stability of misfolded proteins, leading to restoration of normal cellular function. CELL BIOLOGY 2006 PUBLICATIONS Bannykh, S.I., Plutner, H., Matteson, J., Balch, W.E. The role of ARF1 and Rab GTPases in polarization of the Golgi stack. Traffic 6:803, 2005. Chen, C.Y., Balch, W.E. The Hsp90 chaperone complex regulates GDI-dependent Rab recycling. Mol. Biol. Cell 17:3494, 2006. Chen, C.Y., Sakisaka, T., Balch, W.E. Use of Hsp90 inhibitors to disrupt GDIdependent Rab recycling. Methods Enzymol. 403:339, 2005. Fowler, D.M., Koulov, A.V., Alory-Jost, C., Marks, M.S., Balch, W.E., Kelly, J.W. Functional amyloid formation within mammalian tissue. PLoS Biol. 4:e6, 2006. Gurkan, C., Balch, W.E. Recombinant production in baculovirus-infected insect cells and purification of the mammalian Sec13/Sec31 complex. Methods Enzymol. 404:58, 2005. Gurkan, C., Lapp, H., Alory, C., Su, A.I., Hogenesch, J.B., Balch, W.E. Large-scale profiling of Rab GTPase trafficking networks: the membrome. Mol. Biol. Cell 16:3847, 2005. Gurkan, C., Lapp, H., Hogenesch, J.B., Balch, W.E. Exploring trafficking GTPase function by mRNA expression profiling: use of the SymAtlas Web-application and the Membrome datasets. Methods Enzymol. 403:1, 2005. Kelly, J.W., Balch, W.E. The integration of cell and chemical biology in protein folding. Nat. Chem. Biol. 2:224, 2006. Page, L.J., Suk, J.Y., Huff, M.E., Lim, H.J., Venable, J., Yates, J., Kelly, J.W., Balch, W.E. Metalloendoprotease cleavage triggers gelsolin amyloidogenesis. Embo J. 24:4124, 2005. Stagg, S.M., Gurkan, C., Fowler, D.M., LaPointe, P., Foss, T.R., Potter, C.S., Carragher, B., Balch, W.E. Structure of the Sec13/31 COPII coat cage. Nature 439:234, 2006. Suk, J.Y., Zhang, F., Balch, W.E., Linhardt, R.J., Kelly, J.W. Heparin accelerates gelsolin amyloidogenesis. Biochemistry 45:2234, 2006. Wang, X., Venable, J., LaPointe, P., Hutt, D.M., Koulov, A.V., Coppinger, J., Gurkan, C., Kellner, W., Matteson, J., Plutner, H., Riordan, J.R., Kellly, J.W., Yates, J.R. III, Balch, W.E. Hsp90 cochaperone rescue of misfolding disease. Cell, in press. Wiseman, R.L., Balch, W.E. A new pharmacology: drugging stressed folding pathways. Trends Mol. Med. 11:347, 2005. Molecular Mechanisms of Olfactory Perception and Neural Circuit Formation K.K. Baldwin, S. Tate, B. Fields, S. Ghosh n mammals, the sense of smell is critical for survival. Scents trigger suckling at birth, distinguish food from poison, provide warning of predators, and identify attractive mates. A primary goal of neurobiology is to discover how neural circuits link these types of sensory inputs to appropriate behavioral outputs. Surprisingly little is known about how neural circuits specific to one set of inputs are organized or built. We take advantage of the unique architecture and genetic tractability of the mouse olfactory system to study specific olfactory circuits at the first 2 levels of pro- I THE SCRIPPS RESEARCH INSTITUTE 27 cessing. Our goal is to genetically label the neurons that respond to specific odors in the nose and the olfactory bulb, to trace the projections of the neurons into the cortical regions where inputs converge, and to identify the molecular mechanisms that govern the formation of specific neural circuits. We anticipate that our findings will reveal mechanisms common to neural circuit formation throughout the brain and provide insight into genetic bases of human cognitive and behavioral disorders. CLONING MICE FROM NEURONS In contrast to gene activation in other sensory systems, odorant receptor genes are activated by stochastic mechanisms. Stochastic gene activation in the immune system is due to irreversible DNA rearrangements. We searched for chromosomal rearrangements by cloning mice from the nuclei of olfactory sensory neurons. We found that odorant receptor choice is reversible, as is neuronal differentiation. We will now clone mice from other types of neurons to determine whether irreversible chromosomal alterations accompany neuronal diversification in the brain. VISUALIZING OLFACTORY INPUTS TO THE BRAIN A major challenge is to understand how olfactory information is integrated in the olfactory cortex. An important first step is to describe the anatomy of the second-order circuit. This endeavor has been hindered by the lack of specific promoters for the output neurons (mitral cells) of the olfactory bulb. We identified a gene that is expressed specifically in mitral cells and have produced mice in which subsets of these cells express fluorescent proteins. We use confocal and 2-photon microscopy to map the projections of individual mitral cells into the brain. By visualizing the second-order olfactory circuit, we can begin to understand how the brain recognizes odors. N E U R A L D I V E R S I T Y A N D C I R C U I T F O R M AT I O N Although genes that regulate axon guidance and large-scale brain patterning have been identified, the genes that endow neurons with the precise patterns of neuronal synaptic connectivity remain enigmatic. We reasoned that genes expressed in subsets of mitral cells would be good candidates to direct the formation of specific neuronal circuits. Using bioinformatics and single-cell gene profiling, we have identified about 70 genes that can be used to subdivide mitral cells into different classes. We will use gene targeting to test the role of these genes in neural circuit formation. One class of genes that diversifies mitral cells is the large family of approximately 60 clustered genes for pro- 28 CELL BIOLOGY 2006 tocadherins. Protocadherins are expressed throughout the nervous system. Intriguingly, each neuron seems to express a distinct combination of several protocadherin proteins. We have produced mice that do not express one subfamily of protocadherins. These mice have behavioral abnormalities consistent with defects in neuronal function. We are investigating the cellular and physiologic consequences of loss of protocadherin diversity. Automated Molecular Imaging B. Carragher, C.S. Potter, A. Cheng, D. Fellmann, G. Lander, S. Mallick, P. Mercurio, J. Pulokas, J. Quispe , S. Stagg, C. Yoshioka uring the past decade, electron cryomicroscopy has emerged as a powerful method for determining the structure of large macromolecular complexes. Elucidating the structure and mechanism of action of these “molecular machines” is an emerging frontier in understanding how the information in the genome is transformed into cellular activities. Examples of the machines include ribosomes, transcription complexes, track-motor complexes, and membrane-embedded pumps and channels. In electron cryomicroscopy, the macromolecular specimen is preserved in a thin layer of vitreous (glassy) ice and imaged in an electron microscope by using low doses of electrons. The low signal-to-noise ratio of the resulting images means that averaging is required to recover the signal and reconstruct a 3-dimensional map of the structure. In 2002, we established the National Resource for Automated Molecular Microscopy (NRAMM) to develop, test, and apply technology for automating the processes involved in using electron cryomicroscopy to solve macromolecular structures. The goal of automation is not only to facilitate the process of molecular microscopy, although this facilitation is a welcome benefit, but also to expand the scope of accessible problems and push experimental frontiers by making possible investigations deemed too difficult or high risk because of the considerable effort involved in using manual methods. An additional goal of automation is to enable much higher throughput of data and thus improve resolution for single-particle reconstructions by increasing the numbers of particles that contribute to the average 3-dimensional map. Another mission of NRAMM is to D THE SCRIPPS RESEARCH INSTITUTE use the infrastructure developed to open up the sometimes esoteric practices of electron cryomicroscopy to a much wider group of researchers, including investigators in cell biology, x-ray crystallography, and materials science. During the past 3 years, the new techniques and technologies that we developed included a new grid substrate designed to improve quality and throughput for vitreous ice specimens; a prototype of a robotic grid-handling system used for screening; Leginon, an automated system for microscope control and image acquisition; a relational database that tracks and manages data acquired by Leginon and tools for viewing and delivering the data via Web browsers; and ACE, a program for the automated measurement and correction of contrast transfer function. These technologies all contributed in demonstrating the potential for automated high-throughput data acquisition and analysis in an experiment in which images of more than 280,000 particles of GroEL, a molecule involved in protein folding, were acquired in a single 25-hour session at the microscope and subsequently subjected to completely automated procedures to reconstruct a 3-dimensional map to a resolution better than 8 Å (Fig. 1). F i g . 1 . More than 280,000 particles of GroEL were acquired from a single grid by using Leginon in a period of 25 hours. These particles were sorted by ice thickness and used to reconstruct a 3-dimensional density map to a resolution of approximately 8 Å. Visualization by Scott Stagg and Mike Pique. These technological developments have been designed for and used in a number of collaborative research projects, including reconstruction of a minimal coatomer complex II cage and reconstruction of an intact infectious P22 virion. The infrastructure has also, in accordance with our mission, made electron cryomicroscopy accessible to a much wider commu- CELL BIOLOGY 2006 nity, leading to publications from research groups in chemistry, x-ray crystallography, materials science, and industry. NRAMM currently provides support for more than 35 collaborative and service projects, and the Leginon software, including the database, has been distributed to about 30 laboratories outside Scripps Research. We are also distributing ACE, a variety of other software packages, and the novel grid substrates. These efforts are complemented by training activities that include small-group training, a biennial large training course in electron cryomicroscopy, and small workshops focused on various aspects of automation. An additional project, sponsored by the National Science Foundation, is the development of automated data collection techniques for imaging serial sections by using an electron microscope. Understanding the fine structure of cells and cellular components contributes to a more profound understanding of cellular function and intracellular or intercellular interactions. In order to visualize these large, complex structures in 3 dimensions at resolutions sufficient to observe structure on the nanoscale, the cells must be cut into sections and then examined by using a transmission electron microscope. Acquiring high-magnification images of a long series of sections is difficult and extremely labor intensive. The region of interest in each section must be tracked across sections and across grids, a process that requires examining the sections at a variety of scales before acquiring high-magnification images of interesting areas. Multiscale imaging of this sort is not straightforward because the image formed by an electron microscope shifts and rotates as the magnification is changed. The overall task of reconstructing a 3-dimensional volume from a set of serial sections is challenging and time consuming, and the number of large-scale reconstructions has been limited to a few spectacular examples. Our objectives are to design, develop, and implement a software application to automate the task of acquiring high-magnification images of specific regions of the cell across tens to hundreds of serial sections. PUBLICATIONS Cheng, A., Fellmann, D., Pulokas, J., Potter, C.S., Carragher, B. Does contamination buildup limit throughput for automated cryoEM? J. Struct. Biol. 154:303, 2006. Fellmann, D., Banez, R., Carragher, B., Potter, C.S. Temperature monitoring of an EM environment. Microsc. Today 14:24, January 2006. Stagg, S.M., Gurkan, C., Fowler, D.M., LaPointe, P., Foss, T.R., Potter, C.S., Carragher, B., Balch, W.E. Structure of the Sec13/31 COPII coat cage. Nature 439:234, 2006. THE SCRIPPS RESEARCH INSTITUTE 29 Stagg, S.M., Lander, G.C., Pulokas, J., Fellmann, D., Cheng, A., Quispe, J.D., Mallick, S.P., Avila, R.M., Carragher, B., Potter, C.S. Automated cryoEM data acquisition and analysis of 284,742 particles of GroEL. J. Struct. Biol., in press. Suloway, C., Pulokas, J., Fellmann, D., Cheng, A., Guerra, F., Quispe, J., Stagg, S., Potter, C.S., Carragher, B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151:41, 2005. Regulation of Cytomechanochemical Systems G. Danuser, A. Bacconi, J. Dorn, K. Jaqaman, L. Ji, J. Kunken, D. Loerke, M. Machacek, A. Matov, M. Sabouri, K. Thompson, G. Yang e study how force-generating molecular machines are spatially and temporally regulated to mediate complex cell functions, including migration, division, and intracellular transport of organelles and vesicles. Specifically, we investigate the relationships between assembly and contraction of the actin cytoskeleton and the dynamic coupling of actin filaments with other components of the cytoskeleton during cell migration. We also study how assembly and disassembly of microtubules and motor-driven sliding of microtubule bundles are orchestrated to symmetrically segregate replicated DNA from the dividing mother cell into 2 daughter cells. In the past year, we expanded our research program with 2 new, collaborative projects. In the first project, we aim to establish the requirements for local regulation of cortical actin mechanics during endocytosis. In the second, we are analyzing the modes of interaction between microtubule plus end– and minus end–directed motor families in vesicle transport along neuronal axons. To examine molecular systems, we develop computational models to predict the relationship between the dynamics of molecular-level component processes and cellular-level outputs. Subsequently, we validate the models and estimate unknown parameters by fitting the parameters to measurements of cell dynamics. The challenges in such data-driven, multiscale modeling are 2-fold: the precise and complete characterization of cell dynamics in space and time and the implementation of numerical tools for fitting cellularlevel data to models with molecular resolution. In our studies of cell migration, we made 2 major advancements. First, in collaboration with C. WatermanStorer, Department of Cell Biology, we extended fluores- W 30 CELL BIOLOGY 2006 cent speckle microscopy to accomplish an integrated, correlative multiparameter analysis of cytoskeleton dynamics. We can now determine accurately how cell movements depend on molecular processes such as the assembly, disassembly, and transport of cytoskeleton components or adhesions, and we can use biosensor probes to visualize activation of signals. We recently used our analysis framework in studies in which we dissected the roles of several signaling cascades in the regulation of cell motility and the involvement of adhesion molecules in the transient, integrin-mediated coupling of the actin cytoskeleton to the extracellular matrix. A second breakthrough was achieved in our effort to reconstruct intracellular force distributions from light microscopic measurements of cytoskeleton deformation. We established a unique method to probe the relationship between spatially distributed force generation and the resulting cell morphologic outputs, for example, during cell migration. We will use this tool to dissect the mechanism of force regulation by signals and identify feedback interactions between force transduction and signal activation, which are a central element in molecular systems control, not only in cell motility but also in a broad set of other cell functions. To study chromosome segregation, we use fluorescent speckle microscopy to analyze the dynamics of microtubule scaffolds associated with the spindle apparatus in animal cells and 3-dimensional, high-resolution light microscopy to analyze the dynamics of single chromosomes in yeast. The first approach should reveal how microtubule assembly and disassembly across the spindle are coregulated with motor-mediated generation of force. The second approach should allow us to identify the functions of proteins in the kinetochore, a molecular complex that regulates the attachment of chromosomes to spindle microtubules. We have developed fully automated, image-based approaches of unprecedented sensitivity for investigations of phenotype microtubule and chromosome dynamics. In collaboration with E.D. Salmon, University of North Carolina, T. Kapoor, Rockefeller University, and P. Sorger, Massachusetts Institute of Technology, we are using the data obtained to systematically characterize the involvement of spindle- and kinetochore-associated proteins in the regulation of chromosome motion throughout the cell cycle. PUBLICATIONS Cameron, L.A., Yang, G., Cimini, D., Canman, J.C., Kisurina-Evgenieva, O., Khodjakov, A., Danuser, G., Salmon, E.D.S. Kinesin 5-independent poleward flux of kinetochore microtubules in Ptk1 cells. J. Cell Biol. 173:173, 2006. THE SCRIPPS RESEARCH INSTITUTE Danuser, G. Coupling the dynamics of two actin networks: new views on the mechanics of cell protrusion. Biochem. Soc. Trans. 33:1250, 2005. Danuser, G., Waterman-Storer, C.M. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Ann. Rev. Biophys. Biomol. Struct. 35:361, 2006. deRooij, J., Kerstens, A., Danuser, G., Schwartz, M.A., Waterman-Storer, C.M. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J. Cell Biol. 171:153, 2005. Dorn, J.F., Jaqaman, K., Rines, D.R., Jelson, G.S., Sorger, P.K., Danuser, G. Yeast kinetochore microtubule dynamics analyzed by high-resolution three-dimensional microscopy. Biophys. J. 89:2834, 2005. Ji, L., Danuser, G. Tracking quasi-stationary flow of weak fluorescent features by adaptive multi-frame correlation. J. Microsc. 220:150, 2005. Lussi, J.W., Tang, C., Kuenzi, P.A., Staufer, U., Csucs, G., Voros, J., Danuser, G., Hubbell, J.A., Textor, M. Selective molecular assembly patterning at the nanoscale: a novel platform for producing protein patterns by electron-beam lithography on SiO2/indium tin oxide-coated glass substrates. Nanotechnology 16:1781, 2005. Machacek, M., Danuser, G. Morphodynamic profiling of protrusion phenotypes. Biophys. J. 90:1439, 2006. Meijering, E., Smal, I., Danuser, G. Tracking in molecular bioimaging. IEEE Signal Process. Mag. 23:46, May 2006. Ponti, A., Matov, A., Adams, M., Gupton, S., Waterman-Storer, C.M., Danuser, G. Periodic patterns of actin turnover in lamellipodia and lamellae of migrating epithelial cells analyzed by quantitative fluorescent speckle microscopy. Biophys. J. 89:3456, 2005. Shah, S., Yang, G., Danuser, G., Goldstein, L.S.B. Axonal transport: imaging and modeling of a neuronal process. Springer Lecture Notes in Physics. In: The Nobel Symposium. Springer, New York, in press. Yang, G., Matov, A., Danuser, G. Reliable tracking of large scale dense antiparallel particle motion for fluorescence live cell imaging. In: Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05) Workshops. IEEE Computer Society, Washington, DC, 2005, Vol. 3, p. 138. Synthetic Protein Chemistry P.E. Dawson, A. Dirksen, F. Brunel, M. Churchill, F. Hansen, E. Lempens, N. Metanis, T. Shekhter, T. Tiefenbrunn e use chemical synthesis to design and engineer proteins with novel structures and functions. We continue to develop methods to link fully unprotected peptides and carbohydrates via native and nonnative linkages. During the past year, we focused on synthesizing mimics of the HIV envelope protein gp41, the oxidoreductase enzyme glutaredoxin, a glycosylated form of monocyte chemoattractant protein-3, and carbohydrate-binding proteins. We are also using these chemoselective ligation reactions to label proteins such as thrombin and nanoparticle quantum dots. Overall, our goal is to use synthetic chemistry to understand the molecular basis of protein structure and function. W S E L E N O G L U TA R E D O X I N Selenoenzymes have a central role in maintaining cellular redox potential. These enzymes have selenylsul- CELL BIOLOGY 2006 fide bonds in their active sites that catalyze the reduction of peroxides, sulfoxides, and disulfides. The preparation of enzymes containing selenocysteine is experimentally challenging. As a result, little is known about the kinetic role of selenols in enzyme active sites, and the redox potential of a selenylsulfide or diselenide bond in a protein has not been experimentally determined. To fully evaluate the effects of selenocysteine on oxidoreductase redox potential and kinetics, we synthesized glutaredoxin 3 (Grx3; Fig. 1) and all 3 seleno- F i g . 1 . Chemical synthesis of Grx3 by using folding to accelerate the ligation reaction. Selective oxidation of the active-site disulfide allowed alkylation of the cysteine residue at the ligation site. cysteine variants of the enzyme’s conserved 11CXX14C active site. Grx3, Grx3(C11U), and Grx3(C14U) had redox potentials of –193, –259 and –273 mV, respectively. The position of redox equilibrium between Grx3(C11U-C14U) (–308 mV) and thioredoxin (–270 mV) suggests a possible role for diselenide bonds in biological systems. Kinetic analysis indicated that the lower redox potentials of the selenocysteine variants is due primarily to the greater nucleophilicity of the activesite selenium rather than to the role of the selenium as either a leaving group or a “central atom” in the exchange reaction. The 100- to 10,000-fold increase THE SCRIPPS RESEARCH INSTITUTE 31 in the rate of thioredoxin reduction by the seleno-Grx3 analogs indicates that compared with their sulfide counterparts, oxidoreductases containing either selenylsulfide or diselenide bonds can have physiologically compatible redox potentials and enhanced reduction kinetics. H I V VA C C I N E D E S I G N The transmembrane protein gp41 is an attractive target for the development of an HIV vaccine. We are collaborating with M.B. Zwick and D.R. Burton, Department of Immunology, and I.A. Wilson, Department of Molecular Biology, to design peptides that mimic the gp41 epitopes of known neutralizing antibodies. The membrane-proximal external region of gp41 contains several neutralizing epitopes, including 4E10 and Z13e1. On the basis of our previous work on 4E10, we designed and synthesized peptides to map the Z13 epitope and performed an alanine scan to identify key elements within the sequence. Structural constraints are being introduced into the peptides to obtain an antigen capable of eliciting both 4E10- and Z13e1-like antibodies. To better mimic the molecular environment of native gp41, we plan to introduce steric constraints such as polyethylene glycol and carbohydrates. To mimic the viral membrane, we have appended a transmembrane helix and have incorporated the peptide into soluble lipid bilayers. Recently, neutralizing antibodies to the N-heptad repeat of gp41 have been discovered. We designed and synthesized 3-helix bundles to mimic this region of gp41, and we are using them to identify these epitopes and map the key binding interactions. PUBLICATIONS Brunel, F.M., Zwick, M.B., Cardoso, R.M., Nelson, J.D., Wilson, I.A., Burton, D.R., Dawson, P.E. Structure-function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. J. Virol. 80:1680, 2006. Cremeens, M.E., Fujisaki, H., Zhang, Y., Zimmermann, J., Sagle, L.B., Matsuda, S., Dawson, P.E., Straub, J.E., Romesberg, F.E. Efforts toward developing direct probes of protein dynamics. J. Am. Chem. Soc. 128:6028, 2006. Delehanty, J.B., Medintz, I.L., Pons, T., Brunel, F.M., Dawson, P.E., Mattoussi, H. Self-assembled quantum dot-peptide bioconjugates for selective intracellular delivery. Bioconjug. Chem. 17:920, 2006. Medintz, I.L., Clapp, A.R., Brunel, F.M., Tiefenbrunn, T., Uyeda, H.T., Chang, E.L., Deschamps, J.R., Dawson, P.E., Mattoussi, H. Proteolytic activity monitored by fluorescence resonance energy transfer through quantum-dot-peptide conjugates. Nat. Mater. 5:581, 2006. Sagle, L.B., Zimmermann, J., Matsuda, S., Dawson, P.E., Romesberg, F.E. Redoxcoupled dynamics and folding in cytochrome c. J. Am. Chem. Soc. 128:7909. 2006. Yamamoto, N., Takayanagi, A., Sakakibara, T., Dawson, P.E., Kajihara, Y. Highly efficient synthesis of sialylglycopeptides overcoming unexpected aspartimide formation during activation of Fmoc-Asn(undecadisialyloligosaccharide)-OH. Tetrahedron Lett. 47:1341, 2006. 32 CELL BIOLOGY 2006 Regulation of Actin Dynamics in Morphogenesis and Development V.M. Fowler, T. Fath, R.S. Fischer, C. McKeown, J. Moyer, R. Nowak, J. Palomique, K. Weber egulation of actin dynamics at the ends of filaments determines the organization and turnover of actin cytoskeletal structures and is critical for cell motility and architecture and actin-based morphogenetic processes in development. We focus on the tropomodulin family of proteins that cap the pointed ends of actin filaments. Tropomodulins are a conserved family of proteins of about 40 kD that bind to tropomyosin and actin. The tropomodulins are expressed in a tissue-specific and developmentally regulated fashion in vertebrates, flies, and worms. In vertebrates, the tropomodulin 1 isoform is associated with stable architectural arrays of actin filaments such as thin filaments in striated muscle myofibrils and actin filaments in the membrane skeleton of red blood cells (RBCs) and on the lateral membranes of the fiber cells of the eye lens. Previous research indicated that tropomodulin 1 regulates the dynamics of actin pointed ends and thus the length and stability of thin filaments in myofibrils of cultured cardiac muscle cells. Tropomodulin 3, the isoform in the cytoplasm, is associated with dynamic actin filaments in the lamellipodia of crawling endothelial cells, where it is a negative regulator of cell migration. Our goal is to tie the molecular and cellular regulation of the dynamics of actin pointed ends by tropomodulins to the in vivo functions of the proteins in actin-based morphogenetic processes in development. We use mouse genetic models to study the function of tropomodulins in myofibril assembly and cardiac development, the biogenesis and stability of the RBC membrane skeleton, and the morphogenesis and transparency of fiber cells in the eye lens. The structure and function of tropomodulins is best understood for tropomodulin 1, which consists of 2 domains: an unstructured, flexible N-terminal domain and a compact, folded C-terminal domain composed of 5 leucine-rich repeats. The N-terminal domain binds tropomyosin and is regulated by tropomyosin to cap tropomyosin-actin pointed ends with nanomolar affinity. The C-terminal domain caps actin pointed ends with submicromolar affinity and is unaffected by tropomyosin. R THE SCRIPPS RESEARCH INSTITUTE Despite the high level of sequence conservation (~70%) among vertebrate tropomodulins, comparisons of their actin-binding activities reveals that tropomodulin 3, but not tropomodulin 1, binds actin monomers and nucleates actin filament assembly in addition to capping pointed ends. Tropomodulin 3 can be chemically crosslinked to actin in a 1:1 complex, providing a tool to identify the amino acids at the tropomodulin 3–actin binding interface. Initial results from tryptic digestion and mass spectrometry indicate that tropomodulin 3 interacts with actin monomers via a unique interface on the actin and on the tropomodulin 3. Site-directed mutagenesis plus structural and functional interaction studies are in progress to further define the tropomodulin 3–actin binding interface and to develop tropomodulin mutants for studies of cellular functions in vivo. To investigate the in vivo function of tropomodulin 1 in myofibril assembly and cardiac development, we are using mice that lack the gene for this tropomodulin. We showed previously that myofibril assembly in the heart is grossly aberrant in the embryos of these mutants, leading to aborted cardiac development and the death of embryos between days 9 and 10 of development. To investigate the primary defect in myofibril assembly, we examined nascent myofibrils on myocyte membranes in embryos at 4–5 days of development, before the appearance of gross cardiac abnormalities. In wild-type embryos, the earliest myofibrils contain 1–3 sarcomeres in tandem with regularly spaced Z bodies and continuous F-actin, indicative of unregulated filament lengths. Such sarcomere structures are never observed in the absence of tropomodulin 1; instead, α-actinin and F-actin are present in rodlike, aberrant Z disc structures on myocyte membranes. This finding suggests that tropomodulin 1 has a novel early function in the organization of Z discs into sarcomeres. More recently, we found that cardiac development fails specifically at the stage of looping morphogenesis, at an earlier stage than that observed in all other mice that lack genes for contractile proteins. We are testing the hypotheses that defective myofibril assembly in absence of tropomodulin 1 may lead directly or indirectly to aberrant cell-cell contacts or polarity, and/or to defective cell proliferation, leading to failure of looping morphogenesis. To investigate the consequences in RBCs of deleting the gene for tropomodulin 1, we prevented death in the embryos of the mutant mice by expressing a tropomodulin 1 transgene solely in the heart. The result CELL BIOLOGY 2006 was viable mice with no tropomodulin 1 in their RBCs. Hematologic analyses revealed that these mice had a compensated mild hemolytic anemia, with increased reticulocytosis and RBCs that were abnormally variable in size in blood smears. Measurements of mechanical stability and deformability indicated that tropomodulin 1–deficient RBCs were less deformable and more fragile than normal RBCs. Western blotting indicated increased levels of tropomodulin 3 in the tropomodulin 1–deficient RBCs. Using these tropomodulin 1–deficient RBCs, we can test the effects of tropomodulin 3 on the length and dynamics of actin filaments and the consequences for stability of the membrane skeleton, RBC survival, and function in vivo. Mice with the transgene also provide an opportunity to examine the function of tropomodulin 1 in vivo in other tropomodulin 1–expressing cells and tissues such as the eye lens, neurons, and kidney. We are also producing mice that lack the gene for tropomodulin 3 to obtain mice with RBCs deficient in both tropomodulin 1 and tropomodulin 3 to assess the consequences of complete lack of tropomodulin on RBC structure and function. PUBLICATIONS Fowler, V.M., McKeown, C.R., Fischer, R.S. Nebulin: does it measure up as a ruler? Curr. Biol. 16:R18, 2006. Angiogenesis-Dependent Disease and Membrane Protein Topogenesis M. Friedlander, E. Aguilar, E. Banin, F. Barnett, R. Bautchek, M. Dorrell, M. El-Kalay, S.F. Friedlander, S. Hanekamp, R. Jacobson, A. Johnson, V. Machetti, M. Ritter, L. Scheppke, J. Trombley, H. Uusitalo-Jarvinen, V. Marchetti, W. Ruf ANGIOGENESIS-DEPENDENT DISEASE ost diseases that cause catastrophic loss of vision do so as a result of abnormal growth of blood vessels. Similarly, tumors depend on a blood supply for their growth and use these new vessels as an avenue for metastasis. Blood vessels themselves can generate tumors (e.g., hemangiomas) when the growth and organization of vascular endothelial cells is not properly controlled. Our goal is to understand the mechanisms of ocular neovascularization in normal and pathologic situations. M THE SCRIPPS RESEARCH INSTITUTE 33 We use a neonatal mouse retina model to identify regulators of developmental angiogenesis and understand endothelial guidance mechanisms. In addition, in a long-standing collaboration with D.A. Cheresh, University of California, San Diego, we are using this system to evaluate the role of integrins in this process. In collaboration with P.R. Schimmel, Department of Molecular Biology, we found that fragments of tryptophan tRNA synthetase are potent angiostatics that significantly reduce retinal neovascularization. The synthetase fragments are also angiostatic in vivo when delivered by a cell-based method. Most recently, we used combination therapy to show that targeting multiple, distinct angiogenic pathways with fragments of tryptophan tRNA synthetase and antagonists of integrins and vascular endothelial cell growth factor provides highly synergistic, potent angiostatic activity. Although this therapeutic approach should be useful in the treatment of diseases in which complete inhibition of angiogenesis is desirable, it may not be efficacious in the treatment of ischemic retinal disease. In ischemic retinal disease, relief of hypoxia by vascular reconstruction, rather than destruction, may be the desired outcome. To examine possible therapies for diseases of retinal ischemia, we explored the potential usefulness of stem cells derived from the bone marrow of adult mice for cell-based delivery of angiostatic and neurotrophic substances and for the trophic actions of the cells themselves in vascular and neuronal degenerative diseases. We found that both lineage-negative hematopoietic progenitors and CD44 hi -expressing myeloid progenitors specifically target activated retinal astrocytes, incorporate into and around new vessels, and, in a mouse model of retinal degeneration, rescue and stabilize a degenerating retinal vasculature. We also showed that both types of stem cells have a profound neurotrophic effect when injected into eyes of mice with inherited retinal degeneration; not only is the vasculature rescued in these mice but photoreceptors and visual function are also preserved. The stem cells also rescue retinal vasculature subject to hypoxic stress and may be useful in the treatment of ischemic retinal abnormalities such as diabetic retinopathy and retinopathy of prematurity. The mechanism of rescue is not clear, but it is related to high levels of heat-shock proteins found in these populations of cells. We also defined a role for CD44 hi-derived microglia in facilitating vascular recovery in models of retinal ischemia. 34 CELL BIOLOGY 2006 Glioblastoma multiforme is an incurable brain tumor that is usually fatal within 1 year after diagnosis. We are using gene therapy and a rat model of this disease to study the efficacy of an antiangiogenic approach in treating these tumors. Hemangiomas are endothelial tumors that proliferate rapidly and later involute spontaneously. We are using DNA microarrays to study changes in gene expression as hemangiomas progress. Our goal is to identify (1) new targets for therapy for these tumors and (2) novel regulators of angiogenesis. In a collaboration with G.R. Nemerow, Department of Immunology, we used pseudotyped adenovirus to selectively target specific cell types in the retina. By using the appropriate fiber type, we can deliver transgenes to cells, such as photoreceptors, that ordinarily are not targeted by adenovirus. MEMBRANE PROTEIN TOPOGENESIS We are also studying the mechanism whereby proteins are asymmetrically integrated into cell membranes. In addition to studies of membrane protein topogenesis at the molecular level, we are studying defects in protein processing and insertion that occur in several degenerative diseases of the eye. In collaboration with K. Philipson, University of California, Los Angeles, we are investigating the topology of the cardiac sodium-calcium exchanger. On the basis of hydropathy analysis of the amino acid sequence, the exchanger is proposed to contain 12 hydrophobic segments, the first of which is a cleaved signal sequence. Using a variety of reporter domains (glycosylation sites, epitopes, and proteolytic cleavage sites), we analyzed the topology of the exchanger both in vitro and in oocyte expression systems. Because nearly all other polytopic eukaryotic membrane proteins do not have cleaved signal sequences, we are investigating the putative role of such a sequence in the insertion and targeting of these exchangers. Our results indicate that the native, cleaved N-terminal signal sequence is not necessary for insertion of a functional exchanger into the cell membrane. In contrast, the photoreceptor exchanger does not have a cleaved N-terminal signal sequence. If the N-terminal 65 amino acids are deleted, translocation of the N terminus of the protein is disrupted, but the remainder of the exchanger is integrated into the membrane. We are also using large-scale genomic analysis to study transgenic mice in which mutated exchanger is expressed and mice that lack the gene for the exchanger. THE SCRIPPS RESEARCH INSTITUTE PUBLICATIONS Banin, E., Dorrell, M.I., Aguilar, E., Ritter, M.R., Aderman, C.M., Smith, A.C.H., Friedlander, J., Friedlander, M. T2-TrpRS inhibits preretinal neovascularization and enhances physiological vascular regrowth in OIR as assessed by a new method of quantification. Invest. Ophthalmol. Vis. Sci. 47:2125, 2006. Dorrell, M., Uusitalo-Jarvinen, H., Aguilar, E., Friedlander, M. Ocular angiogenesis; basic mechanisms and therapeutic advances. Surv. Ophthalmol., in press. Dorrell, M.I., Friedlander, M. Mechanisms of endothelial cell guidance and vascular patterning in the developing mouse retina. Prog. Retin. Eye Res. 25:277, 2006. Friedlander, M. Stem cells and retinal disease. In: Retina, 4th ed. Ryan, S.J. (Editor-in-Chief). St. Louis, Mosby, 2006, Vol. 1, p 23.* Friedlander, S.F., Ritter, M.R., Friedlander, M. Recent progress in our understanding of the pathogenesis of infantile hemangiomas. Lymphat. Res. Biol. 3:219, 2005. Jin, H., Aiyer, A., Su, J., Borgstrom, P., Stupack, D., Friedlander, M., Varner, J. A homing mechanism for bone marrow-derived progenitor cell recruitment to the neovasculature. J. Clin. Invest. 116:652, 2006. Ritter, M., Aguilar, E., Banin, E., Scheppke, L., Uusitalo-Jarvinen, H., Friedlander, M. Three-dimensional in vivo imaging of the mouse ocular vasculature during development and disease. Invest. Ophthalmol. Vis. Sci. 46:3021, 2005. Ritter, M., Banin, E., Aguilar, E.A., Dorrell, M.I., Moreno, S.K., Friedlander, M. Myeloid progenitors differentiate into microglia and promote vascular repair in a model of ischemic retinopathy. J. Clin. Invest., in press. Ritter, M., Friedlander, M. Integrins in ocular angiogenesis. In: Ocular Angiogenesis: Diseases, Mechanisms, and Therapeutics. Tobran-Tink, J., Barnstable, C. (Eds.). Humana Press. Totowa, NJ, 2006, p. 279. Ritter, M., Reinisch, J., Friedlander, S.F., Friedlander, M. Myeloid cells in infantile hemangioma. Am. J. Pathol. 168:621, 2006. Nucleocytoplasmic Transport and Role of the Nuclear Lamina in Higher Level Nuclear Organization L. Gerace, G. Ambrus-Aikelin, A. Aschrafi, J. Bednenko, A. Bubeck, A. Cassany, B. Chen, E. Choi, K. Datta, T. Guan, M. Huber, K. Kanelakis he nuclear envelope is a specialized domain of the endoplasmic reticulum that forms the boundary of the nucleus in eukaryotic cells. The envelope consists of inner and outer nuclear membranes, the nuclear lamina, and nuclear pore complexes (NPCs). The nuclear lamina, a protein meshwork lining the inner nuclear membrane, provides a structural scaffold for the nuclear envelope and an anchoring site at the nuclear periphery for chromatin. NPCs are large supramolecular assemblies that span the nuclear envelope and serve as channels for molecular transport between the nucleus and the cytoplasm. We are using a combination of biochemical, structural, and functional approaches to investigate NPCs and the lamina. T CELL BIOLOGY 2006 NUCLEOCYTOPLASMIC TRANSPORT MECHANISMS Transport of protein and RNA through NPCs is an energy-dependent process mediated by nucleocytoplasmic shuttling receptors of the karyopherin β family. Karyopherins bind to transport signals on protein or RNA cargo molecules, and the receptor-cargo complexes are translocated through the NPC by receptor binding to a group of NPC proteins (nucleoporins) that contain phenylalanine-glycine amino acid motifs. The directionality of nuclear transport is determined largely by the small GTPase Ran, which directly interacts with karyopherins and thereby regulates cargo binding. Conformational flexibility of karyopherins is thought to be fundamental to their dynamic interactions with cargo, Ran, and nucleoporins. We are using in vitro assays with digitonin-permeabilized cells to analyze the molecular events that specify translocation of cargo-receptor complexes through NPCs. Recently, using site-directed mutagenesis of importin β, the prototypical nuclear import receptor, we characterized 2 distinct binding sites in importin β for nucleoporins containing the phenylalanine-glycine motif and defined mutational hot spots for cargo binding. A major goal is to determine how the conformational dynamics of importin β are linked to discrete transport steps. To this end, we are complementing structure-function studies with analysis involving small-molecule inhibitors. In a related project, we are analyzing nuclear import of the adenovirus genome, which consists of a 36-kb double-stranded DNA molecule. Results from our in vitro transport studies indicate that adenovirus DNA transport is driven by import signals on DNA-associated proteins. Our characterization of multiple import signals in adenovirus protein VII and the tight association of the protein with the genome suggest that this viral protein may be the protein adaptor involved in the DNA import. Nuclear import of protein VII involves several of the major cellular importins, suggesting that adenovirus has evolved to use redundant import pathways to ensure efficient nuclear delivery of its genome. We also are analyzing nuclear export of HIV type 1 mRNA mediated by the viral regulatory protein Rev. Rev polymerizes on a cis-acting sequence of viral mRNAs, providing a platform for assembly of nuclear export factors. We are using proteomics combined with a permeabilized cell assay for Rev-dependent HIV mRNA export to functionally characterize the proteins assembled on the Rev platform. This project is part of a larger collaboration with a research team at Scripps Research to iden- THE SCRIPPS RESEARCH INSTITUTE 35 tify small-molecule inhibitors of Rev transport and function; the goal is to find compounds for developing new drugs to inhibit HIV replication in humans. NUCLEAR LAMINA AND HIGHER LEVEL NUCLEAR O R G A N I Z AT I O N The nuclear lamina in vertebrates contains a polymer of 2–4 related intermediate filament proteins called lamins, which are associated with a number of transmembrane proteins of the inner nuclear membrane. The lamina plays essential roles in nuclear structure and functions, as indicated by the recent findings that more than 15 inherited diseases in humans, including several muscular dystrophies, are caused by mutations in lamins or lamina-associated transmembrane proteins. The involvement of the lamina in disease is thought to be linked to its roles in nuclear integrity, cell signaling, and gene expression. Until recently, only about 12 transmembrane proteins specific to the nuclear envelope had been identified. To determine the full complement of proteins in the nuclear envelope, we carried out a proteomics analysis of the nuclear envelope of rodent liver cells in collaboration with J.R. Yates, Department of Cell Biology. We identified 67 novel putative nuclear envelope transmembrane proteins. Almost all members of this group that we have examined are authentic components of the nuclear envelope. Currently, we are analyzing nuclear envelope transmembrane proteins in muscle, because this is the tissue most sensitive to disruption of lamina function by disease-causing mutations. Using transcriptional profiling of cultured myoblasts, we found that the genes for 6 of the nuclear envelope transmembrane proteins are strongly upregulated in myoblast differentiation. The genes also are highly expressed in muscle in adults, consistent with a role of the genes in muscle differentiation and/or maintenance. We have confirmed that these nuclear envelope transmembrane proteins are authentic nuclear envelope proteins; we are using gene silencing approaches to analyze their requirement in muscle cell function. Our goals are to identify novel genes that may have a role in human muscular dystrophies and to further elucidate how the protein network consisting of lamins and associated transmembrane proteins directs nuclear structure and functions. PUBLICATIONS Ospina, J.K., Gonsalvez, G.B., Bednenko, J., Darzynkiewicz, E., Gerace, L., Matera, A.G. Cross-talk between snurportin1 subdomains. Mol. Biol. Cell 16:4660, 2005. Schirmer, E.C., Gerace, L. The nuclear membrane proteome: extending the envelope. Trends Biochem. Sci. 30:551, 2005. 36 CELL BIOLOGY 2006 Wodrich, H., Cassany, A., D’Angelo, M.A., Guan, T., Nemerow, G., Gerace, L. Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. J. Virol. 80:9608, 2006. Organization and Function of the Neuronal Cytoskeleton S. Halpain, J. Braga, B. Calabrese, L. Dehmelt, E. Hwang, J. Koehler, K. Spencer uring the past year, we made significant progress in research on the development and regeneration of neurons. In 2 main projects, we focused on cytoskeletal proteins of nerve cells, key proteins that underlie the structure and morphologic flexibility required by neurons for transmitting, storing, and processing synaptic signals. We used biochemical, molecular biological, and microscopy-based approaches to understand the function of these molecules. Fluorescence time-lapse imaging of living neurons is an important tool that we used to uncover structure-function relationships for cytoskeletal proteins and the consequences of the dysfunction of the proteins. The results of these projects have contributed to our understanding of molecular events in normal brain development and in regeneration of neuronal structure after injury and disease. D M I C R O T U B U L E - A S S O C I AT E D P R O T E I N S One project concerns microtubule-associated proteins (MAPs). These proteins are important in regulating the assembly and stability of microtubules and the interactions of microtubules with other components of the cytoskeleton. We found that one microtubule-binding protein, MAP2, also directly binds actin filaments and induces filament bundling. Using fluorescence-based time-lapse imaging and high-resolution confocal microscopy, we tracked the behaviors of microtubules and actin filaments in living neuronal cells with normal and mutant forms of MAP2. Recently, we found that the microtubule-based molecular motor dynein plays a key role in transporting microtubules toward the cell periphery. This dynein-dependent activity provides a key force that pushes the cell membrane outward during neurite initiation. Currently, we are using proteomic approaches and high-content, microscopy-based screening technology to identify other cytoskeletal proteins and signal transduction pathways crucial to the initiation of neurites. DENDRITIC SPINES A second project concerns the regulation of dendritic spines, specialized cellular protrusions that form THE SCRIPPS RESEARCH INSTITUTE the receptive, postsynaptic element at glutamate synapses. Spines become lost or dysmorphic in many types of mental retardation and in psychiatric conditions such as chronic depression and schizophrenia. Furthermore, spines are vulnerable to injury in diseases such as stroke and epilepsy, in which excessive release of glutamate can induce neuronal injury and subsequent cell death (a condition termed excitotoxicity). Understanding how spines form, what regulates their stability, and how they recover from injury is therefore of therapeutic interest for several neurologic conditions. Our most recent results suggest a neuroprotective role for spines, because preventing the collapse of dendritic spines attenuates neuronal cell death induced by a subsequent lethal stimulus. The spine cytoskeleton is composed mainly of actin filaments. We discovered that actin filaments in spines are rapidly broken down within minutes of an injury-inducing stimulus. However, this damage to the spine can be rapidly reversed within minutes under appropriate conditions, indicating for the first time that spines can regrow after they collapse. We also discovered that the membrane-associated protein myristoylated alanine-rich C kinase substrate (MARCKS), a major substrate of the signaling enzyme protein kinase C, is a key molecule in the regulation of spine shape and stability. Alterations in MARCKS are implicated in Alzheimer’s disease and in major depression. We have extensively characterized the effects of either depleting MARCKS or overexpressing various mutant forms of MARCKS in hippocampal neurons. The results revealed several key insights into MARCKS function and novel forms of synaptic plasticity in young neurons. PUBLICATIONS Calabrese, B., Wilson, M.S., Halpain, S. Development and regulation of dendritic spine synapses. Physiology (Bethesda) 21:38 2006. Halpain, S., Dehmelt, L. MAP1 family proteins. Genome Biol., in press. Function of Nuclear Receptors in Stress and Mitochondrial Homeostasis A. Kralli, J. Cardenas, B. Hazen, M.B. Hock, F. Jaramillo, C. Tiraby-Nguyen, J. Villena W e are interested in the molecular mechanisms that relay metabolic stress signals to a network of transcriptional regulators and the CELL BIOLOGY 2006 ensuing transcriptional outputs that mediate adaptive metabolic responses to the stress signals. In particular, we focus on the coactivators peroxisome proliferator– activated receptor γ coactivator-1α (PGC-1α) and PGC-1β and the orphan nuclear receptors of the estrogen-related receptor (ERR) subfamily, which control mitochondrial biogenesis and energy homeostasis. Our goals are to elucidate the biology of this specific transcriptional network, understand how deregulation of the network leads to disease, and ultimately identify the components of the network that are most suitable for drug intervention to counteract metabolic disease. R E G U L AT I O N O F T H E P G C - 1 / E R R N E T W O R K Levels of PGC-1α and PGC-1β change in response to signals that relay metabolic needs. The coactivators then relay such signals, via interactions with ERRs and other factors, to regulate the expression of specific target genes. We are interested in the mechanisms that regulate PGC-1s at the postranslational level via covalent modifications of or interaction with other proteins, and thereby control the properties of the PGC-1/ERR network. This past year, in collaboration with M. Stallcup, University of Southern California, we showed that PGC-1α is methylated by the protein arginine methyltransferase 1 and that this methylation increases the activity of PGC-1α. As a result, protein arginine methyltransferase 1 and PGC-1α act cooperatively to activate ERRα and to induce ERRα target genes with roles in mitochondrial biogenesis. Currently, we are investigating the molecular mechanisms by which methylation regulates PGC-1α activity. ROLE OF ERRα IN MITOCHONDRIAL FUNCTION Our previous studies suggested that the effects of PGC-1α and PGC-1β on mitochondrial biogenesis are mediated primarily by ERRα. To determine the physiologic relevance of ERRα for mitochondrial function, we are studying brown adipose tissue. This tissue is rich in mitochondria, it expresses high levels of ERRα, and its function is readily assayed in the context of the whole organism. Compared with wild-type mice, mice lacking ERRα have a decrease in the expression of genes associated with oxidative phosphorylation, lipid oxidation, and the tricarboxylic acid cycle. Morphologic analysis by electron microscopy revealed that brown adipose tissue from mice lacking ERRα has reduced mitochondrial density and increased lipid accumulation. When exposed to cold, mice lacking ERRα had impaired adaptive thermogenesis, despite normal induction of the uncoupling protein UCP-1. Adipocytes isolated from THE SCRIPPS RESEARCH INSTITUTE 37 the mice had reduced oxidative capacity, suggesting that the effect of ERRα on mitochondrial function is cell autonomous. This research establishes for the first time that ERRα is an important component of the regulatory network that supports high levels of mitochondrial biogenesis and oxidative metabolism in vivo. Interestingly, whereas ERRα is an important downstream effector of PGC-1α in the stimulation of mitochondrial biogenesis and oxidative capacity, it counteracts the ability of PGC-1α to induce gluconeogenic genes in hepatocytes. Notably, it is required for proper suppression of gluconeogenic enzymes in the liver of fed mice. Mitochondrial dysfunction has been implicated as an underlying cause of insulin resistance and type 2 diabetes. Moreover, derepression of hepatic gluconeogenesis contributes to high glucose levels in diabetes. The opposing effects of ERRα on genes important for mitochondrial oxidative capacity vs genes important in gluconeogenesis suggest that enhancing ERRα activity could have beneficial effects on glucose metabolism in patients with diabetes via 2 distinct mechanisms: increasing mitochondrial oxidative capacity in peripheral tissues and suppressing inappropriate glucose production in the liver. PUBLICATIONS Cartoni, R., Léger, B., Hock, M.B., Praz, M., Crettenand, A., Pich, S., Ziltener, J., Luthi, F., Dériaz, O., Zorzano, A., Gobelet, C., Kralli, A., Russell A.P. Mitofusins 1/2 and ERRα expression are increased in human skeletal muscle after physical exercise. J. Physiol. 567(Pt. 1):349, 2005. Herzog, B., Cardenas, J., Hall, R.K., Villena, J.A., Budge, P.J., Giguère, V., Granner, D.K., Kralli, A. Estrogen-related receptor α is a repressor of phosphoenolpyruvate carboxykinase gene transcription. J. Biol. Chem. 281:99, 2006. Teyssier, C., Ma, H., Emter, R., Kralli, A., Stallcup, M.R. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes Dev. 19:1466, 2005. Structural and Functional Proteomics P. Kuhn, J. Nieva,* E. Abola, A. Brooun, R. Bruce, C. Chen, J. Chrencik, P. Clark, S. Coon, J. Dupuy, S. Foster, J. Joseph, L. Kim, A. Kolatkar, M. Kraus, N. Lazarus, M. Leach, D. Marrinucci, E. Rayon, K. Saikatendu, V. Subramanian, A. Tang, M. Yadav * Department of Molecular and Experimental Medicine, Scripps Research uring the past 3 years, we have focused on 4 major research programs: detection of cancer cells in circulation, structural proteomics of cancer drug targets, structural and functional proteomics D 38 CELL BIOLOGY 2006 THE SCRIPPS RESEARCH INSTITUTE of the coronavirus that causes severe acute respiratory syndrome (SARS-CoV), and development of novel approaches in miniaturization, integration, and automation to lower the overall cost of moving from the synthesis of genes to examination of the structures encoded by the genes. These programs involve collaborations with R.C. Stevens, Department of Molecular Biology; other researchers at Scripps Research; and scientists at the Palo Alto Research Center and Lyncean Technologies in Palo Alto, California, the Novartis Institutes for Biomedical Research, Cambridge, Massachusetts, and deCODE biostructures, Bainbridge Island, Washington. D E T E C T I N G R A R E C E L L S I N T H E C I R C U L AT I O N Many clinically important cells in blood occur at frequencies of less than 1 cell per 1 million cells. Detecting and characterizing these rare cells requires the development of new technology that operates with exceptional specificity. Scientists at the Scripps-PARC Institute for Advanced Biomedical Sciences have developed an instrument, based on fiber-optic array scanning technology (FAST), that provides rapid and accurate identification of rare cells in the circulation in humans. Potential clinical applications include finding circulating tumor cells, circulating endothelial cells, and circulating fetal cells in the maternal circulation. Malignant cells from solid tumors begin to circulate at the earliest stages in cancer formation. Because the circulating cells are quite rare, technology to detect and characterize them can be valuable in screening for cancer and in guiding individualized cancer therapy. During the past year, we used FAST to accurately enumerate circulating cancer cells in blood samples obtained from patients with metastatic breast or lung cancer (Fig. 1). In collaboration with J. Kroener, Scripps Clinic, we found that accurate detection of circulating tumor cells by FAST can be used to predict prognosis in patients with metastatic breast cancer. We are developing molecular tools to characterize these rare tumor cells after they have been detected. Our goal is to determine their tissue of origin, their potential anatomic destination, and their metastatic potential. STRUCTURAL PROTEOMICS AND DRUG DISCOVERY We are collaborating with the Novartis Institutes for Biomedical Research, Cambridge, Massachusetts, in a project to understand and modulate therapeutically relevant protein-protein interactions. Analysis of an initial subset of 5 therapeutically relevant protein-protein interaction pairs will provide the guiding principles for the selection of a feasible set of drug targets to be examined. We used a high-throughput approach to F i g . 1 . An image of a single circulating tumor cell from a patient with breast cancer identified from a background of 50 million peripheral blood mononuclear cells by using a FAST cytometer. rapidly identify ideal constructs for expression, crystallization, and biophysical studies. The results are providing general insights into the range of different physical interactions for a given protein-protein interaction. The Eph-ephrin interaction is particularly interesting, because it has been implicated in cancer progression and in pathologic forms of angiogenesis. We have solved the crystal structure of the ligand-binding domain of EphB4 in complex with an antagonistic peptide that inhibits ephrin binding and has antitumorigenic properties in vivo. We have also solved the cocrystal structure of an EphB4–ephrin-B2 complex. Structural and biophysical analysis are providing the first insights into how we can modulate pathways involved in tumorigenesis and angiogenesis that rely on EphB4–ephrin-B2 signaling.These results will deepen our understanding of the basic biology behind protein-protein interactions and aid in the development of novel therapeutic approaches for modulating protein-protein interactions. STRUCTURAL AND FUNCTIONAL PROTEOMICS A N A LY S I S O F S A R S - C o V We are generating a structure-function-interaction map of the SARS-CoV proteome and its interactions with the host cell to provide a comprehensive set of targets for rational, structure-based drug and vaccine design. We use bioinformatics, structural biology, genetic methods, and functional assays. CELL BIOLOGY 2006 So far, we have determined the structures of 7 SARS-CoV proteins. We used crystallography for 3 nonstructural proteins, nuclear magnetic resonance for 3 other nonstructural proteins, and electron cryomicroscopy for a large surface glycoprotein spike. These studies are providing important information on the formation of the replicase complex, transcription, and RNA-processing events unique to the SARS-CoV life cycle. A total of 7 other proteins have been successfully expressed in soluble, folded forms, and their structures are being determined. Together, the results from these studies should enable identification of compounds that may be effective agents for treatment of infections caused by SARS-CoV. A C C E L E R AT E D T E C H N O L O G I E S C E N T E R F O R G E N E TO 3D STRUCTURE Scientists at the Accelerated Technologies Center for Gene to 3D Structure are simultaneously developing, operating, and deploying 3 key technologies to improve the costs of using x-ray crystallography to determine the structure of experimental proteins. The first technology, computer-aided design of expressionoptimized synthetic genes and protein constructs for crystallography, improves the success rate for gene isolation and allows researchers to engineer the gene sequence of interest to be optimized for protein production in a desired heterologous expression system. The second technology, microfluidic plug-based nanovolume protein crystallization in microcapillaries for in situ xray screening and data collection, is economical and greatly broadens the range of useful quantities of proteins required for crystal growth. This technology also allows for fine control over chemical gradients in crystal growth, thereby expanding the coverage of crystallization space without consuming large quantities of protein. The third technology, the compact light source, is a tunable laboratory x-ray source with peak intensity at x-ray wavelengths that span selenium anomalous absorbance. Having a tunable laboratory x-ray source in the same facility where a crystal inventory is held will greatly enhance the ability to efficiently solve new protein crystal structures. The future integration of such technologies in a single facility at Scripps Research will enable efficient gene design for improved protein production, small-volume crystallization with in situ x-ray diffraction screening, and tunable x-ray data collection in a single laboratory. Our target focus is the structural elucidation of human proteins of biomedical relevance (or eukaryotic homo- THE SCRIPPS RESEARCH INSTITUTE 39 logs thereof) that belong to superfamilies of integral membrane proteins and transcription factors. PUBLICATIONS Chrencik, J.E., Brooun, A., Kraus, M.L., Recht, M.I., Kolatkar, A.R., Han, G.W., Seifert, J.M., Widmer, H., Auer, M., Kuhn, P. Structural and biophysical characterization of the EphB4–ephrinB2 protein-protein interaction and receptor specificity. J. Biol. Chem. 281:28185, 2006. Chrencik, J.E., Brooun, A., Recht, M.I., Kraus, M.L., Koolpe, M., Kolatkar, A.R., Bruce, R.H., Martiny-Baron, G., Widmer, H., Pasquale, E.B., Kuhn, P. Structure and thermodynamic characterization of the EphB4/ephrin-B2 antagonist peptide complex reveals the determinants for receptor specificity. Structure 14:321, 2006. Hsieh, H.B., Marrinucci, D., Bethel, K., Curry, D.N., Humphrey, M., Krivacic, R.T., Kroener, J., Kroener, L., Ladanyi, A., Lazarus, N., Kuhn, P., Bruce, R.H., Nieva, J. High speed detection of circulating tumor cells. Biosens. Bioelectron. 21:1893, 2006. Joseph, J.S., Saikatendu, K.S., Subramanian, V., Neuman, B.W., Brooun, A., Griffith, M., Moy, K., Yadav, M.K., Velasquez, J., Buchmeier, M.J., Stevens, R.C., Kuhn, P. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J. Virol. 80:7894, 2006. Marrinucci, D.C., Bethel, K., Bruce, R.H., Curry, D.N., Hsieh, B., Humphrey, M., Krivacic, R.T., Kroener, J., Kroener, L., Ladanyi, A., Lazarus, N.H., Nieva, J., Kuhn, P. Case study of the morphologic variation of circulating tumor cells. Hum. Pathol., in press. Neuman, B.W., Stein, D.A., Kroeker, A.D., Chruchill, M.J., Kim, A.M., Kuhn, P., Dawson, P., Moulton, H.M., Bestwick, R.K., Iverson, P.L., Buchmeier, M.J. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 79:9665, 2005. Peti, W., Johnson, M.A., Herrmann, T., Neuman, B.W., Buchmeier, M.J., Nelson, M., Joseph, J., Page, R., Stevens, R.C., Kuhn, P., Wüthrich, K. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J. Virol. 79:12905, 2005. Ratia, K., Saikatendu, K.S., Santarsiero, B.D., Barretto, N., Baker, S.C., Stevens, R.C., Mesecar, A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 103:5717, 2006. Saikatendu, K.S., Joseph, J.S., Subramanian, V., Clayton, T., Griffith, M., Moy, K., Velasquez, J., Neuman, B.W., Buchmeier, M.J., Stevens, R.C., Kuhn, P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1′′-phosphate dephosphorylation by a conserved domain of nsP3. Structure 13:1665, 2005. Yadav, M.K., Gerdts, C.J., Sanishvili, R., Smith, W.W., Roach L.S., Roach, R.F., Ismagilov, F., Kuhn, P., Stevens, R.C. In situ data collection and structure refinement from microcapillary protein crystallization. J. Appl. Crystallogr. 38:900, 2005. Vascular Imaging and Tumor Targeting With Virus-Based Nanoparticles M. Manchester, G. Destito, M. Estrada, M.J. Gonzalez, K. Koudelka, E. Powell, C. Rae, P. Singh, D. Thomas urrent treatment of cancer typically involves chemotherapies that have severe adverse effects. The requirement that patients must withstand the toxic effects of treatment often limits the effectiveness of the therapy. Further, many promising anticancer C 40 CELL BIOLOGY 2006 compounds that are highly effective in vitro are too toxic to be used in vivo. The ability to specifically target therapies to the site of a developing tumor while avoiding healthy tissue is an important goal for cancer research. Similarly, a tremendous need exists to identify, image, and monitor tumors, particularly at early stages and during treatment. Recently, “smart” nanoparticles, which combine these multiple targeting, imaging, and drug delivery functions, have been developed. Therapies based on nanoparticles have tremendous potential to increase the sensitivity and specificity of diagnostic imaging and treatment. Many different classes of nanoparticles are currently in development, including dendrimers, liposomes, paramagnetic nanoparticles, and quantum dots. We focus on virus-based nanoparticles as platforms for the development of tissue-specific targeting and imaging agents in vivo. Two of the viruses we study are cowpea mosaic virus (CPMV) and canine parvovirus. CPMV AS A NOVEL BIOMOLECULAR SENSOR FOR VA S C U L A R I M A G I N G CPMV is an icosahedral, 31-nm particle that is produced easily and inexpensively in black-eyed pea plants. In contrast to the structure of most other nanomaterials, the structure of the CPMV capsid is defined and can be engineered to display peptides or proteins in controlled orientations on particle surfaces via either genetic manipulation of the viral genome or by chemical attachment to the particle surface. CPMV is bioavailable and nontoxic, and the capsids are highly stable to temperature, pH, and the conditions required for chemical reactions. By conjugation to surface lysine residues, CPMV can be labeled with fluorophores at high densities, resulting in an extremely bright, nontoxic material that is an outstanding tool for imaging vasculature in live animals. Working with H. Stuhlmann, Department of Cell Biology, we showed that CPMV can be used to effectively image the complete vasculature in the embryos of several species and that it is superior to other imaging particles such as lectins, fluorescent dextrans, or polystyrene microspheres. CPMV particles have also been highly useful in highlighting angiogenesis in developing tumors. Uptake of particles into endothelial cells occurs, yielding a bright imaging signal that can be used to differentiate between arterial and venous vessels. Such endothelial uptake is mediated by a cellular membrane protein, and uptake can be observed in the endothelium of a variety of THE SCRIPPS RESEARCH INSTITUTE healthy tissues as well as in disease states such as atherosclerosis. T U M O R TA R G E T I N G W I T H V I R U S - B A S E D NANOPAR TICLES We have designed virus-based nanoparticles that can specifically target tumors in vivo. In collaboration with M.G. Finn, Department of Chemistry, we bioconjugated CPMV to tumor ligands such as transferrin and folic acid, whose receptors are upregulated on metabolically active tumor cells. The targeted particles had a high degree of specificity for the tumor ligand and for uptake by tumor cells. In a separate study, we showed that canine parvovirus, which has a natural affinity for the transferrin receptor and thus for tumor cells, could specifically deliver small molecules to tumor cells. These studies will allow the further design of antitumor agents that can provide localized, highly specific imaging and therapy in vivo. Use of virus-based nanoparticles may help us visualize and eliminate small tumors before the tumors have a chance to metastasize. In addition, the ability of the particles to focus toxic effects to the site of the malignant cells, thereby expanding the range of effective therapies that can be used in vivo, holds great promise for reducing cancer-related morbidity and mortality. PUBLICATIONS Hsu, C., Singh, P., Ochoa, W., Manayani, D.J., Manchester, M., Schneemann, A., Reddy, V.S. Characterization of polymorphism displayed by the coat protein mutants of tomato bushy stunt virus. Virology 349:222, 2006. Lewis, J.D., Destito, G., Zijlstra, A., Gonzalez, M.J., Quigley, J.P., Manchester, M., Stuhlmann, H. Viral nanoparticles as tools for intravital vascular imaging. Nat. Med. 12:354, 2006. Manchester, M., Singh, P. Virus-based nanoparticles (VNPs): platform technologies for diagnostic imaging. Adv. Drug Dev. Rev., in press. Rae, C.S., Khor, I.W., Wang, Q., Destito, G., Gonzalez, M.J., Singh, P., Thomas, D.M., Estrada, M.N., Powell, E., Finn, M.G., Manchester, M. Systemic trafficking of plant virus nanoparticles in mice via the oral route. Virology 343:224, 2005. Scobie, H.M., Thomas, D., Marlett, J.M., Destito, G., Wigelsworth, D.J., Collier R.J., Young J.A.T., Manchester, M. A soluble receptor decoy protects rats against anthrax lethal toxin challenge. J. Infect. Dis. 192:1047, 2005. Sen Gupta, S., Kuzelka, J., Singh, P., Lewis, W.G., Manchester, M., Finn, M.G. Accelerated bioorthogonal conjugation: a practical method for the ligation of diverse functional molecules to a polyvalent virus scaffold. Bioconjug. Chem. 16:1572, 2005. Singh, P., Destito, G., Schneemann, A., Manchester, M. Canine parvovirus-like particles, a novel nanomaterial for tumor targeting. J. Nanobiotechnol. 4:2, 2006. Singh, P., Gonzalez, M.J., Manchester, M. Viruses and their uses in nanotechnology. Drug Dev. Res., in press. CELL BIOLOGY 2006 Translational Regulation in Chloroplasts and Expression of Human Monoclonal Antibodies in Eukaryotic Algae THE SCRIPPS RESEARCH INSTITUTE 41 Compared with bacterial 70S ribosomes, the chloroplast ribosome has unique structural domains located primarily on the small ribosomal subunit (Fig. 1). This S.P. Mayfield, D. Barnes, A. Manuell, M. Beligni, J. Marin-Navarro, M. Muto, M. Tran, D. Siefker, R. Henry ene expression in chloroplasts is primarily controlled during the translation of plastid mRNAs into proteins, and understanding how this process is regulated is key to understanding plant development and function. Controlling chloroplast translation is also an essential component in optimizing the production of human therapeutic proteins in algae. Using proteomics and bioinformatics analyses, we identified the set of proteins that function in chloroplast translation. These studies indicated that the core translational apparatus of chloroplasts is highly related to that of bacteria but that chloroplasts have incorporated additional protein components that allow more complex regulatory mechanisms. Some of these additional components are ribosomal proteins; others are protein translation factors. Chloroplast mRNAs also contain a number of RNA regulatory elements that are not found in bacteria, as well as conserved RNA elements, such as ribosome-binding sequences, but even these conserved elements appear to function in chloroplast translation differently than in bacterial translation. The unique components of chloroplast translation provide the opportunity for regulation of chloroplast translation, for example in response to exposure to light, that cannot be achieved in simpler bacterial systems. To better understand translation in plants, we are examining the structure of both the chloroplast and cytoplasmic ribosomes from Chlamydomonas reinhardtii, a unicellular photosynthetic eukaryote. Using electron cryomicroscopy and single-particle reconstruction, we determined the structure of the C reinhardtii cytoplasmic 80S ribosome and found that it is nearly identical to ribosomes from animals, including the human 80S ribosome. Parallel proteomics analysis supported this finding. We also determined the structure of the chloroplast ribosome to 20 Å and found that although it is conserved with bacterial 70S ribosomes, it has large unique structural domains, as predicted by our proteomics analysis. G F i g . 1 . Structure of the chloroplast ribosome from C reinhardtii calculated to 20-Å resolution. Structures identified as chloroplast unique through comparison with bacterial ribosomes are labeled. The chloroplast-unique structures are found on the path of mRNA through the ribosome, which is labeled as mRNA entrance and exit tunnels. These structures are positioned for interaction with mRNAs before and during translation; such interactions most likely are used to select and position mRNA for initiation of translation and to increase the rate and fidelity of mRNA translation. finding is supported by our proteomics results that indicated that the mass of the small (30S) subunit of the chloroplast ribosome is 25% larger than the bacterial 30S subunit. Chloroplast-unique structures are found on the solvent side of the small subunit; the large subunit interacting face is similar to that in bacterial ribosomes. The largest of the chloroplast-unique domains occurs in the vicinity of the mRNA exit tunnel but also extends up alongside the head and down across the platform. This structure has multiple lobes and possibly spans the entire solvent-exposed face of the chloroplast small ribosomal subunit, connecting with a small region of chloroplast-unique density below the shoulder. Another distinct region of chloroplast-unique density is located in the beak region of the ribosome, near the mRNA entrance tunnel. These chloroplast-unique ribosomal structures are poised to interact with chloroplast mRNAs early in message recognition, a key point for translational regulation. These studies have revealed the structural basis from which we can pursue identification of the molecular and biochemical interactions of mRNAs, translation factors, and the chloroplast ribosome that result in regulated translation of chloroplast mRNAs. 42 CELL BIOLOGY 2006 In addition to these basic studies on translation, we have developed a system for the expression of recombinant proteins, including human therapeutic proteins, in C reinhardtii chloroplasts. We have expressed a number of mammalian proteins, including human monoclonal antibodies. We recently expressed 83K7C, a human monoclonal antibody that binds to protective antigen 83 of anthrax toxin. We showed that 83K7C assembles in the cell to form functional antibodies that bind the antigen and thus potentially could block the toxic effects of Bacillus anthracis during infection. We also showed that this algal-based system can be used to produce high levels of a number of other proteins with potential human therapeutic value. These studies indicate that eukaryotic algae have tremendous potential for the expression of recombinant human therapeutic proteins, because algae can be grown economically at large scale. Our continued genetic, biochemical, and structural studies should lead to a greater understanding of the mechanism of chloroplast translation and enable us to design appropriate transgenes to achieve higher levels of expression of therapeutic proteins. PUBLICATIONS Barnes, D., Franklin, S., Schultz, J., Henry, R., Brown, E., Coragliotti, A., Mayfield, S.P. Contribution of 5′- and 3′-untranslated regions of plastid mRNAs to the expression of Chlamydomonas reinhardtii chloroplast genes. Mol. Genet. Genomics 274:625, 2005. Fletcher, S.P., Muto, M., Mayfield, S.P. Optimization of recombinant protein expression in the chloroplast of green algae. In: Transgenic Microalgae as Green Cell Factories. León, R., Gaván, A., Fernández, E. (Eds.). Landes Bioscience, Austin, TX, in press. Manuell, A.L., Mayfield, S.P. A bright future for Chlamydomonas. Genome Biol. 7:327, 2006. THE SCRIPPS RESEARCH INSTITUTE childhood forms of mental retardation to psychiatric disorders such as schizophrenia with onsets in late adolescence and early adulthood to diseases of aging such as Alzheimer’s disease. We use genetic manipulation in mice to investigate the molecular events involved in learning and memory. CALCIUM SIGNALING AND MEMORY We know relatively little at a molecular level about how the brain stores new information. One hypothesis, which we tested, is that calcium-regulated changes in the strength of synaptic connections between nerve cells can store information. The enzyme calcium/calmodulin-dependent protein kinase is abundant at synapses and when activated by calcium can strengthen synaptic connections. We used genetic manipulations in mice to indiscriminately activate this kinase at all synapses in the entorhinal cortex, a part of the brain that is important for memory and is affected in the earliest stages of Alzheimer’s disease in humans. We found not only that the formation of new memories is impaired but also that previously established memories could be erased. If memories are stored as precise patterns of synaptic weights, then the indiscriminate strengthening of synapses might be expected to erase memories in a manner similar to the way writing all 1’s in computer memory will erase previously stored information. We also examined where calcium/calmodulin-dependent protein kinase functions within cells. We found that the synthesis of this kinase from RNA located specifically at synapses is necessary for the stabilization of memories that last several months. GENETIC MODELS OF DISEASE Manuell, A.L., Yamaguchi, K., Haynes, P.A., Milligan, R.A., Mayfield, S.P. Composition and structure of the 80S ribosome from the green alga Chlamydomonas reinhardtii: 80S ribosomes are conserved in plants and animals. J. Mol. Biol. 351:266, 2005. Molecular Basis of Cognitive Function and Dysfunction M. Mayford, E. Korzus, G.J. Reijmers, M. Yasuda, R. Yasuda, S. Miller, N. Matsuo he ability to remember is perhaps the most significant and distinctive feature of our cognitive life. We are who we are in large part because of what we have learned and what we remember. Impairments in learning and memory are a component of disorders that affect human beings throughout life, from T The determination of the complete sequences of the mouse and human genomes indicates that humans are highly similar to mice at the genetic level. One approach for investigating a genetic disease in humans is to introduce the same mutations into mice to produce a model of the disease for better understanding of the molecular pathology and for testing possible treatments. Rubenstein-Taybi syndrome is a developmental and cognitive disorder that results from mutation in the gene CBP. We produced a strain of mice with a defect in CBP and found that the mice were impaired in several learning and memory tasks. More important, we showed that these impairments were not due to problems in development of the brain because they could be reversed by providing a normally functioning CBP gene to adult mice. The protein encoded by CBP chemically modifies histones to allow the expression of a large variety of other CELL BIOLOGY 2006 genes. We found that the memory deficits in the mice could be reversed by treatment with a drug that targets this histone-modifying function, suggesting a possible treatment for Rubenstein-Taybi syndrome and possibly other cognitive disorders. M O L E C U L A R A N AT O M Y O F M E M O R Y When humans learn new information, they use only a tiny fraction of the neurons in the brain. One of the difficulties in studying memory is an inability to identify and specifically manipulate those neurons that participate in a particular memory trace. We developed a genetic technique for use in mice that enables us to specifically introduce genetic changes into neurons that are activated by behavioral stimuli. We are using this approach to introduce marker proteins that enable us to see the connections between neurons that have been activated during learning. We have used this approach to study extinction, a process used in the treatment of phobias by which memories are weakened by repeated exposure to a relevant stimulus. We found that the neurons originally activated by a fearful stimulus were no longer activated after extinction. This finding suggests that extinction training actually erases or interferes with some component of the original memory trace. PUBLICATIONS Colvis, C.M., Pollock, J.D., Goodman, R.H., Impey, S., Dunn, J., Mandel, G., Champagne, F.A., Mayford, M., Korzus, E., Kumar, A., Renthal, W., Theobald, D.E., Nestler, E.J. Epigenetic mechanisms and gene networks in the nervous system. J. Neurosci. 25:10379, 2005. Reijmers, L.G., Coats, J.K., Pletcher, M.T., Wiltshire, T., Tarantino, L.M., Mayford, M. A mutant mouse with a highly specific contextual fear-conditioning deficit found in an N-ethyl-N-nitrosourea (ENU) mutagenesis screen. Learn. Mem. 13:143, 2006. Yasuda, M., Mayford, M.R. CaMKII activation in the entorhinal cortex disrupts previously encoded spatial memory. Neuron 50:309, 2006. Regulation of the Plasminogen Activation System L.A. Miles, A. Baik, J.W. Mitchell, H. Bai, F.J. Castellino,* R.J. Parmer** * University of Notre Dame, Notre Dame, Indiana ** University of California, San Diego, California ssembly of plasminogen and plasminogen activators on cell surfaces is a key control point for positive regulation of cell-surface proteolytic activity necessary in physiologic and pathologic processes. Plasminogen-binding sites are markedly upreg- A THE SCRIPPS RESEARCH INSTITUTE 43 ulated when monocytoid cells undergo apoptosis. Therefore, we are investigating the ability of plasminogen to modulate monocyte apoptosis. We cultured monocytoid cells (freshly isolated human monocytes and U937 cells) in plasminogen-deficient serum and induced the cells to undergo apoptosis by using either TNF-α or cycloheximide. When induced in the presence of increasing concentrations of plasminogen, apoptosis was inhibited in a dose-dependent manner; full inhibition occurred at a concentration of plasminogen equal to its normal physiologic concentration. Treatment with plasminogen also markedly reduced intranucleosomal DNA fragmentation and the active caspase-3, caspase-8, and caspase-9 induced by TNF-α or by cycloheximide. Because monocytoid cells synthesize plasminogen activators, we examined the role of plasmin proteolytic activity in the antiapoptotic effects of plasminogen. A plasminogen active-site mutant did not recapitulate the cytoprotective effect of wild-type plasminogen. In addition, the antiapoptotic activity of plasminogen was blocked by increasing concentrations of α2-antiplasmin, with full reversal at a 2-µM concentration of α2-antiplasmin, suggesting that the cytoprotective effect of plasminogen requires activation of plasminogen to plasmin. Furthermore, antibodies against protease-activated receptor 1 blocked the antiapoptotic effects of plasminogen. Our results suggest that plasminogen protects monocytic cells from apoptosis via a mechanism that requires the proteolytic activity of plasmin and that the protection is mediated by protease-activated receptor 1. Because monocyte apoptosis regulates inflammation and atherosclerosis, these results provide insight into a novel role for plasminogen in these processes. An emerging area of research has indicated a novel role for the plasminogen activation system in regulating the release of neurotransmitters. Prohormones, secreted by cells within the sympathoadrenal system, are processed by plasmin to bioactive peptides that mediate feedback inhibition of secretagogue-stimulated release of neurotransmitters. Catecholaminergic cells of the sympathoadrenal system are prototypic prohormonesecreting cells. Processing of prohormones by plasmin is enhanced in the presence of catecholaminergic cells, and the enhancement requires binding of plasmin(ogen) to cellular receptors. Consequently, modulation of the local cellular fibrinolytic system of catecholaminergic cells results in substantial changes in catecholamine release. However, mechanisms for enhancing prohor- 44 CELL BIOLOGY 2006 mone processing and cell-surface molecules that mediate the enhancement on catecholaminergic cells have not been investigated. We found that plasminogen activation was enhanced more than 6.5-fold on catecholaminergic cells. Treatment with carboxypeptidase B decreased cell-dependent plasminogen activation by almost 90%, suggesting that the binding of plasminogen to proteins exposing C-terminal lysines on the cell surface is required to promote plasminogen activation. Using a novel strategy of targeted specific proteolysis with carboxypeptidase B combined with a proteomics approach involving 2-dimensional gel electrophoresis, radioligand blotting, and tandem mass spectrometry, we identified catecholaminergic plasminogen receptors required for enhancing plasminogen activation. Two major plasminogen-binding proteins that exposed C-terminal lysines on the cell surface contained amino acid sequences corresponding to β- and γ-actin. A monoclonal antibody to actin inhibited cell-dependent plasminogen activation and also enhanced nicotinedependent catecholamine release. Our results suggest that forms of actin expressed on the cell surface bind plasminogen, thereby promoting plasminogen activation and increased prohormone processing leading to inhibition of neurotransmitter release. PUBLICATIONS Mitchell, J.W., Baik, N., Castellino, F.J., Miles, L.A. Plasminogen inhibits TNFα apoptosis in monocytes. Blood 107:4383, 2006. Structure and Action of Molecular Machines R.A. Milligan, J. Chappie, P. Chowdhury, R. Coleman, T. Dang, S. Falke,* E. Gogol,** M.B. Lee, S. Mulligan, M. Reedy,*** M.K. Reedy,*** A.B. Ward, E.M. Wilson-Kubalek, C. Yoshioka * William Jewel College, Liberty, Missouri ** University of Missouri, Kansas City, Missouri *** Duke University Medical Center, Durham, North Carolina acromolecular assemblies may be composed of from 2 to perhaps scores of proteins and are the functional units—the molecular machines—of the cell. We use electron cryomicroscopy and image analysis to study the structure and mechanism of action of several of these machines. We combine the 3-dimensional maps calculated from electron images of the machines with biochemical data and high-resolution x-ray structures of the individual components to pro- M THE SCRIPPS RESEARCH INSTITUTE vide insight into the operation of the machines. During the past year, we continued our work on members of the myosin and kinesin superfamilies, microtubule-stabilizing proteins, and membrane proteins. Although the mechanism of plus end–directed, processive motion by conventional kinesins is now well understood, the mechanism by which members of the kinesin 14 class move toward the minus ends of microtubules is not. Likewise, in the myosin superfamily, how nucleotide-mediated conformational changes in the motor domain of class VI myosins result in “backward” motility is not known. We are elucidating the molecular mechanisms of these more unusual members of the myosin and kinesin superfamilies. (Movies showing the motions of conventional myosin and kinesin can be viewed at www.scripps.edu/milligan/projects.html.) The kinesin Ncd belongs to the kinesin 14 class of motor proteins. Compared with the situation with plus end–directed kinesins, the nature and timing of the structural changes that underlie the motility of kinesin 14 motors are poorly understood. We used electron cryomicroscopy and image analysis to calculate 3-dimensional maps of Ncd bound to microtubules in various stages in its mechanochemical cycle. The maps revealed a minus end–directed rotation of approximately 70° of a coiled coil mechanical element of microtubule-bound Ncd upon ATP binding. In parallel with these structural studies, our collaborators, N. Endres and R. Vale at the University of California, San Francisco, showed that extending or shortening this mechanical element respectively increases or decreases movement velocity, without affecting ATPase activity. These results indicate that as with other kinesins, the force-producing conformational change of Ncd occurs upon ATP binding but, unlike the situation with other kinesins, involves the swing of a rigid, lever arm–like mechanical element similar to that described for myosins. Whereas most kinesins move along intact microtubules, members of the kinesin 13 class, such as KinI, destabilize and depolymerize microtubules and do not appear to have motile properties. We found that a KinI fragment consisting of only the conserved motor core is necessary and sufficient for ATP-dependent depolymerization. The motor core binds along microtubules in all nucleotide states, but in the presence of a nonhydrolyzable ATP analog, depolymerization also occurs. Structural characterization of the analoginduced depolymerization products provided a snapshot of the disassembly machine at the microtubule ends. CELL BIOLOGY 2006 Our data indicate that whereas conventional kinesins use the energy of ATP binding to execute a power stroke that results in unidirectional motion along the microtubule surface, KinIs at the ends of microtubules use the energy to bend the underlying protofilament, thereby destabilizing the microtubule lattice and leading to microtubule depolymerization. Furthermore, when the motor core is associated with the microtubule wall, the core is stalled in a weakly bound, nucleotide-free state. Progression to the strongly bound, ATP-containing state is possible only when the KinI encounters a microtubule end, where it can catalyze deformation of protofilaments and disassembly of microtubules. The unusual mechanochemical coupling of this kinesin provides an elegant mechanistic basis for its microtubule-depolymerizing activity. Our current research focuses on understanding the role of the second head in these dimeric molecules. The protein doublecortin is expressed in migrating and differentiating neurons. In humans, mutations in this protein disrupt brain development, causing lissencephaly. Although doublecortin is associated with and stabilizes the microtubule cytoskeleton, it has no homology with other microtubule-binding proteins such as MAP2 or tau. We found that doublecortin preferentially nucleates and binds to 13-protofilament microtubules. This specificity was explained when we discovered that the protein binds in the valleys between the protofilaments of the microtubule wall. This binding site is unique and appears to be ideally located for microtubule stabilization. In this location, doublecortin most likely contributes to both the longitudinal and the lateral interactions that stabilize the microtubule wall. We are now investigating the binding of proteins that track with the tips of dynamic microtubules. In collaboration with G. Chang, Department of Molecular Biology, we have grown well-ordered arrays of several membrane proteins that are involved in multidrug resistance. These arrays, helical tubes and 2dimensional crystals of membrane-embedded proteins, are suitable for structural studies via electron microscopy. In one instance, we trapped a drug transporter in various stages of its mechanistic cycle and with substrates bound. We anticipate that 3-dimensional electron microscopy maps of membrane-embedded transporters in various states, together with the highresolution x-ray structures of the detergent-solubilized protein, will provide insights into the mechanisms used to transport metabolites and drugs across membranes. In other studies, we developed a general method for helical crystallization of proteins on lipid tubules THE SCRIPPS RESEARCH INSTITUTE 45 that we are using to study the virulence factor perfringolysin O from Clostridium perfringens. Perfringolysin O is a cytolysin, an important class of proteins that oligomerize and embed within membranes as part of the proteins’ lytic functions. We obtained helical crystals of wild-type and several mutant forms of the cytolysin on nickel-lipid tubules. Three-dimensional maps of these proteins derived from images of the helical crystals will be used to complement our studies of pore formation by perfringolysin O on lipid layers. These studies will provide a better understanding of the pathogenic function of cytolysins. Additional studies involving tubular crystallization of membrane proteins and other bacterial toxins are opening up promising new areas for future research. PUBLICATIONS Dang, T.X., Milligan, R.A., Tweten, R.K., Wilson-Kubalek, E.M. Helical crystallization on nickel-lipid nanotubes: perfringolysin O as a model protein. J. Struct. Biol. 152:129, 2005. Endres, N.F., Yoshioka, C., Milligan, R.A., Vale, R.D. A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Nature 439:875, 2006. Manuell, A.L., Yamaguchi, K., Haynes, P.A., Milligan, R.A., Mayfield, S.P. Composition and structure of the 80S ribosome from the green alga Chlamydomonas reinhardtii: 80S ribosomes are conserved in plants and animals. J. Mol. Biol. 351:266, 2005. CNS Development and Mechanosensory Perception U. Müller, C. Barros, R. Belvindrah, F. Conti, S. Franco, N. Grillet, S. Hankel, P. Kazmierczak, R. Radakovits, C. Ramos, A. Reynolds, A. Sczaniecka, M. Schwander, S. Webb disproportionately large number of genes in the genomes of vertebrates encode cell recognition molecules that mediate cell-cell interactions and interactions between cells and the extracellular matrix. This finding most likely reflects an evolutionary trend toward increasingly more complex cellular interactions in higher metazoans. The highest diversity of such interactions occurs in the CNS, where thousands of different neuronal subtypes are connected into defined neuronal circuits. We use mouse genetics, genomics, cell biology, biochemistry, and imaging technology to analyze the function of cell recognition molecules during the development of neuronal circuits in the CNS. In another project, we are elucidating the mechanisms by which cell recognition molecules contribute to mechanosensory perception. A 46 CELL BIOLOGY 2006 F O R M AT I O N O F C O R T I C A L S T R U C T U R E S I N T H E C N S The establishment of the 3-dimensional cytoarchitecture of the nervous system depends on interactions of receptors on neuronal cells with molecules presented within the extracellular matrix and by neighboring cells. Integrins are a class of neuronal receptors that mediate interactions with glycoproteins secreted by the extracellular matrix and with membrane-anchored counterreceptors. Recently, we found that integrins cooperate with secreted signaling molecules such as sonic hedgehog and Reelin to regulate important steps during CNS development, such as cell proliferation and formation of neuronal layers during the development of the cerebral and cerebellar cortex. We are identifying the downstream signaling pathways activated by integrins during cortical development. We are also studying signaling interactions between integrins and other receptors such as receptor tyrosine kinases. Finally, we have extended our studies to the analysis of integrin functions in the CNS in adults. CELL RECOGNITION MOLECULES, MECHANOSENSORY THE SCRIPPS RESEARCH INSTITUTE component of the so called tip-link, which has been predicted to transmit force onto mechanically gated ion channels in the stereocilia of hair cells. We are analyzing the function of cadherin 23, proteins that interact with this cell adhesion molecule, and proteins encoded by additional “deafness” genes for mechanotransduction. We are also doing genetic screens in mice to identify novel recessive deafness traits. Using this strategy, we have already identified several novel genes that may be associated with deafness. PUBLICATIONS Barros, C., Müller, U. Cell adhesion in nervous system development: integrin functions in glial cells. In: Integrins and Development. Danen, E.H.J. (Ed.). Landes Bioscience, Austin, TX, 2006, p. 185. Belvindrah, R., Müller, U. Integrin signaling and central nervous system development. In: Extracellular Matrix in Development and Disease. Miner, J.H. (Ed.). Elsevier, St. Louis, 2005, p. 153. Advances in Developmental Biology and Biochemistry; Vol. 15. Belvindrah, R., Nalbant, P., Chuanyue, W., Bokoch, G.M., Müller, U. Integrinlinked kinase regulates Bergmann glial differentiation during cerebellar development. Mol. Cell. Neurosci., in press. Escher, P., Lacazette, E., Courtet, M., Blindenbacher, A., Landmann, L., Bezakova, G., Lloyd, K., Müller, U., Brenner H.R. Synapses form in skeletal muscles lacking neuregulin receptors. Science 308:1920, 2005. PERCEPTION, AND DEAFNESS Mechanosensation, the transduction of mechanical force into an electrochemical signal, allows living organisms to detect touch, hear, register movement and gravity, and sense changes in cell volume and shape. In mammals, the hair cells of the inner ear are the principle mechanosensors for the detection of sound and movement. Hair cells elaborate stereocilia that contain mechanosensitive ion channels. The stereocilia of a hair cell are interconnected by extracellular bridges into a bundle and are situated next to specialized extracellular matrix assemblies. Sound waves or head movements lead to deflection of the stereocilia bundle, changes in the ion permeability of the mechanosensitive channels, and depolarization of the hair cells. The molecules that regulate development and function of hair cells are poorly defined. Because defects in hair cells cause inherited forms of deafness, we use human and mouse genetics as a guideline to identify and study molecules that regulate the development and function of mechanosensory hair cells. Currently, about 70 genes have been identified in which mutations lead to deafness. Many of these genes encode molecules secreted into the extracellular matrix and membrane-anchored cell adhesion molecules. Mutations in the genes for the cell adhesion molecule cadherin 23 in mice and humans cause deafness. Our findings provide strong evidence that cadherin 23 is a Naylor, M.J., Li, N., Cheung, J., Lowe, E.T., Lambert, E., Marlow, R., Wang, P., Schatzmann, F., Wintermantel, T., Schuetz, G., Clarke, A.R., Müller, U., Hynes, N.E., Streuli, C.H. Ablation of β1 integrin in mammary epithelium reveals a key role for integrin in glandular morphogenesis and differentiation. J. Cell Biol. 171:717, 2005. Senften, M., Schwander, M., Kazmierczak, P., Lillo, C., Shin, J.B., Hasson, T., Geleoc, G.S.G., Gillespie, P.G., Williams, D., Holt, J.R., Müller, U. Physical and functional interaction between protocadherin 15 and myosin VIIa in mechanosensory hair cells. J. Neurosci. 26:2060, 2006. Molecular Mechanisms of Thermosensation A. Patapoutian, M. Bandell, A. Dhaka, A. Dubin, T. Earley, J. Grandl, H. Hu, L. Macpherson, T. Miyamoto, V. Uzzell e are interested in the molecular description of the function of sensory neurons. Of the 5 popularly characterized senses—sight, hearing, taste, smell, and touch—touch is among the most varied and least understood. Within this sense is the ability to sense mechanical forces, chemical stimuli, and temperature, and the molecules that mediate this ability have been a long-standing mystery. Temperature sensation in particular has received relatively little attention from biologists and yet is critical for interactions with the environment. We recently discovered proteins that may enable sensory neurons to convey information about tempera- W CELL BIOLOGY 2006 ture. These proteins are ion channels activated by specific changes in temperature; thus they act as the molecular thermometers of the body. Specifically, our results have led to the identification and characterization of 1 novel warm-activated transient-receptor potential (TRP) channel, TRPV3 (33°C threshold) and 2 novel cold-activated TRP channels, TRPM8 (25°C threshold) and TRPA1 (ANKTM1, 17°C threshold). We found that TRPM8 is also the receptor for the compound menthol, providing a molecular explanation of why mint flavors are typically perceived as cooling. Furthermore, we discovered that TRPA1 is activated by cinnamaldehyde, allicin (garlic), and other compounds with a burning sensory quality, consistent with a role of TRPA1 in the detection of noxious cold sensations. Together these temperature-activated channels represent a new subfamily of TRP channels that we have dubbed thermoTRPs. In agreement with a role in initiating temperature sensation, most of the thermoTRPs are normally found in subsets of neurons in dorsal root ganglia. A surprisingly distinct expression pattern was observed for TRPV3, the warm receptor. In mice, high levels of TRPV3 occur solely in skin keratinocytes, suggesting that skin cells might be able to “sense” temperature and then communicate this information to neurons in dorsal root ganglia. How temperature information is coded from the skin to the spinal cord is not well understood, and we are using a variety of approaches to answer this question. For example, data from mice lacking the gene for TRPV3 suggest that TRPV3 is indeed required for proper heat sensation in vivo, reinforcing a role of skin in thermosensation. All organisms have a need for thermosensation. Because some invertebrate species are more amenable to genetic studies than mammals are, we asked whether nonvertebrates also use thermoTRPs to sense temperature. We showed that the Drosophila ortholog of TRPA1 is an ion channel activated by warm temperatures, suggesting an evolutionarily conserved role of TRP channels in temperature sensing. In collaborative efforts with P. Garrity, Massachusetts Institute of Technology, Cambridge, Massachusetts, and W. Shafer, University of California, San Diego, we are using genetic studies to examine the role of TRPA family members in invertebrate species. Another key question is what makes thermoTRPs temperature sensitive whereas other TRPs are not? Answering this question requires insight into the fundamental biophysical mechanism of how temperature activates ion channels. Our ongoing structure-function experiments, including mutagenesis and chimeric protein THE SCRIPPS RESEARCH INSTITUTE 47 analysis of the thermoTRPs, will provide important clues about how cold or heat activates these ion channels. Our long-term goal is to synthesize an integrated picture of sensory neuron function. By identifying the proteins that initiate the molecular cascade leading to temperature perception, we have provided the basis for probing the foundation of the sense of temperature. We now have the opportunity to extend these insights into important areas of human health, such as pain pathophysiology. For example, TRPA1 is a potential target for treating pain, and we are identifying small-molecule inhibitors of TRPA1 in collaboration with scientists at the Genomics Institute of the Novartis Research Foundation, San Diego, California. Therefore, the approaches we are using will yield insights into the basic biology of the peripheral nervous system and may also have an effect on novel treatments for pain. PUBLICATIONS Bandell, M., Dubin, A.E., Petrus, M.J., Orth, A., Mathur, J., Hwang, S.W., Patapoutian, A. High-throughput random mutagenesis screen reveals TRPM8 residues specifically required for activation by menthol. Nat. Neurosci. 9:493, 2006. Dhaka, A., Viswanath, V., Patapoutian, A. TRP ion channels and temperature sensation. Annu. Rev. Neurosci. 29:135, 2006. Macpherson, L.J., Geierstanger, B.H., Viswanath, V., Bandell, M., Eid, S.R., Hwang, S.W., Patapoutian, A. The pungency of garlic: activation of TRPA1 and TRPV1 in response to allicin. Curr. Biol. 15:929, 2005. Functional Proteins in Tumor Metastasis and Angiogenesis J.P. Quigley, E.I. Deryugina, A. Zijlstra, J.P. Partridge, T. Kupriyanova, M. Madsen, V. Ardi, M.C. Subauste e have established a number of in vivo model systems that can recapitulate the major cellular and tissue events that occur during tumor metastasis and angiogenesis. The model systems allow quantitative measurements, microscopic analysis in real time, biochemical and immunologic probing, and direct molecular and therapeutic interventions. Recently, use of small interfering RNA molecules directed against specific expressed genes and applied directly into the models provided insights into the contributory role of the gene products in tumor dissemination and neovascularization. In addition, use of subtractive immunization, which is used to generate unique function-blocking monoclonal antibodies, in combination with immunoproteomics enables us to identify specific antigenic molecules that are functionally active in metas- W 48 CELL BIOLOGY 2006 tasis and angiogenesis. Finally, use of activity-based protein profiling, in collaboration with B.F. Cravatt, Department of Cell Biology, enables us to detect, isolate, and identify active proteolytic enzymes that are distinctively and differentially activated during metastasis and angiogenesis. M E TA S TA S I S Selected human tumor cells inoculated onto the chorioallantoic membrane of developing chick embryos form primary tumors on the membrane in 4–7 days. A small percentage of the cells in the primary tumor disseminate through the vasculature and within 3–4 days arrest and proliferate in secondary organs of the embryo. Measuring a small number of early-arriving metastatic cells (<200) growing and expanding in the secondary organ has always been technically difficult. We now use an approach in which unique regions of human DNA, known as Alu repeat sequences, are amplified by polymerase chain reaction from the total DNA extracted from various organs of the tumor-bearing chick embryo. Chicken DNA contains no Alu sequences, so any product generated by the polymerase chain reaction indicates that human tumor cells are present in the chick embryo organ and would have arrived there via the known sequential steps in metastasis. We can now detect as few as 25–50 human tumor cells present in the entire chick embryo lung, liver, or brain and can measure the expansion of these metastatic cells by using real-time polymerase chain reaction. We are using various screening procedures in this model system to identify molecules that enhance, or conversely inhibit, the appearance of metastatic human tumor cells in organs of chick embryos. The screening procedures include direct inoculation of primary tumor cells that have been transfected with various small interfering RNA constructs to silence specific genes that might contribute to metastatic dissemination. Inoculating monoclonal antibodies directly into the tumor-bearing embryo and monitoring the influence of the antibodies on metastasis are also part of our screening procedures. We are also using a more conventional method of monitoring human tumor metastasis in specific immunodeficient mice. However, compared with our chick embryo metastasis assay, this method is less quantitative, requires more time (3–5 weeks), and is more difficult to use for inhibitor screening and molecular intervention. We are using the mouse metastasis assay to take advantage of mouse genetics and to confirm the efficacy of various effector molecules and inhibitors that initially are identified in the chick embryo metastasis assay. THE SCRIPPS RESEARCH INSTITUTE ANGIOGENESIS One of the most commonly used in vivo assays for angiogenesis is the chick embryo chorioallantoic membrane assay. We developed a quantitative variation of this assay that enables us to detect and measure the newly sprouting blood vessels responding to an angiogenic stimulus such as a specific growth factor or a growing tumor. A highly specific metalloproteinase, MMP-13, has been implicated in the tissue remodeling that occurs during the formation of the new blood vessels. We characterized this specific proteolytic event and found that specific collagen-cleaving metalloproteinases are implicated directly in the outgrowth of new vessels. We also found that another metalloproteinase, MMP-9 (gelatinase B), most likely is involved in angiogenic tissue remodeling. The proteolytic activity of this enzyme also appears to be necessary for a full angiogenic response. Interestingly, these 2 critical enzymes are actively imported into the vascular/stromal tissue by distinct inflammatory cells responding to the angiogenic stimulation. Neutrophil-like heterophils rapidly and almost immediately import MMP-9 into the tissue, whereas monocyte/macrophages actively deliver MMP13 1–2 days later, possibly in response to specific secreted products of the early-arriving heterophils. Thus, normal angiogenesis and tumor angiogenesis are closely linked to an accompanying host inflammatory response that contributes critical functional molecules to the angiogenic process. We are dissecting out and identifying the specific molecules and cells that link the inflammatory response to the angiogenic process and to the progression of malignant neoplasms. We are also trying to decipher whether the relevant functional molecules are derived from host cells or tumor cells. I N T R AVA S AT I O N We are also investigating intravasation, the entry of primary tumor cells into the host vasculature, often the vasculature that is newly formed during tumor angiogenesis. Intravasation appears to be the least-studied process in the metastatic cascade but most likely is a rate-limiting step in tumor dissemination. We recently isolated 2 isogenic variants of a human fibrosarcoma cell line that differ 100-fold in their ability to enter the vasculature in vivo and in their ability to metastasize. We are using array technology, proteomics approaches, and intravital microscopy in the cellular and molecular analysis of these 2 variants. The results should indi- CELL BIOLOGY 2006 cate specific molecules that are functionally important in interactions between tumor cells and the vasculature and contribute to intravasation. Using activitybased protein profiling, we found that the proteolytic enzyme urokinase is differentially activated during intravasation and catalytically contributes to the enhanced entry of tumor cells into the vasculature. PUBLICATIONS Deryugina, E.I., Zijlstra, A., Partridge, J.J., Kupriyanova, T.A., Madsen, M.A., Papagiannakopoulos, T., Quigley, J.P. Unexpected effect of matrix metalloproteinase down-regulation on vascular intravasation and metastasis of human fibrosarcoma cells selected in vivo for high rates of dissemination. Cancer Res. 65:10959, 2005. Lewis, J.D., Destito, G., Zijlstra, A., Gonzalez, M.J., Quigley, J.P., Manchester, M., Stuhlmann, H. Viral nanoparticles as tools for intravital vascular imaging. Nat. Med. 12:354, 2006. Madsen, M.A., Deryugina, E.I., Niessen, S., Cravatt, B.F., Quigley, J.P. Activitybased protein profiling implicates urokinase activation as a key step in human fibrosarcoma intravasation. J. Biol. Chem. 281:15997, 2006. Zijlstra, A., Seandel, M., Kupriyanova, T.A., Partridge, J.J., Madsen, M.A., HahnDantona, E.A., Quigley, J.P., Deryugina, E.I. Proangiogenic role of neutrophil-like inflammatory heterophils during neovascularization induced by growth factors and human tumor cells. Blood 107:317, 2006. Regulators of Clathrin-Mediated Endocytosis S.L. Schmid, J. Chappie, S.D. Conner, M. Ishido, M. Leonard, R. Ramachandran, F. Soulet, B.D. Song, M.C. Surka, D. Yarar lathrin-mediated endocytosis is essential for the efficient uptake of nutrients and other macromolecules into cells and for the regulation of signaling by cell-surface receptors. The process occurs at clathrin-coated pits, which concentrate receptorligand complexes, deform the membrane, invaginate, and eventually pinch off, forming clathrin-coated vesicles (CCVs). The major components involved in formation of CCVs are clathrin, adaptor proteins, and dynamin, an atypical GTPase. Clathrin self-assembles into a polygonal lattice and serves as a scaffold for the formation of coated pits. Adaptor protein-2 is a heterotetrameric protein that triggers clathrin assembly at the plasma membrane and interacts directly with the cytoplasmic tails of surface receptors to concentrate the receptors into the assembling coated pit. We view dynamin as the master regulator of endocytosis. Previously, we developed a cell-free assay in which CCVs are reconstituted from sheets of purified plasma membranes from rat liver. Using this in vitro reconstitu- C THE SCRIPPS RESEARCH INSTITUTE 49 tion system and a new biochemical complementation assay, we explored the limiting cytosolic requirements for endocytosis of the low-density lipoprotein receptor–related protein (LRP) from isolated plasma membranes. LRP, also known as a scavenger receptor, binds multiple, distinct ligands and participates in constitutive endocytosis and signal transduction. We found that clathrin, adaptor protein-2, and dynamin do not support efficient LRP uptake; additional factors present in a 30% ammonium sulfate supernatant fraction of bovine brain cytosol are required. Fractionation of the supernatant revealed that multiple and redundant factors are required to support LRP endocytosis. Our data suggest that LRP, which has several distinct endocytic motifs in its cytoplasmic domain, may use multiple pathways for endocytosis in vitro. The factors we identified, 70-kD heat-shock cognate protein, synaptojanin, and collapsin response mediator protein-2, are all implicated in clathrin-mediated endocytosis in vivo, thus validating the assay. However, we found that these factors were sufficient but not necessary for formation of CCVs. Thus, functional redundancy and complexity make this assay biochemically intractable, and we have chosen, at least for the moment, to abandon it. We continue to analyze the structure and function of dynamin. Dynamin is a multidomain protein consisting of an N-terminal GTPase domain, a middle domain of previously unknown function, a PH domain that binds phosphatidylinositol 4,5-biphosphate, a GTPase effector domain (GED) required for dynamin self-assembly that functions as an assembly-dependent GTPase-activating protein for dynamin, and a C-terminal proline-arginine–rich domain. Dynamin self-assembles in vitro into spirals on liposomes containing phosphatidylinositol 4,5-biphosphate to tubulate the liposomes, resulting in a greater than 100-fold stimulation of GTPase activity. Self-assembly and assembly-stimulated GTPase activity at the necks of deeply invaginated coated pits are thought to drive conformational changes that mediate membrane fission. Our collaborators recently solved the crystal structure of the isolated, unoccupied GTPase domain of rat dynamin-1. Unlike the situation in other GTPases, the normally unstructured switch 1 and switch 2 regions that surround the unoccupied GTP binding pocket were ordered. On the basis of the structure, 2 arginines near the active site were predicted to be required for catalysis, but our enzymatic analysis of lysine and alanine 50 CELL BIOLOGY 2006 substitutions of these residues suggested otherwise. Thus, the mechanism of GTP hydrolysis remains obscure. However, the structure revealed the existence of a hydrophobic groove created by the N- and C-terminal α-helices of the GTPase domain, which was suggested to be a docking site for the GED. In a recent study, we identified mutations in the GED that resulted in reduced GTPase activity without affecting self-assembly. Because these mutations mapped to a predicted amphipathic helix at the extreme C terminus of GED, we speculated that this GED helix formed a 3-helical bundle with the GTPase domain helices. To test this hypothesis, we have generated a construct consisting of the GTPase of dynamin together with a C-terminal extension composed of a short sequence of turn-preferring residues followed by the 20 amino acid C-terminal GED helix. This construct, unlike GTPase domain or GED constructs is largely soluble when expressed in Escherichia coli and has basal GTPase activity equivalent to that of full-length dynamin. These exciting results suggest that we have fully reconstituted the GED-GTPase interactions. We are currently expressing the GTPase-GED peptide construct for high-resolution x-ray crystallography studies, and mutagenesis is under way to probe the mechanisms of GED-stimulated GTPase activity. We have also identified a new class of mutants in the middle domain that alter the quaternary structure of dynamin. Native dynamin exists as a tetramer, but analytical ultracentrifugation and gel filtration chromatography coupled to multiangle light scattering have confirmed that the middle domain mutants are dimeric. Kinetic studies established that the basal GTPase activity of dynamin requires a highly cooperative, GTP-dependent conformational change in dynamin tetramers. The dimeric dynamin mutants are defective in both activation in the basal state and self-assembly into higher order structures. Finally, in following up our earlier discovery that actin dynamics are required for multiple stages of clathrin-mediated endocytosis, we found that the protein sorting nexin 9 is a molecular link between dynamin and actin assembly. Sorting nexin 9 binds dynamin through the nexin’s SH3 domain and activates neural Wiskott-Aldrich syndrome protein to trigger actin-related protein 2/3–dependent actin assembly into branched filaments. Thus, sorting nexin 9 may trigger the burst of actin assembly that accompanies the scission of coated vesicles. THE SCRIPPS RESEARCH INSTITUTE PUBLICATIONS Leonard, M., Song, B.D., Ramachandran, R., Schmid, S.L. Robust colorimetric assays for dynamin’s basal and stimulated GTPase activities. Methods Enzymol. 404:490, 2005. Miwako, I., Schmid, S.L. A cell-free biochemical complementation assay reveals complex and redundant cytosolic requirements for LRP endocytosis. Exp. Cell Res. 312:1335, 2006. Miwako, I., Schmid, S.L. Clathrin-coated vesicle formation from isolated plasma membranes. Methods Enzymol. 404:503, 2005. Reubold, T.F., Eschenburg, S., Becker, A., Leonard, M., Schmid, S.L., Vallee, R.B., Kull, F.J., Manstein, D.J. Crystal structure of the GTPase domain of rat dynamin 1. Proc. Natl. Acad. Sci. U. S. A. 102:13093, 2005. Molecular Biology of Olfaction L. Stowers, I.S. Bharati, P. Chamero, J. Cruz, K. Flanagan, J. Lin, D. Logan, T. Marton, C. Ramos very breath samples the environment for olfactory chemical information, determining the quality of food, warning of danger, and confirming safety. The neurons that mediate olfaction are of 2 types: those that mediate an evocative perception that varies with each individual’s experience and those that regulate stereotyped innate social behaviors such as aggression and mating. Neurons that elicit odorant perception reside in the olfactory epithelium and relay chemical information through activation of cAMP-responsive channels. Recently, we showed that behavior-generating neurons are located in the vomeronasal organ and respond to pheromones through a cascade that ultimately activates C-type transient-receptor potential 2 (TRP2) channels. We are using a molecular genetic approach to characterize the function of these pheromone-responsive neurons. Through electrophysiologic recordings, we have shown that mutant mice lacking C-type TRP2 channels do not depolarize in response to natural sources of pheromones. Behavioral assays with these animals revealed that this pheromone response is necessary for both intermale aggression and gender recognition. We are identifying other unique molecular subpopulations of pheromone-responsive neurons, and through genetic ablation, biochemistry, and electrophysiology, we are assigning biological function to each neuron type. A full characterization of the repertoire of chemosensory neurons will be essential in understanding the logic of olfactory information coding. To this end, we are investigating a novel class of olfactory neurons that lack both the cAMP and C-type TRP2 signaling components. Analysis of these neurons by transcriptional E CELL BIOLOGY 2006 profiling and then molecular genetics and biochemistry is being used to identify their role in olfactory function. Elucidation of the function of specific neural circuits that regulate mammalian behavior has been hindered because the pheromone compounds that signal each behavior have not been identified from their complex natural sources. To obtain these important molecules, we are fractionating natural sources of pheromones and then using biobehavioral assays to identify the molecules that initiate activity. The results will enable us to both activate specific neural circuits and analyze the natural production and regulation of the signaling ligands. In total, we expect to define the pheromone response pathway of mice and to reveal general principles of neurons that govern complex social behavior. THE SCRIPPS RESEARCH INSTITUTE 51 in vitro differentiation of embryonic stem cells into embryoid bodies. Our results indicate that Vezf1 affects vascular differentiation by regulating cell proliferation, differentiation, and deposition of extracellular matrix. We are examining the molecular pathways of Vezf1 function. In collaborative studies with L. Benjamin, Beth Israel Deaconess Medical Center, Boston, we found that VEZF1, the protein encoded by Vezf1, interacts with Rho GTPases to modulate the function of Rho in the endothelium. Microarray cDNA analysis with RNA from wild-type embryos and embryos lacking Vezf1 suggested that genes for fibrinogen and claudin and several genes involved in metabolite transport are target genes for Vezf1. A N E A R LY M A R K E R F O R E N D O T H E L I A L C E L L S A N D THEIR PROGENITORS Molecular Regulation of Vascular System Development in Mammals H. Stuhlmann, M.J. Fitch, Z. Zou, S. Chitnis, J.D. Lewis, A. Durrans, W. LeVine stablishment of a functional circulatory system during development is crucial for the delivery of nutrients and oxygen to embryos. Defects in the development of blood vessels result in death before birth or in congenital cardiovascular abnormalities. We examine the molecular and genetic pathways that regulate the 3 principal processes of vascular development: determination of vascular lineage, vasculogenesis, and angiogenesis. We focus on the mouse model because of the ready availability of genetic information on mice and experimental tools and because of similarities between mice and humans. Using an expression-based “gene trap” screen in mouse embryonic stem cells and embryos, we identified 2 novel genes involved in these processes: Vezf1 and Egfl7. E Expression of a second endothelial gene identified in our screen, Egfl7, is restricted to the vascular endothelium and endothelial progenitors in the yolk sac mesoderm. Egfl7 is also expressed in multipotent stem cells in embryos, in primordial germ cells, and during spermatogenesis. In the quiescent vasculature in adults, overall Egfl7 expression is downregulated. During physiologic angiogenesis in the uterus during pregnancy and in the regenerating endothelium after vascular injury, expression of Egfl7 is upregulated. EGFL7, the protein encoded by Egfl7, is partially secreted and acts as a chemoattractant on both endothelial cells and embryonic fibroblasts in in vitro migration assays. EGFL7 is a compact 278 amino acid protein with an amino-terminal signal peptide; an EMI domain; and 2 central epidermal growth factor–like domains, one of which contains a putative Notch interaction domain. Importantly, recent collaborative studies with J. Kitajewski, Columbia University, New York City, provide support for our hypothesis that EGFL7 binds to Notch receptors and acts as an agonist for Notch signaling during vascular development. D E V E L O P M E N T O F M U LT I VA L E N T V I R A L N A N O PA R T I C L E S F O R I N V I V O VA S C U L A R A ZINC FINGER GENE ESSENTIAL FOR NORMAL TA R G E T I N G A N D I M A G I N G VA S C U L A R A N D LY M P H AT I C D E V E L O P M E N T In collaboration with M. Manchester, Department of Cell Biology, we have developed viral nanoparticles based on the cowpea mosaic virus (CPMV) for noninvasive imaging and targeting of the mammalian cardiovascular system. CPMV can be fluorescently labeled to high densities with no quenching, resulting in bright particles that allow high-resolution intravital imaging of the vascular endothelium and blood flow deep inside Vezf1 is the gene for an early zinc finger transcription factor that controls the development of blood vessels and the lymphatic system in mice. Using functional genetic studies, we previously showed that Vezf1 plays an essential and dosage-dependent role in the proliferation, remodeling, and integrity of the developing vasculature. We recently extended these studies by using 52 CELL BIOLOGY 2006 mouse or chick embryos for up to 72 hours. Using a human fibrosarcoma model for tumor angiogenesis, we found that fluorescent CPMV can be used to distinguish between arterial and venous vessels and to monitor the neovascularization of the tumor microenvironment. We are extending these studies by conjugating peptides to the CPMV capsid to target the peptides to the vasculature, both during development and in disease models. PUBLICATIONS Lewis, J.D., Destito, G., Gonzalez, M.J., Zijlstra, A., Quigley, J.P., Manchester, M., Stuhlmann, H. Viral nanoparticles as tools for intravital vascular imaging. Nat. Med. 12:354, 2006. Kuhnert, F., Stuhlmann, H. Role of Vezf1 during vascular and lymphatic development. In: Endothelial Biomedicine. Aird, W.C. (Ed.), Cambridge University Press, New York, in press. Zijlstra, A., Lewis, J.D., DeGryse, B., Stuhlmann, H., Quigley, J.P. Inhibition of tumor cell intravasation and subsequent metastasis through the regulation of CD151-mediated in vivo tumor cell motility. Cancer Cell, in press. Ion Channels and Fast Synaptic Transmission N. Unwin on channels play a central role in the rapid transmission of electrical signals throughout the nervous system. To determine how these membrane proteins work, my colleagues and I are using electron microscopy to analyze the structures of the proteins trapped in different physiologic states. Current studies center on the nicotinic acetylcholine receptor at the nervemuscle synapse. We wish to find out how this ion channel achieves its ion selectivity and high transport rate and how it opens and desensitizes in response to acetylcholine released into the synaptic cleft. For our studies, we use postsynaptic membranes isolated from the (muscle-derived) electric organ of the Torpedo ray, which form tubular crystals of acetylcholine receptors. The acetylcholine receptor is a member of a superfamily of transmitter-gated ion channels, which includes the receptors for serotonin 5-HT3, γ-aminobutyric-acids A and C, and glycine. It has a cation-selective pore, delineated by a ring of 5 similar subunits, that opens upon binding of acetylcholine to the 2 ligand-binding (α) subunits at the subunit interfaces. Recently, we obtained a refined atomic model of the acetylcholine receptor in the closed-channel form. We found that the individual subunits in the N-terminal ligand-binding domain are organized around 2 sets I THE SCRIPPS RESEARCH INSTITUTE of β-sheets packed in a curled β-sandwich, as in the related soluble pentameric acetylcholine-binding protein. Each of the subunits in the membrane-spanning domain is made from 4 α-helical segments. The helical segments arrange symmetrically, forming an inner ring of helices that shape a water-filled pore and an outer shell of helices that coil around each other and shield the inner ring from the lipids. In the closed channel, the helices in the inner ring come together near the middle of the membrane and make a constricting hydrophobic girdle. This girdle, which is about 50 Å from the acetylcholine-binding sites, constitutes an energetic barrier to ion permeation and functions as the gate of the channel. These details, together with those obtained earlier from studies of the receptor trapped in the open-channel form, have enabled us to understand in outline the structural mechanism by which acetylcholine opens the pore. In the absence of acetylcholine, the pore is normally closed. When acetylcholine enters the binding sites, localized rearrangements in the α-subunits occur that stabilize an alternative extended conformation of the channel in which the inner sets of β-sheets are rotated by about 10° about axes perpendicular to the membrane plane, relative to the orientations of the sheets in the closed channel. These rotations are communicated through the inner membrane-spanning helices and open the pore by breaking the hydrophobic girdle apart. Improvements in resolution of the 3-dimensional structure, in both the closed- and open-channel forms, are now being attempted so that the structural mechanism of gating of the channel can be described in greater detail. The knowledge gained from the refined structure of the locations of amino acid residues, in relation to the ion pathway, is also being used to develop quantitative explanations of how the high cation selectivity and high conduction rates of this channel are achieved. These studies are yielding crucial insight into the nature of a number of neuromuscular disorders, including several well-characterized congenital myasthenic syndromes. They are also providing important 3-dimensional information about the binding sites for drugs that affect the brain by modulating the function the related γ-aminobutyric acid, serotonin, glycine, and neuronal acetylcholine receptors. PUBLICATIONS O’Brien, J., Unwin, N. Organization of spines on the dendrites of Purkinje cells. Proc. Natl. Acad. Sci. U. S. A. 103:1575, 2006. CELL BIOLOGY 2006 Microscopes and Motility: Systems Integration in Cell Migration C.M. Waterman-Storer, O. Rodriguez, S.L. Gupton, K. Kita, R. Littlefield, K. Hu, A. Wheeler, W. Shin, M.L. Gardel, I.C. Schnieder, A. Parapera, J. Lim ell migration is critical to development, the immune response, and wound healing. In cancer cells, loss of regulation of cell motility results in deadly metastasis. The locomotion of vertebrate tissue cells is thought to require complex and dynamic interactions between the microtubule and actin cytoskeletal polymers, the endomembrane trafficking system, and focal adhesions to the extracellular environment. We develop quantitative light microscopy methods to analyze the dynamic interactions between these complex macromolecular systems in living cells to understand how the systems are spatiotemporally coordinated to drive directed cell movement. We then use these microscopic assays to analyze cells with specific perturbations of cytoskeletal, membrane, or adhesive proteins to dissect the molecular mechanisms of the regulation of the proteins and their contribution to cell morphogenesis and migration. We pioneered fluorescent speckle microscopy, a powerful method that allows quantitative analysis of the dynamics of macromolecular assemblies in living cells. Recently, we extended the technology to multispectral total internal fluorescence reflection fluorescence microscopy, allowing analysis of the integration of proteins within focal adhesions with the actin cytoskeleton during cell migration. In collaboration with G. Danuser, Department of Cell Biology, we developed correlational fluorescent speckle microscopy to measure the coupling of focal adhesion proteins to the actin cytoskeleton. We found that different classes of focal adhesion structural and regulatory molecules have different degrees of correlated motions with actin filaments, indicating differential transmission of actomyosin motion through focal adhesions. Our results suggest that transient interactions between focal adhesion proteins and actin filaments constitute a friction clutch between the cytoskeleton and the extracellular environment that is regulated during the morphodynamic transitions of cell migration. On the basis of a mathematical model, researchers predicted 15 years ago that migration speed would C THE SCRIPPS RESEARCH INSTITUTE 53 have a biphasic response to increasing strength of cell adhesion, with slow migration occurring at low and high strengths and fast migration at intermediate strength. The assumption of the model was that migrating cells have an asymmetry in adhesion strength from the front part of the mass of cells to the rear part, with the cells connected by symmetric contractile elements, but dynamic organizational states of F-actin and focal adhesions were not considered. This biphasic dependence of migration velocity on increasing adhesion strength has since been supported experimentally, and a front-to-rear gradient in cell adhesion strength has also been shown. We sought to determine if distinct organizational states of F-actin, myosin II, and focal adhesions accompany adhesion-dependent changes in velocity. We characterized a unique phenotype for optimal migration at intermediate adhesion strength, entailing rapid flow convergence and local depolymerization of F-actin, local activation of myosin II, rapid renewal of the components of focal adhesions, and intermediate lifetime and turnover rates of focal adhesions. We recapitulated this phenotype and fast migration at a nonoptimal adhesion strength by manipulating the activity of myosin II. In contrast to the results with simple models, we found that a complex spatiotemporal integration and feedback between F-actin, myosin II, and focal adhesions mediates the classically observed biphasic migration velocity response to increasing adhesion strength, so that a specific balance between adhesion and contraction induces maximal migration velocity. The microtubule and actin cytoskeletons interact in cells to promote a coordinated effort to drive protrusion of the leading edge of cells during cell migration. The tumor suppressor protein adenomatous polyposis coli and its binding partner EB1 accumulate on the ends of microtubules. The tumor suppressor protein specifically collects on microtubules in cell protrusions, suggesting that it may promote cell protrusion. We simultaneously visualized dynamics of the suppressor protein and microtubules in living cells. We found that the association of the protein with the ends of microtubules correlates with the increased growth stability of the microtubules and that this stabilization can occur independent of the association of the protein with EB1. We also found that the protein and EB1 associate with the ends of microtubules by distinct mechanisms. Thus, cancer-causing mutations in this tumor suppressor protein may arise from defects in microtubule stability and cell migration. 54 CELL BIOLOGY 2006 PUBLICATIONS Danuser, G., Waterman-Storer, C.M. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct. 35:361, 2006. deRooij, J., Kerstens, A., Danuser, G., Schwartz, M.A., Waterman-Storer, C.M. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J. Cell Biol. 171:153, 2005. Gupton, S.L., Waterman-Storer, C.L. Live-cell fluorescent speckle microscopy (FSM) of actin cytoskeletal dynamics and their perturbation by drug perfusion. In: Cell Biology: A Laboratory Handbook, 3rd ed. Celis, J., et al. (Eds.). Academic Press, San Diego, 2005, Vol. 3, p. 137. Gupton, S.L., Waterman-Storer, C.L. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell 125:1361, 2006. Kita, K., Wittmann, T., Nathke, I.S., Waterman-Storer, C.L. Adenomatous polyposis coli on microtubule plus ends in cell extensions can promote microtubule net growth with or without EB1. Mol. Biol. Cell 17:2331, 2006. Ponti, A., Matov, A., Adams, M., Gupton, S., Waterman-Storer, C.M., Danuser, G. Periodic patterns of actin turnover in lamellipodia and lamellae of migrating epithelial cells analyzed by quantitative fluorescent speckle microscopy. Biophys. J. 89:3456, 2005. Systems Biology and Malaria E.A. Winzeler, C. Kidgell, V. Ramachandran, N. Kato, K. Henson, J. Young, J. Johnson espite the widespread impact of malaria on the world’s health and economies, relatively little is known about the function of the majority of the 5300 genes in the genome of Plasmodium falciparum, the causative agent of the most severe form of malaria in humans. This lack of knowledge retards the development of drugs and vaccines against the parasite. We use systematic discovery-based approaches to predict the function of uncharacterized Plasmodium genes; our goal is to facilitate the discovery of new treatments. We are using mRNA and protein expression to reveal genetic regulatory networks and to suggest protein-protein interactions. We are also developing new methods that can be used in systems biology research. In addition to our past work on blood-stage parasites, we have characterized the expression program of the sexual stages of malarial parasites. These stages, which are essential for the mosquito transmission of the disease, are the focus of the development of drugs and vaccines that block transmission. To better understand genes important to sexual development, we used a fullgenome high-density oligonucleotide microarray to profile the transcriptomes of P falciparum gametocytes. To interpret this transcriptional data, we developed and used a novel knowledge-based data-mining algorithm termed ontology-based pattern identification. With this algorithm, published or custom gene classifications D THE SCRIPPS RESEARCH INSTITUTE are used to optimize normalization methods and cluster boundaries so that the largest number of any given gene type is found in the smallest cluster size. This analysis resulted in the identification of a sexual development cluster containing 246 genes, of which approximately 75% were unclassified and which contained most known sexual stage genes. These genes had highly correlated, gametocyte-specific expression patterns. Statistical analysis of the upstream promoter regions of these 246 genes revealed putative cis regulatory elements. In addition, we extended the ontologybased pattern identification by using current annotations provided by the Gene Ontology Consortium to identify 380 statistically significant clusters containing genes with expression patterns characteristic of various biological processes, cellular components, and molecular functions. We are also studying genetic diversity by using hybridization-based approaches to further characterize parasite genes. By performing a full genome scan of allelic variability of 14 field and laboratory strains of P falciparum, we showed that 10% of the genome has higher than neutral rates of diversity at tens of thousands of loci. We found that whereas many genes are exceptionally well conserved across parasite isolates, paralog genes (i.e., genes related by duplication within a genome that have different functions), genes near the ends of chromosomes, genes that encode proteins that are trafficked to the surface of the infected red cell, and genes that encode known and potential drug targets are exceptionally diverse. These data suggest that rates of mitotic recombination are elevated among genes with paralogs and that selection pressure on those without paralogs is strong. We also revealed gene amplification events, including one associated with pfmdr1, the gene for multidrug resistance in P falciparum, and a previously uncharacterized amplification centered on the gene for GTP cyclohydrolase, the first enzyme in the folate biosynthesis pathway. Although GTP cyclohydrolase is not the known target of any current drugs, downstream members of the pathway are targeted by several widely used antimalarial agents. We propose that amplification of the GTP cyclohydrolase enzyme in the folate biosynthesis pathway may facilitate increased flux through this pathway and increase resistance to antifolate drugs. These data and recent publications indicating that 90% of a small eukaryote’s genetic variation can be captured in a single microarray hybridization, suggest that population genomics will be a fruitful approach CELL BIOLOGY 2006 THE SCRIPPS RESEARCH INSTITUTE 55 ass spectrometry has emerged as a powerful technique for cellular proteomics, complementing traditional gene-by-gene approaches with a comprehensive description of the molecular factors that contribute to a biologically relevant system. We remain at the forefront of this field, developing new strategies to address more sophisticated scientific questions through proteomics, such as how to measure global changes in protein abundance and how to characterize complex posttranslational modifications. hypothesized that a subset of insulin-signaling targets, specifically, daf-2 (the gene for an insulin receptor) and daf-16 (the gene for a transcription factor negatively regulated by daf-2) mutants, might be differentially expressed in wild-type C elegans and thus could be identified by using our quantitative proteomics approach. We identified 104 proteins that were present in higher or lower levels in daf-2 mutants than in wild-type and daf-16 mutant organisms, including the known targets superoxide dismutase 3 and catalases. Gene ontology analysis revealed that the upregulated proteins in daf-2 mutants were overrepresented in the metabolism of reactive oxygen species, metabolism of carbohydrates, and biosynthesis of amino acids; the downregulated proteins were enriched in proteins involved in translation and lipid transport. We confirmed by genetics analysis that some of the possible insulin-signaling components identified in the study play a role in regulating life span and/or formation of dauer larvae, both of which are regulated by C elegans insulins. Among the confirmed targets is a protein phosphatase. Using a green fluorescent protein as a label, we found that the phosphatase was upregulated in worms in which daf-2 was inactivated via RNA interference, consistent with the results of mass spectrometry. Further genetic analysis indicated that this phosphatase acted upstream of and/or in parallel to the protein DAF16 in regulation of aging and dauer formation. Taken together, our data suggest that this protein phosphatase is both a downstream target and a regulator of insulin signaling. Therefore it may be part of a feedback loop. This study illustrates the effectiveness of combining quantitative mass spectrometry and C elegans genetics. Such an approach can be extended to other studies beyond insulin signaling. Q U A N T I TAT I V E P R O T E O M I C A N A LY S I S O F I N S U L I N P R O T E I N S U M O Y L AT I O N SIGNALING The characterization of posttranslational modifications is also an emerging application of mass spectrometry–based proteomics. We are developing proteomic tools to study the family of small ubiquitin-like modifiers (SUMOs). Recently, we focused on Smt3p, the budding yeast homolog of a human SUMO. Although genetic studies have indicated that Smt3p is required for proper regulation of a variety of different cellular processes, including transcription, intracellular transport, progression of the cell cycle, and the maintenance of genome integrity, the mechanisms by which it does so remain largely unknown. Using proteomic approaches, we addressed 2 major areas in protein sumoylation: the large-scale identifi- for discovering new determinants of drug resistance in a variety of infectious agents. PUBLICATIONS Carret, C.K., Horrocks, P., Konfortov, B., Winzeler, E., Qureshi, M., Newbold, C., Ivens, A. Microarray-based comparative genomic analyses of the human malaria parasite Plasmodium falciparum using Affymetrix arrays. Mol. Biochem. Parasitol. 144:177, 2005. Kidgell, C., Winzeler, E.A. Using the genome to dissect the molecular basis of drug resistance. Future Microbiol. 1:185, 2006. Winzeler, E.A. Applied systems biology and malaria. Nat. Rev. Microbiol. 4:145, 2006. Young, J.A., Fivelman, Q.L., Blair, P.L., de la Vega, P., Le Roch, K.G., Zhou, Y., Carucci, D.J., Baker, D.A., Winzeler, E.A. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol. Biochem. Parasitol. 143:67, 2005. Advancing Applications in Mass Spectrometry–Based Proteomics J.R. Yates III, A.O. Bailey, G.T. Cantin, E. Chen, D. Cociorva, J. Coppinger, C. Delahunty, M.Q. Dong, J. Hewel, J.R. Johnson, L. Liao, B.W. Lu, I. McLeod, D. McClatchy, R. Park, E. Romijn, H. Prieto, C.I. Ruse, J. Venable, J. Wohlschlegel, C. Wong, T. Xu M Quantitative mass spectrometry–based proteomics relies on internal isotopic standards. Metabolic labeling with nitrogen 15 is a preferred method of introducing internal standards. It produces a standard for every peptide or protein to be characterized. In addition, it is stable, nonradioactive, less error-prone than chemical labeling, and relatively inexpensive. We have used metabolic labeling with nitrogen 15 and quantitative mass spectrometry to study insulin signaling in the worm Caenorhabditis elegans. Insulin regulates a wide range of processes, including metabolism, development, and aging, but only a handful of its downstream targets are known. We 56 CELL BIOLOGY 2006 cation of sumoylation targets and the development of strategies for mapping SUMO modification sites. By combining different affinity chromatography strategies with our multidimensional protein identification platform, we identified 271 new SUMO targets. These substrates play roles in a diverse set of biological processes and greatly expand the known scope of SUMO regulation in eukaryotic cells. This research also revealed coordinated SUMO modification of multiple proteins in well-defined macromolecular complexes. This intriguing result suggests that sumoylation may target protein complexes rather than individual proteins. We are also characterizing the mechanism that underlies this observation. Characterization of many of the new substrates identified in our study was limited by difficulties in identifying SUMO attachment sites in the target of interest. To solve this problem, we recently developed a method for the rapid and efficient identification of SUMO attachment sites in cellular proteins. In this method, different SUMO mutants are used in combination with various protease digestion strategies, and then mass spectrometry is used to directly and specifically map the locations of the modified lysine residues. We showed the usefulness of this method by identifying SUMO modification sites in an assortment of model SUMO substrates and in complex mixtures. The development of a method for identifying SUMO attachment sites will be a powerful tool for the characterization of the new substrates identified in our initial global analysis. PUBLICATIONS Cantin, G.T., Venable, J.D., Cociorva, D., Yates, J.R. III. Quantitative phosphoproteomic analysis of the tumor necrosis factor pathway. J. Proteome Res. 5:127, 2006. Chen, E.I., Hewel, J., Felding-Habermann, B., Yates, J.R. III. Large scale protein profiling by combination of protein fractionation and multidimensional protein identification technology (MudPIT). Mol. Cell. Proteomics 5:53, 2006. Sadygov, R., Wohlschlegel, J., Park, S.K., Xu, T., Yates, J.R. III. Central limit theorem as an approximation for intensity-based scoring function. Anal. Chem. 78:89, 2006.. Venable, J.D., Xu, T., Cociorva, D., Yates, J.R. III. Cross-correlation algorithm for calculation of peptide molecular weight from tandem mass spectra. Anal. Chem. 78:1921, 2006. Wohlschlegel, J.A., Johnson, E.S., Reed, S.I., Yates, J.R. III. Improved identification of SUMO attachment sites using C-terminal SUMO mutants and tailored protease digestion strategies. J. Proteome Res. 5:761, 2006. THE SCRIPPS RESEARCH INSTITUTE Macromolecular Assemblies Visualized by Electron Cryomicroscopy and Image Analysis: Membrane Proteins and Viruses M. Yeager, R. Abagyan,**** B.D. Adair, K. Baker, K. Altieri, A. Cheng, M.J. Daniels, K.A. Dryden, B. Ganser, J. Harless, Y. Hua, M. Matho, F.A. Palida, M.A. Arnaout,* A.R. Bellamy,** N. Ben-Tal,*** M.J. Buchmeier,**** F.V. Chisari,**** K. Coombs,***** H.B. Greenberg,† J.E. Johnson,**** S. Matsui,† L.H. Philipson,†† T.D. Pollard,††† A. Rein,†††† A. Schneemann,**** J.A. Tainer,**** J.A. Taylor,** V.M. Unger††† * Harvard Medical School, Boston, Massachusetts ** University of Auckland, Auckland, New Zealand *** Tel-Aviv University, Tel-Aviv, Israel **** Scripps Research ***** University of Manitoba, Winnipeg, Manitoba † Stanford University, Stanford, California †† University of Chicago, Chicago, Illinois ††† Yale University, New Haven, Connecticut †††† National Cancer Institute, Frederick, Maryland he ultimate goal of our studies is to gain a deeper understanding of the molecular basis of important human diseases, such as sudden death, heart attacks, and HIV infection, that cause substantial mortality and suffering. The structural details revealed by our research may provide clues for the design of more effective and safer medicines. At the basic science level, we are intrigued by questions at the interface between cell biology and structural biology: How do membrane proteins fold? How do membrane channels open and close? How are signals transmitted across a cellular membrane when an extracellular ligand binds to a membrane receptor? How do viruses attach to and enter host cells, replicate, and assemble infectious particles? To explore such problems, we use high-resolution electron cryomicroscopy and computer image processing. With this approach, we can examine the molecular architecture of supramolecular assemblies such as membrane proteins and viruses. In electron cryomicroscopy, biological specimens are quick frozen in a physiologic state to preserve T CELL BIOLOGY 2006 their native structure and functional properties. A special advantage of this method is that we can capture dynamic states of functioning macromolecular assemblies, such as open and closed states of membrane channels and viruses actively transcribing RNA. Threedimensional density maps are obtained by digital image processing of the high-resolution electron micrographs. The rich detail in the density maps exemplifies the power of this approach to reveal the structural organization of complex biological systems that can be related to the functional properties of such assemblies. Ongoing research projects include the structure analysis of (1) membrane proteins involved in cell-tocell communication (gap junctions), water transport (aquaporins), ion transport (potassium channels), transmembrane signaling (integrins), and viral recognition (rotavirus NSP4); (2) viruses responsible for significant human diseases (retroviruses, hepatitis B virus [HBV], rotavirus, astrovirus); and (3) viruses used as model systems to understand mechanisms of pathogenesis (arenaviruses, reoviruses, nodaviruses, tetraviruses and sobemoviruses). The following sections highlight selected projects that exemplify the themes of our research program. THE SCRIPPS RESEARCH INSTITUTE 57 F i g . 1 . Intercellular gap junction channels have a diameter of about 65 Å and are formed by the end-to-end docking of 2 hemichannels, each composed of a hexamer of connexin subunits. A Cα model (ribbons) for the membrane-spanning α-helices of the hemichannels was derived by combining the information from a computational analysis of connexin sequences, the results of more than a decade of biochemical studies, and the constraints provided by a 3-dimensional map derived by using electron cryocrystallography. Although individually, none of these approaches provided high-reso- GAP JUNCTION MEMBRANE CHANNELS lution information, their sum yielded an atomic model that predicts Gap junction channels connect the cytoplasms of adjacent cells by means of an intercellular conduit formed by the end-to-end docking of 2 hexameric hemichannels called connexons. Gap junctions play an essential functional role by mediating metabolic and electrical communication within tissues. For instance, in the heart, gap junction channels organize the pattern of current flow to allow a coordinated contraction of the muscle. We expressed a recombinant cardiac gap junction protein, termed connexin 43, and produced 2-dimensional crystals suitable for electron cryocrystallography. Our previous findings indicated that each hexameric connexon is formed by 24 closely packed α-helices. We have now extended this analysis to 5.7-Å in-plane and 19.8-Å vertical resolution, a step that enables us to identify the positions and tilt angles for the 24 α-helices within each hemichannel (Fig. 1). The 4 hydrophobic segments in connexin sequences were assigned to the α-helices in the map on the basis of biochemical and phylogenetic data. Evolutionary conservation and an analysis of compensatory mutations in connexin evolution were used to identify the packing interfaces between the helices. The final model, which specifies the coordi- how connexin mutations (spheres), which result in diseases such as nonsyndromic deafness and Charcot-Marie-Tooth disease, may interfere with formation of functional channels by disrupting helixhelix packing. nates of Cα atoms in the transmembrane domain, provides a structural basis for understanding the different physiologic effects of almost 30 mutations and polymorphisms in terms of structural deformations at the interfaces between helices, revealing an intimate connection between molecular structure and disease. INTEGRINS Integrins are a large family of heterodimeric transmembrane receptor proteins that modulate important biological processes such as development, cell adhesion, angiogenesis, wound healing, and neoplastic transformation. The ectodomain of the integrin αvβ3 crystallizes in a bent, genuflexed conformation, which is considered to be inactive (i.e., unable to bind physiologic ligands in solution) unless it is fully extended by activating stimuli. To assess whether the bent integrin can bind physiologic ligands, we collaborated with M.A. Arnaout, Harvard Medical School, Boston, Massachusetts, to generate a stable, soluble complex of the manganese-bound αvβ3 ectodomain with a frag- 58 CELL BIOLOGY 2006 ment of fibronectin containing type III domains 7–10 and the EDB domain. Electron microscopy and singleparticle image analysis were used to determine the 3-dimensional structure of this complex (Fig. 2). THE SCRIPPS RESEARCH INSTITUTE core protein has been studied intensively. However, little is known about the structure and assembly of native capsids present in infected cells, and even less is known about the structure of mature virions. We used electron cryomicroscopy and image analysis to examine HBV virions (also called Dane particles) isolated from the serum of a patient with hepatitis B and capsids positive and negative for HBV DNA isolated from the livers of transgenic mice (Fig. 3). F i g . 2 . The 3-dimensional density map (grayscale transparency) of the integrin αvβ3 in a complex with fibronectin was determined by using electron microscopy and image analysis. The x-ray structures of the αv and β3 proteins have been docked into the electron microscopy density envelope. Additional density (lower right) can accommodate fibronectin domain 10 adjacent to the ligand-binding site as well as domain 9 at the synergy site. The complex is shown adjacent to the white box, which represents the 30-Å-thick hydrophobic part of the cellular membrane across which signals are transmitted. Most αvβ3 particles, whether unliganded or bound to fibronectin, had compact, triangular shapes. A difference map comparing ligand-free and fibronectin-bound integrin revealed density that could accommodate the fibronectin type III domain 10 containing arginine– glycine–aspartic acid in proximity to the ligand-binding site of β3, with domain 9 just adjacent to the synergy site binding region of αv. This study suggests that the ectodomain of αvβ3 has a bent conformation that can stably bind a physiologic ligand in solution. These results are relevant for understanding how binding of ligands to the extracellular domain leads to conformational changes that transmit signals across the plasma membranes of cells, culminating in changes in gene transcription in the nucleus. F i g . 3 . A model of the HBV virion (diameter ~450 Å) based on electron cryomicroscopy and image analysis. A, The double-stranded DNA genome is encapsidated by an icosahedral capsid shell composed of 120 spikes. The surface is studded with glycoproteins spaced about 60 Å apart that bind to membrane receptors on liver cells. B, In the close-up view, the x-ray crystal structure of a recombinant capsid has been docked into the electron cryomicroscopy density map of the virion capsid. The core spikes are in close apposition but do not penetrate the envelope. H E PAT I T I S B V I R U S HBV currently infects more than 350 million people, of which 1 million will die every year. The infectious virion is an enveloped capsid containing the viral polymerase and the double-stranded DNA genome. The structure of the capsid assembled in vitro from expressed Both types of capsids assembled as icosahedral particles indistinguishable from previous image reconstructions of capsids. Likewise, the virions contained capsids with either T = 3 or T = 4 icosahedral symmetry. Projections extending from the lipid envelope CELL BIOLOGY 2006 were attributed to surface glycoproteins. The packing of the projections was unexpectedly nonicosahedral but conformed to an ordered lattice. These structural features distinguish HBV from other enveloped viruses. PUBLICATIONS Daniels, M.J., Wood, M.R., Yeager, M. In vivo functional assay of a recombinant aquaporin in Pichia pastoris. Appl. Environ. Microbiol. 72:1507, 2006. Daniels, M.J., Yeager, M. Phosphorylation of aquaporin PvTIP3;1 defined by mass spectrometry and molecular modeling. Biochemistry 44:14443, 2005. Dryden, K.A., Wieland, S.F., Whitten-Bauer, C., Chisari, F.V., Yeager, M. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol. Cell 23:843, 2006. Greig, S.L., Berriman, J.A., O’Brien, J.A., Taylor, J.A., Bellamy, A.R., Yeager, M., Mitra, A.K. Structural determinants of rotavirus subgroup specificity mapped by cryo-electron microscopy. J. Mol. Biol. 356:209, 2006. Wiedenheft, B., Mosolf, J., Willits, D., Yeager, M., Dryden, K.A., Young, M., Douglas, T. An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc. Natl. Acad. Sci. U. S. A. 102:10551, 2005. THE SCRIPPS RESEARCH INSTITUTE 59