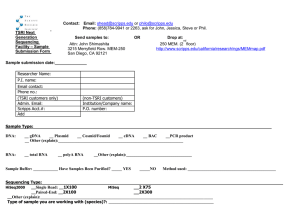
S C R I P P S Published by TSRI Press .
advertisement

SCRIPPS FLORIDA Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Biochemistry Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Scott Simanski, Research Assistant, and Nagi G. Ayad, Ph.D., Assistant Professor, Department of Biochemistry Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. BIOCHEMISTRY 2005 S TA F F S C I E N T I S T S Anthony D. Smith, Ph.D. Michael J. Chalmers, Ph.D. Achim M. Sorg, Ph.D. Qitao Yu, Ph.D. Sahba Tabrizifard, Ph.D. Steve A. Kay* Professor and Chairman Jiu Zhao, Ph.D. Mariola Tortosa, Ph.D. Nagi G. Ayad, Ph.D. Assistant Professor R E S E A R C H A S S O C I AT E S DEPAR TMENT OF BIOCHEMISTRY S TA F F Alessandra Cervino, Ph.D.** Assistant Professor Michael D. Conkright, Ph.D. Assistant Professor Patrick R. Griffin, Ph.D.** Professor John B. Hogenesch, Ph.D.*** Professor Malcolm A. Leissring, Ph.D. Assistant Professor Phillip LoGrasso, Ph.D.** Associate Professor Kendall W. Nettles, Ph.D.** Assistant Professor Howard T. Petrie, Ph.D. Professor Mathew T. Pletcher, Ph.D.** Assistant Professor Teresa M. Reyes, Ph.D. Assistant Professor William R. Roush, Ph.D.**** Professor Associate Dean, Kellogg School of Science and Technology, Florida Oliver Umland, Ph.D. Juliette C. Walker, Ph.D. Julie E. Baggs, Ph.D. Amie L. Williams, Ph.D. * Joint appointments in the Department of Cell Biology and the Institute for Childhood and Neglected Diseases ** Joint appointment in the Translational Research Institute *** Joint appointments in the Translational Research Institute and Molecular and Integrative Neurosciences Department **** Joint appointments in the Translational Research Institute and the Department of Chemistry Kristin Clarke, Ph.D. Etzer Darout, Ph.D. S C I E N T I F I C A S S O C I AT E S Amy C. DeBaillie, Ph.D. Yi An Lu, Ph.D. Joshua R. Dunetz, Ph.D. Qi Tao Yu, Ph.D. Masaki Handa, Ph.D. Dympna Harmey, Ph.D. Kevin R. Hayes, Ph.D. Tamara Hopkins, Ph.D. Jelena Janjic, Ph.D. Rebecca J. Kocerha, Ph.D. Brooke H. Miller, Ph.D. William Mwangi, Ph.D. Hiroshi Nakase, Ph.D. Michael H. Ober, Ph.D. Swati Prasad, Ph.D. Troy D. Ryba, Ph.D. James Tam, Ph.D. Professor S E C T I O N C O V E R F O R T H E D E P A R T M E N T O F B O C H E M I S T R Y : A combination of Claes Wahlestedt, M.D., Ph.D.** Professor structural approaches, including x-ray crystallography, in the laboratory of K.W. Nettles, Ph.D., and amide hydrogen-deuterium exchange mass spectrometry, in the laboratory of P.R. Griffin, Ph.D., were used to dissect how diverse ligands control the allosteric function of the nuclear receptor ligandbinding domain. The ligand-binding domain of the estrogen receptor is shown bound to an estrogen derivative, trifluoromethylphenylvinyl-estradiol. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 347 348 BIOCHEMISTRY 2005 INVESTIGATORS’ R EPORTS Role of Ubiquitin-Mediated Proteolysis in Irreversible Transitions During the Cell Cycle N.G. Ayad, A. Smith, D. Harmey, S. Simanski e are interested in the cell biological basis of irreversible transitions that occur during the eukaryotic cell cycle. Ubiquitin-mediated proteolytic pathways ensure that irreversibility is achieved by targeting specific inhibitors of these transitions for proteasomal degradation. One of the most important ubiquitn E3 ligases is the anaphase-promoting complex (APC). The complex is active during G1 and in fully differentiated cells. Furthermore, our recent studies indicate that the APC is required to initiate differentiation of neuronal precursor cells. This finding is especially important because it suggests that the complex is controlling an essential step in cell-cycle exit or differentiation, a control that is both biologically and medically relevant. Although we understand a great deal about the role of the APC during the cell cycle, its role in initiating exit from the cell cycle is virtually unknown. We are uncovering the mechanism of APC activation and the proteins turned over via the APC during differentiation. For these studies, we are using both PC12 cells and the primary cerebellar granule cell system to probe the role of the APC during differentiation. In addition, in collaboration with J. Hogenesch, Scripps Florida, we are using cellbased screening technologies to identify novel activators and inhibitors of the APC. For this research, we have developed a high-throughput luciferase-based measure of APC activity. In this method, the N terminus of the APC substrate cyclin B is fused with luciferase, so an increase in luciferase corresponds to increased levels of cyclin B, a finding that indicates lower APC activity. We recently began screening 15,000 human cDNAs to identify novel APC regulators. Identification of these regulators most likely will illuminate both temporal and spatial control of APC activity during development. In addition to identifying novel regulators and substrates of the APC, we have concentrated on one of the known APC substrates, the cytosolic protein trigger of mitotic entry 1. This protein is required for degradation of the mitosis-inhibitory kinase wee1 and entry into W Steve A. Kay, Ph.D. Chairman’s Overview he Department of Biochemistry at Scripps Research was recently created to span both the La Jolla and Florida campuses. The overall theme of the department centers on the need to understand physiologic processes from the molecular level through the whole organism. Our faculty members are generally multidisciplinary biologists who wield cutting-edge tools of structural biology, protein dynamics, biological chemistry, genetics and genomics, pathway analysis, and computational approaches to understand how organisms maintain homeostasis and respond to stress. We have broad interests; we seek to answer contemporary questions in neurobiology, metabolic control, immunology, and cancer biology. By taking integrative approaches to substantial problems in modern biology, we will affect the understanding of a wide variety of diseases such as diabetes, cancer and CNS disorders. T Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. BIOCHEMISTRY 2005 mitosis in both frogs and humans. We are determining the intracellular location of wee1 degradation in humans and the role of phosphorylation in wee1 degradation. In collaboration with J. Busby, Scripps Florida, we are using mass spectrometry to identify wee1 phosphorylation sites. This multidisciplinary approach will provide greater insight into proteolysis, the cell cycle, and neurogenesis and may be useful in cancer research and nerve regeneration studies. PUBLICATIONS Ayad, N.G. CDKs give Cdc6 a license to drive into S phase. Cell 122:815, 2005. Ayad, N.G., Rankin, S., Ooi, D., Rape, M., Kirschner, M.W. Identification of ubiquitin ligase substrates by in vitro expression cloning. Methods Enzymol., in press. Rankin, S., Ayad, N.G., Kirschner, M.W. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates [published correction appears in Mol. Cell 18:609, 2005]. Mol. Cell 18:185, 2005. Song, M.S., Song, S.J., Ayad, N.G., Chang, J.S., Lee, J.H., Hong, H.K., Choi, N., Kim, J., Kim, H., Kim, J.W., Choi, E.J., Kirschner, M.W., Lim, D.S. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat. Cell Biol. 6:129, 2004. Wei, W., Ayad, N.G., Wan, Y., Zhang, G., Kirschner, M.W., Kaelin, W.G., Jr. Destruction of the SCF component Skp2 in G1 by the anaphase-promoting complex. Nature 428:194, 2004. 349 molecule libraries, to identify all of the proteins that make up and regulate the cAMP pathway. Using this technology, we successfully identified proteins called transducers of regulated CREB activity, a novel family of CREB coactivators. Ascertaining the factors involved in the cAMP signaling pathway will be paramount in delineating why the biological function of CREB differs so drastically between tissues. PUBLICATIONS Conkright, M.D., Montminy, M. CREB: the unindicted cancer co-conspirator. Trends Cell Biol. 15:457, 2005. Screaton, R.A., Conkright, M.D., Katoh, Y., Best, J.L., Canettieri, G., Jeffries, S., Guzman, E., Niessen, S., Yates, J.R. III, Takemori, H., Okamoto, M., Montminy, M. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119:61, 2004. Zhang, X., Odom, D.T., Koo, S.H., Conkright, M.D., Canettieri, G., Best, J., Chen, H., Jenner, R., Herbolsheimer, E., Jacobsen, E., Kadam, S., Ecker, J.R., Emerson, B., Hogenesch, J.B., Unterman, T., Young, R.A., Montminy, M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U. S. A. 102:4459, 2005. Molecular Mechanisms of cAMP-Mediated Transcription Ligand Interactions and Allosteric Control in Nuclear Receptors: Use of Hydrogen-Deuterium Exchange Mass Spectrometry M.D. Conkright, B.A. Mercer P.R. Griffin, S.A. Busby, M.J. Chalmers, S. Prasad gamut of biological functions depend on the cAMP signaling cascade, including long-term memory, survival of beta cells, glucose metabolism, cardiomyopathy, and proliferation of chondrocytes. We study the molecular mechanisms involved in the conversion of these signals to transcriptional events. Increases in cellular levels of cAMP stimulate the expression of numerous genes by liberating the catalytic subunits of protein kinase A, which phosphorylates the transcription factor cAMP-responsive element binding protein (CREB). Phosphorylation of CREB promotes the recruitment of the coactivator CREB-binding protein/p300 and the initiation of transcription. The diversity of biological functions associated with CREB and the cAMP pathway will be impossible to fully understand until all of the components involved in the pathway have been identified and characterized. Currently, we are using high-throughput cell-based screening technologies, including cDNA expression libraries, small interfering RNA libraries, and small- eroxisome proliferator-activated receptor γ (PPARγ), a ligand-dependent transcription factor and member of the nuclear hormone superfamily, is the molecular target of the drug class known as the glitazones. These compounds are used to treat muscle insulin resistance, a sign or cause of type 2 diabetes, and are referred to as insulin sensitizers. The glitazones are widely prescribed for treatment of type 2 diabetes. However, they have limited usefulness in patients with mild insulin resistance or a history of cardiovascular disease because the drugs are associated with specific receptor-mediated side affects, such as weight gain, fluid retention, and plasma volume expansion. Unfortunately, the incidence of cardiovascular disease is high in patients with type 2 diabetes. In addition, a clear link exists between body mass index and the incidence of insulin resistance and type 2 diabetes. Thus, the search continues for a PPARγ modulator that increases muscle insulin sensitivity without causing weight gain and plasma volume expansion. A Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. P 350 BIOCHEMISTRY 2005 Recent studies with animal models of insulin resistance indicated that weight gain and plasma volume expansion can be minimized without loss of insulin sensitization by using partial agonists of PPARγ, although the mechanism of this dissociation of efficacy from unwanted events is unclear. Studies with preadipocytes revealed that these partial agonists do not induce adipogenesis as the full agonists do, and expression profiling has shown that the gene expression patterns are different for the different functional classes of activators. Currently, we are studying the mechanism of activation of PPARγ by partial agonists and the role of conformation-mediated recruitment of coactivators in adipogenesis. In addition, we are developing a rapid, sensitive structure-based approach to aid in selecting conformational-specific modulators of PPARγ. Amide hydrogen-deuterium exchange coupled with proteolysis and mass spectrometry has become a powerful technique for studying protein structure and dynamics, protein-ligand interactions, and protein-protein interactions. With this method, we can measure changes in the solvent accessibility and stability of a protein in the presence and absence of ligands. We are using hydrogen-deuterium exchange mass spectrometry to detect compound-specific conformational stabilization within the ligand-binding domain of PPARγ. Ligand binding to the domain alters amide hydrogen-deuterium exchange rates in specific regions of the protein with differential magnitude and direction, depending on the chemical structure of the ligand. We have shown that perturbations in hydrogendeuterium exchange can be used to classify agonists, partial agonists, and antagonists of PPARγ. More importantly, these hydrogen-deuterium exchange profiles indicate that the mechanism of activation of PPARγ by full agonists, such as the glitazones, which involves recruitment of coactivators via stabilization of helix-12 and the AF2 surface of the ligand-binding domain, is different for certain classes of partial agonists. We are continuing our investigations of the mechanism of activation of PPARγ by diverse chemotypes of PPARγ partial agonists. Our overall goal is to determine the relationship between ligand-induced receptor conformation and pharmacologic response in rodent models of type 2 diabetes. To date, we have identified localized regions of PPARγ that provide sensors specific for binding and conformational induction for specific chemotypes. Differential recruitment of coactivators by various chemotypes of PPARγ modulators could be integral to Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. the mechanism of activation of PPARγ and its associated physiologic response. Using 2-hybrid assays, we are screening libraries of nuclear receptor modulators against a library of coactivator genes to identify the molecular determinants of coactivator selectivity for these compounds. We have also used hydrogen-deuterium exchange mass spectrometry to study other biological systems. In collaboration with D. Silverman, University of Florida, Gainesville, Florida, we are examining differences in exchange rates between the dimer and tetramer interfaces of manganese superoxide dismutase. Our goal is to understand how this protein regulates the conversion of superoxide to oxygen and hydrogen peroxide to protect cells against oxidative damage. In other studies with G. Fields, Florida Atlantic University, Boca Raton, Florida, we are using hydrogen-deuterium exchange to examine the mechanisms of both unwinding and cleaving of triple helical peptides by matrix metalloprotease 1, an enzyme involved in the metastasis of breast cancer and the progression of arthritis. Finally, in collaboration with W. Cance at the University of Florida, we are using hydrogen-deuterium exchange to characterize interactions for peptides that bind to focal adhesion kinase and have anticancer activity in cell cultures. Research is also under way to improve the technical aspects of the hydrogen-deuterium exchange method. We have developed a fully automated system for hydrogen-deuterium exchange experiments. An autosampler is used to mix solutions and control the online enzymatic digestion of the protein being analyzed and the subsequent separation of the products. Importantly, we have written unique software that enables us to measure subsecond hydrogen-deuterium exchange rates. To increase the dynamic range of the proteins and complexes that we can study by using hydrogen-deuterium exchange, we are collaborating with A.G. Marshall at the National High Magnetic Field Laboratory at Florida State University, Tallahassee, Florida. Dr. Marshall and his colleagues are widely acknowledged as the world leaders in the development of Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS). FT-ICR MS provides the highest resolving power and mass accuracy of any mass analyzer (100-fold greater than our current ion-trap mass spectrometer). The resolution of FT-ICR MS enables us to follow the deuterium uptake of more proteolytic peptides during hydrogendeuterium exchange, and the improved mass accuracy BIOCHEMISTRY 2005 351 increases the likelihood that the peptide-sequence assignments are correct. The Scripps Florida robot used for automated preparation of samples for hydrogen-deuterium exchange was interfaced to a 14.5-T FT-ICR mass spectrometer at the National High Magnetic Field Laboratory. The custombuilt 14.5-T magnet is the highest field magnet ever constructed for use in FT-ICR MS. Using this system, we determined the hydrogen-deuterium exchange of a complex consisting of the ligand-binding domains of PPARγ and retinoic X receptor α in the presence and absence of a PPARγ full agonist and retinoic acid. The results are the first known hydrogen-deuterium exchange analysis of how drug binding affects the structure of an entire protein complex, rather than a single protein. The automation of the hydrogen-deuterium exchange method creates an absolute requirement for an integrated data management and analysis system. A single study may generate up to 40 Gb of raw data, which must be archived and processed. No such commercial solution exists. In collaboration with N. Tsinoremas, C. Mader, B. Pascal, and J. Zysmann, Scripps Florida, we are developing integrated data analysis software and computing infrastructure to control workflow, storage, and analysis of data obtained in hydrogen-deuterium exchange experiments. This system is fully integrated with our laboratory information management system. When completed, the software, named HD Desktop, will be offered to academic laboratories as an open-source solution. Cell-Based Screening and the Mammalian Circadian Clock J.B. Hogenesch, T.K. Sato, J.E. Baggs, K.R. Hayes, J.M. Geskes, J.N. Harada ompletion of the sequencing of several mammalian genomes has revealed a surprisingly sparse cohort of genes that encode proteins. Despite this unexpectedly limited repertoire, most of these genes have never been reported in the literature. To bridge this gap between gene identification and an understanding of their functions, we developed high-throughput methods to study the function of genes in cellular pathways. These methods work on the basis of 2 principles: gain of function and loss of function. For gain-of-function screens, we overexpress 16,000+ distinct mouse and human cDNAs individually in cells and study the consequential pheno- C Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. type of interest. For loss-of-function screens, we use short interfering RNAs to suppress expression of specific mRNAs and then analyze the resultant cellular phenotypes. We currently have a library of 28,000 short interfering RNAs designed to reduce transcript levels of 7000 human genes that can be targeted by pharmacologic drugs or protein therapeutic agents. Because of the generic nature of the screens, many different types of cell-based screens can be performed, including cell proliferation, use of easily measured output of light, and even high-content imaging, in which individual cells are photographed and scored for distinct phenotypes. Thus, we can study the function of a substantial part of the mammalian genome by using powerful cell-based assays and can determine the cellular roles for many previously poorly or unannotated genes. We are using these methods to study the circadian clock in mammals. Circadian rhythms are conserved from cyanobacteria to Neurospora to Drosophila to mice to humans; each organism regulates aspects of its physiology to be in tune with the environment. In mammals, complex behaviors such as locomotor activity and physiologic events such as heme biosynthesis, hormonal signaling, and temperature rhythms are regulated by the clock. Interestingly, in all of the organisms mentioned, the clock itself is regulated at the level of transcription. Several transcription factors conserved between flies, mice, and humans have been identified as clock components. Along with some accessory factors, the genes for these components work in concert to generate molecular rhythms of transcription with a period length of 24 hours. The entire pathway exists not only in the brain, the site of the master circadian oscillator, but also in peripheral tissues and even individual cells. We have adapted a cell line model of clock function in order to use molecular genetic tools long applied in research on bacteria and yeasts. We are using gain- and loss-of-function alleles of the proteins Clock, Bmal, and cryptochrome in screens to examine clock function. Evaluation of the alleles in cellular assays revealed the absolute requirement of transcriptional repression for clock function. The power of mammalian cellular genetics has rapidly matured during the past several years. We have used these tools and techniques not only to study the circadian clock in mammals but also to understand the function of noncoding RNAs, numerous cellular signaling pathways, and basic cellular processes such as proliferation and differentiation. Potentially, these tools can be 352 BIOCHEMISTRY 2005 used to uncover the functions of many important and understudied genes and can lead the way to the next generation of therapeutic drug targets. As such, the tools are an important addition to the existing repertoire used in studies of gene function. PUBLICATIONS Harada, J.N., Bower, K.E., Orth, A.P., Callaway, S., Nelson, C.G., Laris, C., Hogenesch, J.B., Vogt, P.K., Chanda, S.K. Identification of novel mammalian growth regulatory factors by genome-scale quantitative image analysis. Genome Res. 15:1136, 2005. Sato, T.K., Panda, S., Miraglia, L.J., Reyes, T.M., Rudic, R.D., McNamara, P., Naik, K.A., FitzGerald, G.A., Kay, S.A., Hogenesch, J.B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43:527, 2004. Su, A.I., Wiltshire, T., Batalov, S., Lapp, H., Ching, K.A., Block, D., Zhang, J., Soden, R., Hayakawa, M., Kreiman, G., Cooke, M.P., Walker, J.R., Hogenesch, J.B. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 101:6062, 2004. Willingham, A.T., Orth, A.P., Batalov, S., Peters, E.C., Wen, B.G., Aza-Blanc, P., Hogenesch, J.B., Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309:1570, 2005. Genetic Determinants of the Efficacy of Selective Serotonin Reuptake Inhibitors M.T. Pletcher, B.H. Miller, B.M. Young, G.M. Zastrow linical depression is a mood disorder of high morbidity and mortality that is estimated to occur in more than 15% of the adult population in the United States. Depression has a wide range of symptoms, including loss of energy, changes in weight, diminished interest or pleasure in daily activities, insomnia or excessive sleep, anxiety, slowness of movement, feelings of worthlessness, difficulty concentrating, and thoughts of death. Depression can be a one-time occurrence but is often an ongoing problem throughout a person’s lifetime. A total 15% of patients with major depressive disorders die of suicide. The onset of depression is often linked to environmental factors such as life events that greatly increase stress. Despite this association, depression has a strong genetic component. Genetics accounts for 40%–50% of the risk for of depression in a person’s lifetime, and the risk for members of a family does not change if they are raised separately. We are using cell-based screening technology and a mouse model to identify the genes and pathways that contribute to depressive behavior. Currently, we are con- C Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. ducting a survey in 30 strains of mice for differences in molecular, biochemical, and behavioral traits that represent endophenotypes (i.e., single well-defined symptoms or traits of a much more complex disorder) associated with depression. In cataloging the variations in quantities of neurotransmitters and stress hormones in the blood, gene expression in regions of the brain such as the hippocampus and hypothalamus, and performance in behavioral models such as the tail suspension and open field tests, we are producing the data necessary to identify the genetic controls for each of these traits. Using the in silico genetic mapping method, we can correlate the natural variation for these measurements in the inbred lines of mice with the underlying haplotype structure of the mice, thereby pinpointing biologically important genes. In parallel, we are developing a cell-based assay to monitor the function of the serotonin transporter, the gene target of the most common pharmaceutical agents for depression: selective serotonin reuptake inhibitors. Using this assay, we can screen the 18,000 full-length clone cDNA library of Scripps Florida for genes that modify the function of the transporter. Additionally, introducing a selective serotonin reuptake inhibitor into the screen will enable us to identify genes that modulate the efficacy of this important class of drugs. Cumulatively, we hope to provide a better understanding of the molecular basis of depression and its treatment to allow improvements in diagnosis and therapies. PUBLICATIONS Bystrykh, L., Weersing, E., Dontje, B., Sutton, S., Pletcher, M.T., Wiltshire, T., Su, A.I., Vellenga, E., Wang, J., Manly, K.F., Lu, L., Chesler, E.J., Alberts, R., Jansen, R.C., Williams, R.W., Cooke, M.P., de Haan, G. Uncovering regulatory pathways that affect hematopoietic stem cell function using “genetical genomics.” Nat. Genet. 37:225, 2005. Kahlem, P., Sultan, M., Herwig, R., Steinfath, M., Balzereit, D., Eppens, B., Saran, N.G., Pletcher, M.T., South, S.T., Stetten, G., Lehrach, H., Reeves, R.H., Yaspo, M.L. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of Down syndrome. Genome Res. 14:1258, 2004. Pletcher, M., Wiltshire, T. Can we find the genes involved in complex traits? Genome Biol. 5:347, 2004. Pletcher, M.T., McClurg, P., Batalov, S., Su, A.I., Barnes, S.W., Lagler, E., Korstanje, R., Wang, X., Nusskern, D., Bogue, M.A., Mural, R.J., Paigen, B., Wiltshire, T. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2:e393, 2004. Sandberg, M.L., Sutton, S.E., Pletcher, M.T., Wiltshire, T., Tarantino, L.M., Hogenesch, J.B., Cooke, M.P. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev. Cell 8:153, 2005. Sussan, T.E., Pletcher, M.T., Murakami, Y., Reeves, R.H. Tumor suppressor in lung cancer 1 (TSLC1) alters tumorigenic growth properties and gene expression. Mol. Cancer 4:28, 2005. Wen, B.G., Pletcher, M.T., Warashina, M., Choe, S.H., Ziaee, N., Wiltshire, T., Sauer, K., Cooke, M.P. Inositol (1,4,5) trisphosphate 3 kinase B controls positive selection of T cells and modulates Erk activity. Proc. Natl. Acad. Sci. U. S. A. 101:5604, 2004. BIOCHEMISTRY 2005 353 Functional Neuroanatomy of Appetite and Metabolism PUBLICATIONS Sato, T.K., Panda, S., Miraglia, L.J., Reyes, T.M., Rudic, R.D., McNamara, P., Naik, K.A., FitzGerald, G.A., Kay, S.A., Hogenesch, J.B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43:527, 2004. T.M. Reyes, K.J. Clarke, C.D. Easson Sawchenko, P.E., Yuan, Z.F., Reyes, T.M. The corticotropin-releasing factor family of signaling molecules and their roles in adaptive responses to stress. J. Histochem. Cytochem. 52:S14, 2004. besity is an important risk factor in the development of heart disease, diabetes, stroke, and metabolic syndrome, and the prevalence of obesity is increasing in all segments of the population. Development and resolution of obesity involve an interaction between genetic, environmental, and behavioral factors, and the role of the CNS in this process is the focus of our research program. We are defining the CNS pathways that regulate appetite and metabolism. Our central goals are to define CNS molecules that respond to and drive alterations in appetite and metabolism and to characterize the expression and regulation of these molecules. To this end, we couple functional neuroanatomic techniques with genomics and high-throughput screening tools in genetic and disease models. One focus is the loss of appetite and alterations in metabolism that occur in acute and chronic infection or inflammation. Plasma cytokines are elevated during infection and inflammation, and these endocrine signaling molecules act on the brain and are associated with a loss of appetite and changes in metabolism. Studies are under way to identify regions of the brain that drive the primary response to cytokines and to identify the neuropeptides and neurotransmitters recruited to effect these changes. The ability to site specifically alter gene expression in the brain is an important tool for these studies. We are developing animals in which the genes for various neuropeptides and adipokines are expressed conditionally in a Cre-LoxP system. Cre recombinase is introduced either through crosses with mice that express Cre or through stereotaxic delivery of lentiviruses that express Cre recombinase. In a related series of studies, we are investigating the link between systemic infection and obesity. Adverse conditions in utero can increase the risk for obesity. Little is known about the mechanisms that underlie this developmental programming. We are using RNA profiling studies to determine whether epigenetic modifications as a result of events in utero (either nutritional or immune) can exacerbate the development of obesity. Our research combines the power of whole-genome profiling with detailed functional neuroanatomic analyses of single molecules. Collectively, these studies should contribute to understanding how the CNS and inflammation interact in the development of obesity. O Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Synthesis of Natural Products, Development of Synthetic Methods, and Medicinal Chemistry W.R. Roush, R. Bates, Y.-T. Chen, T. Dineen,* E. Flamme,* G. Halvorsen, M. Handa, C. Heitzman,* C.-W. Huh, L. Julian,* W. Lambert,* R. Lira, C. Liu,* D. Mergott,* E. Mertz,* J. Methot,* J. Neitz,* C. Nguyen, M. Ober, P. Orahovats,* R. Owen,* R. Pragani, J. Qi,* J.B. Shotwell,* A. Sorg, L. Steffans, K. Takao,** J. Tinsley,* M. Tortosa, T. Trullinger,* P. Va, A. Williams, S. Winbush, N. Zheng* * University of Michigan, Ann Arbor, Michigan ** Keio University, Yokohama, Japan ur research has 2 major themes. One is the synthesis of structurally complex, biologically active natural products (Fig. 1). These efforts in total synthesis are pursued in parallel with stereochemical studies and the development of new synthetic methods. We have been particularly interested in stereochemical aspects of intramolecular and transannular Diels-Alder reactions, in the development of methods for the diastereoselective and enantioselective reactions of allylmetal reagents with carbonyl compounds, and in the development of highly stereoselective methods for synthesis of 2-deoxyglycosides. Recent efforts to develop new synthetic methods included studies on the Diels-Alder reactions of (Z)-dienes, double allylboration reactions of aldehydes with γ-boryl-substituted allylboranes for stereocontrolled synthesis of 1,5-endiols, synthesis of highly substituted tetrahydrofurans via [3 + 2]-annulation reactions of highly functionalized allylsilanes, and use of 2-deoxy-2-iodoglycosyl imidates and 2-deoxy-2-iodoglycosyl fluorides for the highly stereocontrolled synthesis of 2-deoxy-β-glycosides. Natural products of current interest to us include amphidinolides C and F, amphidinol 3, angelmicin B, annonaceous acetogenin analogs, durhamycin A and analogs, integramicin, lomaiviticin A, peloruside A, psymberin, quartromicin D1, reidispongiolide A, spin- O 354 BIOCHEMISTRY 2005 agent of Chagas’ disease, and Plasmodium falciparum, the most virulent of the malaria parasites. This research is performed in collaboration with colleagues at the University of California, San Francisco. We have also participated in the development of proapoptotic benzodiazepines, in collaboration with G.D. Glick, University of Michigan, and the design of novel heterocycles targeting HIV. New projects at Scripps Florida involve discovery of small molecules that affect cancer and other validated targets, studies of the structure-activity relationship of certain natural products, and optimization of the pharmacologic profiles of natural products. PUBLICATIONS Dineen, T.A., Roush, W.R. Stereoselective synthesis of the octahydronaphthalene unit of integramycin via an intramolecular Diels-Alder reaction. Org. Lett. 7:1355, 2005. Dineen, T.A., Roush, W.R. Total synthesis of cochleamycin A. Org. Lett. 6:2043, 2004. Durham, T.B., Blanchard, N., Savall, B.M., Powell, N.A., Roush, W.R. Total synthesis of formamicin. J. Am. Chem. Soc. 126:9307, 2004. Flamme, E.M., Roush, W.R. Synthesis of the C(1)-C(25) fragment of amphidinol 3: application of the double-allylboration reaction for synthesis of 1,5-diols. Org. Lett. 7:1411, 2005. Flamme, E.M., Roush, W.R. Synthesis of 2,6-trans-disubstituted 5,6-dihydropyrans from (Z)-1,5-syn-endiols. Beilstein J. Org. Chem. [serial online] 1:7, 2005. Available at: http://bjoc.beilstein-journals.org/. Heitzman, C.L., Lambert, W.T., Mertz, E., Shotwell, J.B. Tinsley, J.M., Va, P., Roush, W.R. Efficient protiodesilylation of unactivated C(sp3)-SiMe2Ph bonds using tetrabutylammonium fluoride. Org. Lett. 7:2405, 2005. Julian, L.D. Newcom, J.S., Roush, W.R. Total synthesis of (+)-13-deoxytedanolide. J. Am. Chem. Soc. 127:6186, 2005. Liu, C.L. Smith, W.J. III, Gustin, D.J., Roush, W.R. Experimental evidence for chair-like transition states in aldol reactions of methyl ketone lithium enolates: stereoselective synthesis and utilization of a deuterium-labeled enolate as a probe of reaction stereochemistry. J. Am. Chem. Soc. 127:5770, 2005. Mergott, D.J., Frank, S.A., Roush, W.R. Total synthesis of (–)-spinosyn A. Proc. Natl. Acad. Sci. U. S. A. 101:11955, 2004. Mertz, E., Tinsley, J.M., Roush, W.R. [3 + 2]-Annulation reactions of chiral allylsilanes and chiral aldehydes: studies on the synthesis of bis-tetrahydrofuran substructures of annonaceous acetogenins. J. Org. Chem. 70:8035, 2005. F i g . 1 . Structures of recently synthesized natural products. osyn A biosynthetic intermediates, scytophycin C, superstolide A, tedanolide, and tetrafibricin. We selected these molecules as targets because of their biological properties and their interesting and complex structures. We place a significant emphasis on the discovery, development, and/or illustration of new reactions and synthetic methods for achieving high levels of stereochemical control in each of these synthesis efforts. Our second major area of interest focuses on problems in bioorganic chemistry and medicinal chemistry. One long-term project involves the design and synthesis of inhibitors of cysteine proteases isolated from tropical parasites, such as Trypanosoma cruzi, the causative Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Methot, J.L., Roush, W.R. Nucleophilic phosphine organocatalysis. Adv. Synth. Catal. 346:1035, 2004. Narayan, S., Roush, W.R. Studies toward the total synthesis of angelmicin B (hibarimicin B): synthesis of a model CD-D′ arylnaphthoquinone. Org. Lett. 6:3789, 2004. Owen, R.M., Roush, W.R. Stereoselective synthesis of the C(1)-C(11) fragment of peloruside A. Org. Lett. 7:3941, 2005. Roush, W.R., Neitz, R.J. Studies on the synthesis of landomycin A: synthesis of the originally assigned structure of the aglycone, landomycinone, and revision of structure. J. Org. Chem. 69:4906, 2004. Shotwell, J.B., Roush, W.R. Synthesis of the C11-C29 fragment of amphidinolide F. Org. Lett. 6:3865, 2004. Tinsley, J.M., Mertz, E., Chong, P.Y., Rarig, R.-A.F., Roush, W.R. Synthesis of (+)bullatacin via the highly diastereoselective [3 + 2] annulation reaction of a racemic aldehyde and a nonracemic allylsilane. Org. Lett. 7:4245, 2005. Tinsley, J.M., Roush, W.R. Total synthesis of asimicin via highly stereoselective [3 + 2] annulation reactions of substituted allylsilanes. J. Am. Chem. Soc. 127:10818, 2005. BIOCHEMISTRY 2005 Genome Science and CNS Drug Discovery C. Wahlestedt, P. Kenny, P. McDonald, M.A. Faghihi, J. Kocerha, M. Przydzial, H. Thonberg,* H.-Y. Zhang,* T. Andersson,* O. Larsson,* L. Huminiecki,* C. Schéele,* C. Dahlgren,* P. Georgii-Hemming,* K. Wennmalm,* Z. Liang,* J.A. Timmons* * Karolinska Institutet, Stockholm, Sweden I D E N T I F I C AT I O N A N D F U N C T I O N A L A N A LY S I S O F NONCODING AND ANTISENSE TRANSCRIPTS ost likely conventional protein-coding genes account for only a minority of human RNA transcripts. A substantial component of the full-length mouse and human cDNA sets that we and others have analyzed does not contain an annotated protein-coding sequence and most likely corresponds to noncoding RNA. Many of the noncoding sequences constitute natural antisense RNA transcripts. We have shown that the majority of noncoding RNAs identified to date have substantial conservation across species. Moreover, we have shown that many noncoding RNA and antisense transcripts have differential expression under various conditions and can affect conventional gene expression. M RNA INTERFERENCE AND DEVELOPMENT OF HIGHTHROUGHPUT GENOMICS TECHNOLOGY RNA interference has become one of the most important gene manipulation technologies. Short interfering RNA, the inducer of RNA interference in mammals, can be used to elucidate gene functions by rapidly silencing expression of a target gene. Today, short interfering RNAs are widely used as research tools and have potential for becoming therapeutic agents. We have built a portfolio of short interfering RNA technology. In this package we have a powerful short interfering RNA vector system, a validation system, and a design system, all of which are unique. Combining these technologies with the high-throughput chemistry for on-chip DNA synthesis, we have set up a system for constructing short interfering RNAs. Finally, we have also introduced the use of locked nucleic acids in short interfering RNAs and have shown a range of beneficial properties of these agents. G P R O T E I N – C O U P L E D R E C E P T O R S A S D R U G TA R G E T S More than half of known drugs bind to G protein– coupled receptors (GPCRs). We have continued our long-standing work on GPCRs, particularly certain neuropeptide receptors. At Scripps Florida, these efforts Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 355 are forming part of the drug discovery program. Moreover, during the past year, we published on our human GPCR database, which contains polymorphism data, that is, data that point to interindividual differences in GPCR sequences. Information on polymorphisms may be increasingly important in drug discovery and development efforts. HUMAN GENETICS AND PHARMACOGENOMICS We are pursuing drug discovery related to several human disorders that affect the brain. Our goal is to identify biomarkers that are associated with such common disorders. We wish to understand what makes certain individuals susceptible and how their responses to drug treatment may differ (pharmacogenomics). We are involved in genotyping DNA from patients with major depression, alcoholism, diabetes, obesity, Alzheimer’s disease, Parkinson’s disease, and attention deficit/hyperactivity disorder. PUBLICATIONS Carninci, P., Kasukawa, T., Katayama, S., et al. The transcriptional landscape of the mammalian genome. Science 309:1559, 2005. Chalk, A.M., Wahlestedt C., Sonnhammer E.L.L. Improved and automated prediction of effective siRNA. Biochem. Biophys. Res. Commun. 319:264, 2004. Chen, M., Zhang, L., Zhang, H.Y., Xiong, X., Wang, B., Du, Q., Lu, B., Wahlestedt, C., Liang, Z. A universal plasmid library encoding all permutations of small interfering RNA. Proc. Natl. Acad. Sci. U. S. A. 102:2356, 2005. Ding, B., Kull, B., Liu, Z., Mottagui-Tabar, S., Thonberg, H., Gu, H.F., Brookes, A.J., Grundemar, L., Karlsson, C., Hamsten, A., Arner, P., Ostenson, C.G., Efendic, S., Monne, M., von Heijne, G., Eriksson, P., Wahlestedt, C. Human neuropeptide Y signal peptide gain-of-function polymorphism is associated with increased body mass index: possible mode of function. Regul Pept. 127:45, 2005. Du, Q., Thonberg, H., Wang, J., Wahlestedt, C., Liang, Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites [published correction appears in Nucleic Acids Res. 33:3698, 2005]. Nucleic Acids Res. 33:1671, 2005. Du, Q., Thonberg, H., Zhang, H.-Y., Wahlestedt, C., Liang, Z. Validating siRNA using a reporter made from synthetic DNA oligonucleotides. Biochem. Biophys. Res. Commun. 325:243, 2004. Elmén, J., Thonberg, H., Ljungberg, K., Frieden, M., Westergaard, M., Xu, Y., Wahren, B., Liang, Z., Ørum, H., Koch, T., Wahlestedt, C. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 33:439, 2005. Elmén J., Wahlestedt, C., Brytting, M., Wahren, B., Ljungberg, K. SARS virus inhibited by siRNA. Preclinica 2:135, 2004. Elmén, J., Zhang, H.-Y., Zuber, B., Ljungberg, K., Wahren, B., Wahlestedt, C., Liang, Z. Locked nucleic acid containing antisense oligonucleotides enhance inhibition of HIV-1 genome dimerization and inhibit virus replication. FEBS Lett. 578:285, 2004. Faghihi, M.A., Mottagui-Tabar, S., Wahlestedt, C. Genetics of neurological disorders. Expert Rev. Mol. Diagn. 4:317, 2004. Gu, H.F., Abulaiti, A., Östenson, C.-G., Humphreys, K., Wahlestedt, C., Brookes, A.J., Efendic, S. Single nucleotide polymorphisms in the proximal promoter region of the adiponectin (APM1) gene are associated with type 2 diabetes in Swedish Caucasians. Diabetes 53(Suppl.1):S31, 2004. 356 BIOCHEMISTRY 2005 Heilig, M., Zachrisson, O., Thorsell, A., Ehnvall, A., Mottagui-Tabar, S., Sjögren, M., Åsberg, M., Ekman, R., Wahlestedt, C., Ågren, H. Decreased cerebrospinal fluid neuropeptide Y (NPY) in patients with treatment refractory unipolar major depression: preliminary evidence for association with preproNPY gene polymorphism. J. Psychiatr. Res. 38:113, 2004. Isacson, R., Kull, B., Wahlestedt, C., Salmi, P. A 68930 and dihydrexidine inhibit locomotor activity and d-amphetamine-induced hyperactivity in rats: a role of inhibitory dopamine D1/5 receptors in the prefrontal cortex? Neuroscience 124:33, 2004. Katayama, S., Tomaru, Y., Kasukawa, T., Waki, K., Nakanishi, M., Nakamura, M., Nishida, H., Yap, C.C., Suzuki, M., Kawai, J., Suzuki, H., Carninci, P., Hayashizaki, Y., Wells, C., Frith, M., Ravasi, T., Pang, K.C., Hallinan, J., Mattick, J., Hume, D.A., Lipovich, L., Batalov, S., Engstrom, P.G., Mizuno, Y., Faghihi, M.A., Sandelin, A., Chalk, A.M., Mottagui-Tabar, S., Liang, Z., Lenhard, B., Wahlestedt, C.; RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science 309:1564, 2005. Kemmer, D., Faxén, M., Hodges, E., Lim, J., Herzog, E., Ljungström, E., Lundmark, A., Olsen, M.K., Podowski, R., Sonnhammer, E.L.L., Nilsson, P., Reimers, M., Lenhard, B., Roberds, S.L., Wahlestedt, C., Höög, C., Agarwal, P., Wasserman, W.W. Exploring the foundation of genomics: comparative analysis of transcript profiling technologies. Comp. Funct. Genomics 5:584, 2004. Larsson, O., Schéele, C., Liang, Z., Moll, J., Karlsson, C., Wahlestedt, C. Kinetics of senescence-associated changes of gene expression in an epithelial, temperaturesensitive SV40 large T antigen model. Cancer Res. 64:482, 2004. Larsson, O., Wahlestedt, C., Timmons, J.A. Considerations when using the significance analysis of microarrays (SAM) algorithm. BMC Bioinformatics 6:129, 2005. Mottagui-Tabar, S., Faghihi, M.A., Mizuno, Y., Engstrom, P.G., Lenhard, B., Wasserman, W.W., Wahlestedt, C. Identification of functional SNPs in the 5-prime flanking sequences of human genes. BMC Genomics 6:18, 2005. Mottagui-Tabar, S., McCarthy, S., Reinemund, J., Andersson, B., Wahlestedt, C., Heilig, M. Analysis of 5-hydroxytryptamine 2c receptor gene promoter variants as alcohol-dependence risk factors. Alcohol. 39:380, 2004. Pang, K.C., Stephen, S., Engström, P.G., Tajul-Arifin, K., Chen, W., Wahlestedt, C., Lenhard, B., Hayashizaki, Y., Mattick, J.S. RNAdb: a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 33:D125, 2005. Thonberg, H., Dahlgren, C., Wahlestedt, C. Antisense-induced Fas mRNA degradation produces site-specific stable 3′-mRNA fragment by endonuclease cleavage at the complementary sequence. Oligonucleotides 14:221, 2004. Thonberg, H., Schéele, C.C., Dahlgren, C., Wahlestedt, C. Characterization of RNA interference in rat PC12 cells: requirement of GERp95. Biochem. Biophys. Res. Commun. 318:927, 2004. Timmons, J.A., Larsson, O., Jansson, E., Fischer, H., Gustafsson, T., Greenhaff, P.L., Ridden, J., Rachman, J., Peyrard-Janvid, M., Wahlestedt, C., Sundberg, C.J. Human muscle gene expression responses to endurance training provide a novel perspective on Duchenne muscular dystrophy. FASEB J. 19:750, 2005. Wahlestedt, C., Brookes, A.J., Mottagui-Tabar, S. Lower rate of genomic variation identified in the trans-membrane domain of monoamine sub-class of human G-protein coupled receptors: the Human GPCR-DB Database. BMC Genomics 5:91, 2004. Xu, Y., Linde, A., Larsson, O., Thormeyer, D., Elmén, J., Wahlestedt, C., Liang, Z. Functional comparison of single- and double-stranded siRNAs in mammalian cells. Biochem. Biophys. Res. Commun. 316:680, 2004. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Infectology Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Chris Baker, Ph.D., Research Associate, Department of Infectology Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. INFECTOLOGY 2005 DEPAR TMENT OF INFECTOLOGY SENIOR RESEARCH A S S O C I AT E S S TA F F Carlos Coito, Ph.D. Charles Weissmann, M.D., Ph.D. Professor and Chairman Corinne Lasmezas, Ph.D. Professor Vittorio Verzillo, Ph.D. R E S E A R C H A S S O C I AT E S Chris Baker, Ph.D. Donny Strosberg, Ph.D. Professor Tim Tellinghuisen, Ph.D. Assistant Professor Prem Subramaniam, Ph.D. Shawn Browning, Ph.D. S E N I O R S TA F F SCIENTIST Nicole Sales, Ph.D. S TA F F S C I E N T I S T Sukhvir Mahal, Ph.D. S E C T I O N C O V E R F O R T H E D E P A R T M E N T O F I N F E C T O L O G Y : A single, prion- infected neuroblastoma cell labeled with green fluorescent protein is cultured on a feeder layer of uninfected cells. The prions secreted by the single infected cell are quantified by using the scrapie cell assay. The purpose of the project is to determine how prions are propagated in a chronically infected cell culture. Research and photograph by C.A. Baker, Ph.D., Research Associate, in the laboratory of C. Weissmann, M.D., Ph.D. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 359 360 INFECTOLOGY 2005 INVESTIGATORS’ R EPORTS Generation and Transmission of Prions C. Weissmann, C.A. Baker, S.P. Mahal, C. Demczyk, A. Sherman he agents that cause transmissible spongiform encephalopathies such as Creutzfeldt-Jakob disease in humans, bovine spongiform encephalopathy in cattle, and scrapie in sheep are termed prions. The unusual resistance of prions to radiation led early on to the proposal that prions might be devoid of nucleic acid and consist only of protein. The discovery of PrPSc, a protease-resistant protein found only in organisms with transmissible spongiform encephalopathies; the cloning of PrP cDNA and its gene; the recognition that the gene encodes a normal host protein, PrPC, from which PrPSc is derived by conformational rearrangement; and the linkage between the PrP gene and familial prion disease supported the suggestion that an abnormal conformer of PrPC (generically designated as PrP*) is the main or perhaps only constituent of prions. The essential role of PrP in prion diseases was established by the finding that mice lacking the PrP gene were resistant to disease and incapable of propagating prions. The “protein only” hypothesis proposes that the infectious, abnormal conformer of PrP C is propagated autocatalytically; a specific mechanism is suggested by the “seeding hypothesis.” Intriguingly, distinct prion strains, which generate different disease phenotypes, may be associated with the same PrP sequence, suggesting that the phenotypes are encoded by different PrP conformations. Infectivity of prions is classically measured in a bioassay that takes many months to complete and requires large numbers of animals. We created a cell-based assay (scrapie cell assay, SCA) that can be carried out in less than 2 weeks, is at least as sensitive and accurate as the animal assay, and allows the simultaneous processing of hundreds of samples by a semiautomated procedure. It has been proposed that prions consist mainly or entirely of the protease-resistant conformer PrPSc and that they are resistant to heating at 80°C for 60 minutes. Using the SCA, we found, in collaboration with F. Properzi, MRC Prion Unit, London, England, that standard protease digestion or heating at 80°C eliminated T Charles Weissmann, M.D.,Ph.D. Chairman’s Overview he Department of Infectology was founded in 2004, and the first research group focused on prion diseases. These diseases are of interest not only because of the emergence of bovine spongiform encephalopathy (mad cow disease) in the United Kingdom, where transmission to humans has led to more than 150 deaths due to variant Creutzfeldt-Jakob disease, but also because of the unusual properties of the infectious agent, which is unique in lacking a nucleic acid genome. The first priority was to establish a cell-based assay for prions to circumvent the slow, expensive, and inaccurate mousebased bioassay. The cell-based assay is being used to analyze the properties of prions and their propagation, both in cell culture and in cell-free systems. In a second phase, the assay will be adapted for high-throughput screening for drugs capable of blocking prion propagation. Corinne Lasmezas joined the department in the summer of 2005 to direct a second group working on prion diseases. A further research effort is directed toward screening for drugs against leishmaniasis, specifically against J-binding protein, a putative target, that involves Scripps scientists in both California and Florida. A third field of endeavor is hepatitis C; the 2 group leaders in this area are Donny Strosberg and Tim Tellinghuisen, who joined the department in November 2005. The growth of the department is currently restricted by the lack of space; however, the planned construction of a further temporary building should alleviate this bottleneck. T Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. INFECTOLOGY 2005 more than 90% of prion infectivity, indicating that the essential component is a conformer of PrP other than PrP Sc . Again using the SCA, in collaboration with S. Supattapone and N. Deleault, Dartmouth Medical School, Hanover, New Hampshire, we found that a cellfree system containing PrP C and primed with prioninfected brain homogenate increased prion infectivity 6-fold within 8 hours, showing that infectivity can be rapidly amplified in a cell-free system. Only a few cell lines are susceptible to persistent infection by prions, for as yet unknown reasons. The high susceptibility of the subclone N2aPK1 developed for the SCA is unstable, and even after repeated cloning, susceptible populations consist of both highly susceptible and almost resistant cells; susceptibility appears to be determined epigenetically. Similarly, populations of cloned, chronically infected N2aPK1 cells consist of infected and uninfected cells. Interestingly, a nondividing subpopulation is the most prolific in producing and secreting prions, while a dividing subpopulation generates very low levels of prions, perhaps because of the emergence of only a small number of “producer cells” from the population. N2aPK1 cells are, surprisingly, susceptible to a particular murine prion strain (139A) but not to any others tested so far. We are now searching for cell lines that are sensitive to other strains and are striving to identify the features that enable susceptibility to prions in general and to certain strains in particular. A panel of cell lines with distinct susceptibilities will greatly facilitate the currently arduous task of typing of prion strains. 361 maniasis occur in humans: cutaneous, mucocutaneous, and visceral. According to estimates, 1.5 million to 2 million new cases occur annually, mainly in the tropics and subtropics, including the Middle East. An effective, well-tolerated, and inexpensive therapy is greatly needed. The DNA of Leishmania organisms, as well as that of other kinetoplastids, contains a modified base, J (β-D -glucosyl-hydroxymethyluracil), that does not occur in higher eukaryotes. Leishmania species contain a protein, J-binding protein (JBP), that binds to the modified base within the context of a nucleotide sequence and plays a role in the conversion of thymine to J. Deletion of JBP is lethal for the organisms. Therefore, most likely a compound that interferes with the binding of JBP to J would be detrimental to the growth or survival of Leishmania organisms. Because J and JBP do not occur in the host, such a compound might lead to a therapeutic drug. Using a JBP-based approach, members of our broadly based consortium have set out to search for a lead compound, and if proof of principle is achieved, to develop a drug. A fluorescence polarization assay for the binding of a J-containing, fluorescently labeled oligonucleotide to recombinant JBP has been developed and will be optimized for high-throughput screening of libraries of various compounds at Scripps Florida. Host-Virus Protein Interactions in Hepatitis C A.D. Strosberg, C. Coito, S. Kota, D. Willoughby* J-Binding Protein of Leishmania as a Potential Drug Target C. Weissmann, P. Subramaniam, P. Wentworth,* D. Millar,** R. Sabatini,*** P. Borst**** *Department of Chemistry, Scripps Research ** Department of Molecular Biology, Scripps Research *** Marine Biological Laboratory, Woods Hole, Massachusetts **** Netherlands Cancer Institute, Amsterdam, the Netherlands eishmaniasis consists of a group of diseases caused by parasitic protozoans of the genus Leishmania. The pathogens are transmitted by sand flies and can infect skin, mucous membranes, and certain internal organs. Three major types of leish- L Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. * Ocean Ridge Biosciences, Jupiter, Florida epatitis C is caused by a 9.5-kb positive singlestranded RNA virus that encodes 11 proteins and belongs to the family Flaviviridae. Worldwide, an estimated 170 million persons are carriers of hepatitis C virus (HCV), including 3 million in the United States. Liver cirrhosis develops in 20% of those who are persistently infected, and hepatocellular carcinoma develops in up to 2.5% of those who have cirrhosis. No vaccine for HCV is available yet, and the only treatment is the combination of interferon alfa and ribavirin, which is effective in about 50% of all patients. Several alternative drugs in development target the enzymatic activities of the viral protease or the viral RNA-dependent RNA polymerase. However, even if H 362 INFECTOLOGY 2005 these drugs are successful initially, ultimately patients will become resistant to them because of high levels of mutations in the virus, as has occurred with HIV. The development of such resistance would be less likely if drugs acted at the interface of interaction between 2 proteins, especially if one of the proteins was a host cellular protein. Viruses recruit a number of cellular proteins in order to enter cells, integrate cellular organelles (e.g., the endoplasmic reticulum), go into latency, or start assembly and replication inside a cell. At each step, interactions between host and viral proteins are essential, but for HCV, most of these interactions have not yet been elucidated. Previously, a variety of methods, including use of the yeast 2-hybrid system and coprecipitation with antibodies to one of the protein partners, were used by our collaborators to analyze the HCV-HCV and the hepatocyte-HCV interactions. The HCV protein domains involved in several of these interactions were delineated precisely by using multiple fragments of cDNAs that encode various HCV proteins. We are continuing these studies, and we plan to develop compounds that modulate HCVHCV and host-HCV interactions. HCV-HCV PROTEIN INTERACTIONS We have selected several HCV interacting domains for our molecular studies. One of these domains corresponds to the first half of the 191-residue-long capsid protein. This protein polymerizes to form the main structural element to which other HCV proteins and the viral RNA are associated to form complete virions. The N-terminal 84-residue-long domain actually forms homodimers without the need for the rest of the protein. Using either peptide synthesis or expression in Escherichia coli, we prepared forms of these HCV protein domains labeled with biotin, dinitrophenol, glutathione S-transferase, or a short peptide called FLAG. We are now studying interactions between the labeled HCV domains by monitoring energy transfer between fluorescent antibodies that bind to the different labels. The homogeneous time-resolved fluorescence assay that we use for this purpose is sensitive and scalable, so we should be able to measure inhibition of interaction by a variety of peptides and small chemical compounds. From the National Institutes of Health we have obtained a set of 428 synthetic peptides, each of which is 18 residues long, that accounts for the whole HCV proteome. Each peptide will be evaluated for its potential inhibitory capacity. Other, nonpeptidic compounds Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. will be obtained from a variety of sources, including the extensive small-molecule library being assembled at Scripps Florida. Once such HCV inhibitors have been identified and optimized chemically, we will study their effect on HCV grown in hepatoma-derived Huh7.5 and other similar cells. To facilitate titration, we are preparing HCV virions labeled with green flourescent protein or luciferase. KINASES INVOLVED IN HCV PROTEIN INTERACTIONS A N D H C V R E P L I C AT I O N Previous research indicated that a few kinases thought to play a role in HCV replication act on the NS5A viral protein. Using a number of potential kinase inhibitors currently being developed at Scripps Florida and elsewhere, we will study the effects of the molecules on replication of HCV particles in hepatoma cells. The availability of compounds that disrupt protein interactions between viral and human host proteins would help us understand the role of each protein. The compounds might then be used as the basis for drugs to complement direct antiviral and immunomodulatory approaches, thus ultimately providing a broad combination therapy for hepatitis C. PUBLICATIONS Coito, C., Diamond, D.L., Neddermann, P., Korth, M.J., Katze, M.G. High-throughput screening of the yeast kinome: identification of human serine/threonine protein kinases that phosphorylate the hepatitis C virus NS5A protein. J. Virol. 78:3502, 2004. Nouet, S., Amzallag, N., Li, J.M., Louis, S., Seitz, I., Cui, T.X., Alleaume, A.M., Di Benedetto, M., Boden, C., Masson, M., Strosberg, A.D., Horiuchi, M., Couraud, P.O., Nahmias, C. Trans-inactivation of receptor tyrosine kinases by novel angiotensin II AT2 receptor-interacting protein, ATIP. J. Biol. Chem. 279:28989, 2004. Translational Research Institute Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Claes Wahlestedt, Ph.D., Director, CNS Discovery Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. TRANSL ATIONAL RESEARCH INSTITUTE 2005 T R A N S L AT I O N A L RESEARCH INSTITUTE Layton H. Smith, Ph.D. Associate Scientific Director, Discovery Biology R E S E A R C H A S S O C I AT E S I N F O R M AT I C S S TA F F John B. Bruning, Ph.D. Mohammad Fallahi-Sichani Nicholas F. Tsinoremas, Ph.D. Senior Director, Informatics Yen Ting Chen, Ph.D. Mark M. Gosink, Ph.D. Julie Conkright, Ph.D. Christopher C. Mader Claes Wahlestedt, M.D., Ph.D.* Director, CNS Disorders Jeffrey E. Habel, Ph.D. Bruce D. Pascal Yuanjun He, Ph.D. Stephan Schuerer, Ph.D. Rong Jiang, Ph.D. Mark R. Southern S TA F F Patrick R. Griffin, Ph.D.* Head, Drug Discovery Jennifer C. Busby, Ph.D. Associate Scientific Director, Protein Sciences and Proteomics Michael Cameron, Ph.D. Laboratory Head, Drug Metabolism and Pharmacokinetics SENIOR SCIENTISTS Magdalena Przydzial, Ph.D. Thomas D. Bannister, Ph.D. Derek R. Duckett, Ph.D. Alessandra Cervino, Ph.D. Informatics Assistant Professor Marcel Koenig, Ph.D. Louis D. Scampavia, Ph.D. Yangbo Feng, Ph.D. Associate Director, Medicinal Chemistry S E N I O R S TA F F Peter S. Hodder, Ph.D. Associate Director, Lead Identification SCIENTISTS John B. Hogenesch, Ph.D.** Head, Genome Technologies Tomas Vojkovsky, Ph.D. Ted Kamenecka, Ph.D. Associate Director, Medicinal Chemistry Chris Liang, Ph.D. Director, Medicinal Chemistry Phillip LoGrasso, Ph.D.* Director, Discovery Biology Kendall W. Nettles, Ph.D.* Discovery Biology 365 Jeremiah D. Tipton, Ph.D. * Joint appointment in the Department of Biochemistry ** Joint appointments in the Department of Biochemistry and the Molecular and Integrative Neurosciences Department *** Joint appointments in the Department of Biochemistry and the Department of Chemistry Jiu-Xiang Ni, Ph.D. S TA F F S C I E N T I S T S Scott A. Busby, Ph.D. Josephine Harada, Ph.D. Paul J. Kenny, Ph.D. Patricia H. McDonald, Ph.D. Trey Sato, Ph.D. Thomas Schroeter, Ph.D. Mathew T. Pletcher, Ph.D.* Genome Technologies William R. Roush, Ph.D.*** Executive Director, Medicinal Chemistry, Associate Dean, Kellogg School of Science and Technology, Florida Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. S E C T I O N C O V E R F O R T H E T R A N S L A T I O N A L R E S E A R C H I N S T I T U T E : Collection of 3-dimensional chemical structures from top-selling drugs for 2004. The Translational Research Institute brings together high-throughput screening, discovery biology, medicinal chemistry, informatics, and drug metabolism with genomic and proteomic technologies to translate basic research into potential clinical therapeutic agents. Art work by Stephan C. Schuerer, Ph.D., Group Leader, Informatics. 366 TRANSL ATIONAL RESEARCH INSTITUTE 2005 provided by Josephine Harada and Trey Sato, staff scientists, and in genetics, provided by Mat Pletcher, assistant professor of biochemistry. Protein Sciences is headed by Jennifer Busby, associate director. Researchers in the Proteomics Laboratory within Protein Sciences focus on the application of liquid chromatography and mass spectrometry to the identification, quantitation, and characterization of proteins and posttranslational protein modifications. The laboratory is involved in scientific collaborations within Scripps Florida and with several external investigators. Informatics is headed by Nick Tsinoremas, senior director. The goal of the informatics department is to provide computational tools and services to the main research areas of Scripps Florida. Researchers focus on 5 main areas of modern informatics: scientific computing, development of scientific software, computational biology, drug discovery informatics and chemoinformatics, and data mining and statistical sciences. Patrick R. Griffin, Ph.D. Head, Drug Discovery Chairman’s Overview he Translational Research Institute combines the drug discovery efforts at Scripps Florida with advanced technology platforms: genome technologies, protein sciences, and informatics. The goal of the drug discovery operation is to discover and develop small-molecule therapeutic agents for unmet medical needs in inflammation, neurodegeneration, stroke, cancer, and metabolic disorders. The operation is headed by Pat Griffin, professor of biochemistry. Groups include Lead Identification, headed by Peter Hodder, associate director; Medicinal Chemistry, headed by William Roush, executive director and professor of biochemistry; Discovery Biology, headed by Phil LoGrasso, director and associate professor of biochemistry; Drug Metabolism and Pharmacokinetics, headed by Mike Cameron, senior scientist; and CNS Disorders, headed by Claes Wahlestedt, director and professor of biochemistry. Genome Technologies is headed by John Hogenesch, professor of biochemistry. The goal of this department is to provide Scripps investigators and external collaborators cutting-edge technologies in cell-based screening, T Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. TRANSL ATIONAL RESEARCH INSTITUTE 2005 367 INVESTIGATORS’ R EPORTS ties and to determine differential protein binding affinities on the basis of the site of modification. Proteomics Core Facility PUBLICATIONS Hogan, K.T., Sutton, J.N., Chu, K.U., Busby, J.A.C., Shabanowitz, J., Hunt, D.F., Slingluff, C.L., Jr. Use of selected reaction monitoring mass spectrometry for the detection of specific MHC class I peptide antigens on A3 supertype family members. Cancer Immunol. Immunother. 54:359, 2005. J.A. Caldwell Busby, J. Tipton, V. Cavett he Proteomics Core Facility at Scripps Florida has general high-performance liquid chromatography and mass spectrometry capabilities for protein and peptide identification via nanoflow chromatography followed by tandem mass spectrometry and analysis. Current instrumentation includes an ion-trap mass spectrometer, which is used predominantly for protein and peptide identification, and a triple quadrupole mass spectrometer, which is used for relative quantitation experiments. Each mass spectrometer is interfaced to nanoflow electrospray ionization sources and capillary high-performance liquid chromatography columns. Expertise exists for identifying and mapping posttranslational modifications of proteins and peptides. As a core facility, we are responsible for providing collaborative mass spectrometry services to other faculty members at Scripps Florida. These collaborative projects will necessarily determine the development of technology in the facility. Current efforts in technology development include on-line sample cleanup and digestion, relative quantitation of serum proteins, and creation of software for differential analysis of mass spectrometry data. During its lifetime, a protein can have several locations and duties within a cell. Location, current status, and 3-dimensional structures of proteins are all influenced by static and dynamic chemical modifications that occur after translation. These modifications vary, from small methyl groups, which are a part of the histone codes, to large lipid and glycosylation modifications, which act as cellular markers and signaling molecules. Mass spectrometry has the unique advantage of being able to detect both the small and large changes in mass that occur because of these modifications. Phosphorylation is one posttranslational modification that receives attention because of its important role in signaling pathways. Unfortunately, phosphorylation is still relatively rare, and enrichment techniques are needed to detect sites of phosphorylation. In the core facility, we use immobilized metal affinity chromatography to enrich complex samples for phosphorylated peptides. This mapping of phosphorylation sites has been used to identify kinase and phosphatase activi- T Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Roig, J., Groen, A., Caldwell, J., Avruch, J. Active Nercc1 protein kinase concentrates at centrosomes early in mitosis, and is necessary for proper spindle assembly. Mol. Biol. Cell 16:4827, 2005. Stover, D.R, Caldwell, J., Marto, J.M., Root, K.R., Mestan, J., Stumm, M., Ornatsky, O., Orsi, C., Radosevic, N., Liao, L., Fabbro, D., Moran, M. Differential phosphoprofiles of EGF and GFR kinase inhibitor-treated human tumor cells and mouse xenografts. Clin. Proteomics J. 1:69, 2004. Thompson, L.W., Hogan, K.T., Caldwell, J.A., Pierce, R.A., Hendrickson, R.C., Deacon, D.H., Settlage, R.E., Brinkerhoff, L.H., Engelhard, V.H., Shabanowitz, J., Hunt, D.F., Slingluff, C.L., Jr. Preventing the spontaneous modification of an HLAA2-restricted peptide at an N-terminal glutamine or an internal cysteine residue enhances peptide antigenicity. J. Immunother. 27:177, 2004. Drug Discovery P. Hodder, L. Scampavia, T. Spicer, C. Chung, P. Chase, N. Kushner, P. Baillargeon igh-throughput screening requires sophisticated automation, detection, and assay technologies to test large (e.g., several hundred thousands) collections of compounds for biological or biochemical activity. This method typically is used by large pharmaceutical companies in their efforts to discover new drugs. With funding from the State of Florida and Palm Beach County, we are creating a state-of-the-art highthroughput screening operation at Scripps Florida. Our major goals are to screen large libraries of compounds and to identify compounds that may result in new clinical drugs. We intend to accomplish these goals by using novel high-throughput screening technologies. When these objectives are fully realized, we will be able to (1) support research at Scripps that requires high-throughput screening, (2) manage the extensive collection (>600,000 compounds) of compounds at Scripps used for drug screening, (3) provide assistance in the development of assays compatible with highthroughput screening, and (4) explore and implement novel technologies for the advancement of high-throughput screening. Highlights of our work in 2005 include the following. H CONSTRUCTION OF A HIGH-THROUGHPUT S C R E E N I N G L A B O R AT O R Y The Lead Identification Department is building a state-of-the-art high-throughput screening laboratory 368 TRANSL ATIONAL RESEARCH INSTITUTE 2005 at Scripps Florida. In addition to a fully staffed laboratory devoted to the development of high-throughput screening assays, the centerpieces of the laboratory are 2 automated robotic systems. The first robotic system (Fig. 1) is dedicated solely to high-throughput screening. With this system, we can test more than 1 million samples per day. The system has an on-line storage capacity of more than 1 million compounds, can be run 24 hours a day, 7 days a week, and can be used for both biochemical and cell-based assays. Because microtiter plates with 1536 wells (assay volumes of ~10 µL per well), are used in the assays, high-throughput screening can be completed rapidly and economically. F i g . 1 . The Scripps Florida high-throughput screening system will provide fully automated technology to detect compounds that may lead to the discovery of new drugs. The second robot stores a copy online of the entire collection of the compounds used for high-throughput screening. It has the automation to instantly retrieve and reformat any member of the collection to a 384or a 1536-well plate. This robot will be used in studies after a high-throughput screening campaign has been completed, allowing scientists to rapidly select compounds for further pharmacologic analysis. The final installation and testing of the robotic systems were scheduled to be completed in November 2005 and should be ready for use by the start of 2006. ACQUISITION AND MANAGEMENT OF COMPOUNDS FOR SCREENING In addition to setting up the screening facility, we are acquiring a proprietary collection of more than 600,000 druglike compounds to be managed and maintained for high-throughput screening. This collection will include compounds from commercial sources and novel compounds discovered by researchers at the Scripps Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. facilities. In order to support drug-target profiling and facilitate grant writing efforts, smaller collections will be established. Currently, our goal is to have the compounds on hand and ready for high-throughput screening by the first quarter of 2006. R E S E A R C H C O L L A B O R AT I O N S The Scripps research facilities have been designated a member of the Molecular Libraries Screening Center Network, as part of the National Institutes of Health Roadmap Initiative (http://nihroadmap.nih.gov/). In collaboration with H. Rosen, Department of Immunology, Scripps Research, we will support the implementation and execution of high-throughput screening done to meet the goals of the network. The data acquired via high-throughput screening will be made publicly available through PubChem, to aid in the discovery of probes that explore the functions of biological mechanisms in health and disease. As part of our commitment to the State of Florida, we are also setting up processes for Florida academic researchers to access our expertise in high-throughput screening. We are creating a specialized collection of compounds and a screening model tailored to academic research. PUBLICATIONS Hale, J.J., Lynch, C.L., Neway, W., Mills, S.G., Hajdu, R., Keohane, C.A., Rosenbach, M.J., Milligan, J.A., Shei, G.J., Parent, S.A., Chrebet, G., Bergstrom, J., Card, D., Ferrer, M., Hodder, P., Strulovici, B., Rosen, H., Mandala, S. A rational utilization of high-throughput screening affords selective, orally bioavailable 1-benzyl-3-carboxyazetidine sphingosine-1-phosphate-1 receptor agonists. J. Med. Chem. 47:6662, 2004. Hodder, P., Mull, R., Cassaday, J., Berry, K., Strulovici, B. Miniaturization of intracellular calcium functional assays to 1536-well plate format using a fluorimetric imaging plate reader. J. Biomol. Screen. 9:417, 2004. Zuck, P., O’Donnell, G.T., Cassaday, J., Chase, P., Hodder, P., Strulovici, B., Ferrer, M. Miniaturization of absorbance assays using the fluorescent properties of white microplates. Anal. Biochem. 342:254, 2005. Development of Protein Kinase Inhibitors C. Liang, M. Koenig, T. Vojkovsky, Y. He, Y. Feng, T. Kamenecka, T. Schroeter, A. Weiser ur goal is to discover protein kinase inhibitors that can be used as therapeutic agents for the treatment of human diseases such as cancer and arthritis. Protein kinases are a class of enzymes that catalyze the transfer of the γ-phosphate from ATP to protein substrates. These enzymes play critical roles O TRANSL ATIONAL RESEARCH INSTITUTE 2005 in signal transduction for a number of cellular functions. In particular, they regulate most of the hallmarks of cancer: cell proliferation, cell survival, cell motility/metastasis, cell cycle/division, and angiogenesis. Protein kinases are also implicated in inflammatory diseases such as arthritis and asthma. The approval of the tyrosine kinase inhibitors imatinib mesylate (Gleevec) for treatment of chronic myelogenous leukemia and gastrointestinal stromal tumors and bevacizumab (Avastin) for treatment of non–small cell lung cancer validated protein kinase inhibitors/antagonists as effective, noncytotoxic anticancer agents. These drugs provide new hope and models in the long fight against cancer. In addition, later stage clinical data strongly suggest that p38α MAP kinase inhibitors could be effective antiinflammatory agents. For these reasons, protein kinases are being investigated as valuable therapeutic targets by virtually every pharmaceutical company, and according to estimates, about 25% of all current pharmaceutical research is devoted to these targets. We synthesized and evaluated more than 500 potential drug candidates during the past year. In 2 projects, we found compounds that have better in vitro properties than the best known competitors do. Several of these compounds also have good pharmacokinetic properties and in vivo efficacy. Currently, they are being evaluated as potential clinical drug candidates. Recently, we began a collaboration with the NeoRx Corporation, Seattle, Washington, to discover novel multitargeted protein kinase inhibitors for the treatment of cancer. An Integrated Proteomics Environment and the Deuterator System B. Pascal, C. Mader, N.F. Tsinoremas e are building an integrated proteomics informatics environment to support the proteomics research activities of the drug discovery group at Scripps Florida. To date, we have focused on establishing a software infrastructural system to support acquisition of the primary mass spectral data and on developing software for the analysis and presentation of data from hydrogen-deuterium exchange experiments. Scientists at Scripps Florida are conducting solution-phase amide hydrogen-deuterium exchange exper- W Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 369 iments, which require specific software support. Systems have been put in place for high-throughput data analysis and visualization. Hydrogen-deuterium exchange experiments begin with a typical proteomics experiment: protein is digested with an enzyme, and the resulting peptides are analyzed by using liquid chromatography and tandem mass spectrometry. The identity of the peptides is established by using commercially available database search tools such as Sequest. Subsequent solution-phase amide hydrogen-deuterium exchange experiments are performed for predefined periods (8–10 time points spanning 1–10,000 seconds). After incubation with deuterium oxide, the protein is digested, and the resulting peptides are analyzed by using liquid chromatography and tandem mass spectrometry. The increase in peptide mass reveals the extent of hydrogen-deuterium exchange at each time point. Because the incubation with deuterium oxide precedes the digestion of protein, the measured rate of deuterium incorporation for each peptide reveals the hydrogen-deuterium exchange rate for the corresponding region of the intact protein. These hydrogen-deuterium exchange rates can be determined by using our software, which is titled the Deuterator. Additional features allow for differential hydrogen-deuterium exchange analysis between free and ligand-bound protein. NEED FOR A UNIFIED SYSTEM High-throughput analysis for hydrogen-deuterium data of this type is not generally commercially available at this time. Our system provides a complete infrastructure for this analysis, from the acquisition of foundational data to analysis and visualization. When possible, existing components have been used and generic structures built to allow reuse with other projects. Increasing the throughput of hydrogen-deuterium exchange experiments required constructing an informatics infrastructure to support the increase in the production of data. We have built a powerful informatics infrastructure that can support the acquisition, management, and analysis of large data sets (terabytes). T H E I N T E G R AT E D P R O T E O M I C S E N V I R O N M E N T The integrated proteomics informatics environment is used to manage data from the time the data are created by the mass spectrometer through hydrogendeuterium exchange analysis, and in the future, differential analysis via mass spectrometry. Figure 1 shows an overall architecture of the environment. A laboratory information management system (LIMS) has been put in place to acquire process data and act as 370 TRANSL ATIONAL RESEARCH INSTITUTE 2005 We have developed a system called Morphimi that converts raw mass spectrometric data to other formats, usually to mzXML. Morphimi also maintains a repository of and manages the metadata of the converted mzXML data. mzXML is an XML-based common file format for mass spectrometric data, developed at the Institute for Systems Biology. Having the mass spectrometric data in mzXML allows access to a number of current and future tools written for use with the schema (Fig. 1). The mass spectrometer produces binary RAW files, which are then submitted to the LIMS. Morphimi then retrieves these files and converts them to mzXML and places them in the Morphimi archive. Sequest sequence searches are performed from the RAW files, and the results are submitted into the LIMS. The Deuterator then retrieves the search result file from the LIMS and the mzXML file from the Morphimi archive and proceeds with peak picking and centroid calculations. The resulting data set is made available via a Web interface, and the reviewed results are sent to the statistical analysis and visualization tools, which are also available through the Web interface (Fig. 2). F i g . 1 . Overview of the integrated proteomics environment. 1. Mass spectrometer (MS) produces raw spectral data. 2. The LIMS system captures and organizes project information and maintains an archive of the raw MS data. 3. Morphimi MBean is used to manage conversion of raw MS data to mzXML. 4. Morphimi maintains an index and archive of mzXML data. 5. Peak Picking toolkit filters and prepares mzXML data for analysis. 6. The Deuterator performs hydrogen-deuterium exchange analysis. 7. The Diff MS application suite will provide tools for differential analysis of MS data. 8. Various open source libraries are used to analyze the data. 9. Various open source libraries are used to visualize the data. 10. Protein database search tools (Sequest and MASCOT are currently supported) are used to identify the peptides. a repository for the raw data produced by the mass spectrometers. This solution has been provided by Genologics, Victoria, British Columbia, and is called ProteusLIMS. This system is being used to set up projects and upload raw and associated data files, which immediately become available to the downstream analysis software. The Institute for Systems Biology, Seattle, Washington, provides a variety of open source software tools for analysis and organization of data obtained in proteomics research. The goal of the Sashimi (http://sashimi .sourceforge.net) project begun at the institute is to provide the scientific community free open source software tools for the analysis of mass spectrometric data. We have incorporated a number of components from the institute within our system. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. F i g . 2 . Deuterator work flow diagram. H/D indicates hydrogen- deuterium. The peptide set obtained from the database search is initially grouped according to sequence and charge (multiple occurrences of the same peptide ion often occur within the raw data set). Redundant data are removed, and the retention time for each peptide ion becomes the mean of all measured values. A retention time window is then created for each peptide within the set. This range is used by the software to go into the raw data and calculate the average of all spectral points for all scans located within this region. The monoisotopic mass of each peptide is then calculated. Accounting for the TRANSL ATIONAL RESEARCH INSTITUTE 2005 371 charge state of the given peptide, the 0% and the 100% deuterium incorporation mass-to-charge ratio limits for the given peptide are calculated. The intensity-weighted mean mass-to-charge ratio between these values is calculated, defined as the centroid mass-to-charge ratio, and the values for each peptide set are presented in the main interface page. On the basis of queries to the LIMS, the user interface enables drilling down from the project level to the sample level and finally to the individual time point (Fig. 3). All processed time points are immediately available through this interface. F i g . 5 . Spectral viewer. file format. The extracted ion graph displays the ion intensities for a given mass across all scans. This graph is used to zone in to the areas with the best data for a given peptide’s centroid mass (Fig. 5). FUTURE DEVELOPMENT F i g . 3 . Project, sample, and time point selection. The main analysis page has 3 sections. The grid presents all of the calculations and allows users to select a peptide for viewing (Fig. 4). By using the tab browser functionality in the Firefox browser, many time points can be seen simultaneously. F i g . 4 . Selection of a peptide for viewing. The spectral viewer initially displays the raw spectral data for the calculated range of the selected peptide (Fig. 5), and navigation controls can be used to further refine the range. The centroid is automatically recalculated for each refinement. When the user is satisfied, the results can be saved into the database. At any time, the entire grid data can be exported to a common Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Rendering components will be built that provide extended graphing capabilities, 2- and 3-dimensional color gradient maps, and data grids that show changes in deuterium incorporation between different experiments. Analysis tools will be built that will calculate the theoretical isotopic envelope and fit it onto the spectral output, as well as deconvolute overlapping peptides. These components will be tightly integrated into the system.