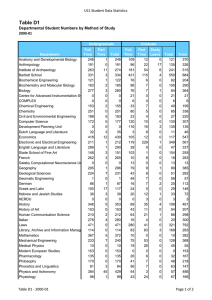
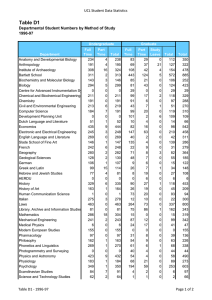
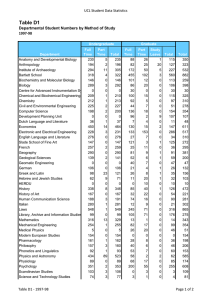
Cell Biology Published by TSRI Press . Copyright 2005,
advertisement

Cell Biology Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Lindsey MacPherson, Graduate Student, and Ardem Patapoutian, Ph.D., Associate Professor, Department of Cell Biology Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. CELL BIOLOGY 2005 19 DEPAR TMENT OF CELL BIOLOGY S TA F F Sandra L. Schmid, Ph.D.* Professor and Chairman Francisco Asturias, Ph.D.** Associate Professor William E. Balch, Ph.D.* Professor Bridget Carragher, Ph.D.** Associate Professor Benjamin Cravatt, Ph.D.*** Professor Gaudenz Danuser, Ph.D.** Assistant Professor Philip E. Dawson, Ph.D.**** Associate Professor Stephen P. Mayfield, Ph.D.**** Associate Professor Associate Dean, Kellogg School of Science and Technology Mark Mayford, Ph.D.***** Associate Professor Lindsey Miles, Ph.D. Associate Professor Alan Bell, B.S.C.S. Xerox Palo Alto Research Center Palo Alto, California Ronald A. Milligan, Ph.D.** Professor Ulrich Müller, Ph.D.***** Associate Professor Ardem Patapoutian, Ph.D.††† Associate Professor Clinton Potter, B.S.** Associate Professor Martin Friedlander, M.D., Ph.D. Professor Lisa Stowers, Ph.D. †††† Assistant Professor Steve Kay, Ph.D. † Professor Natasha Kralli, Ph.D. Assistant Professor Peter Kuhn, Ph.D.** Associate Professor David Loskutoff, Ph.D. Professor Emeritus Mark J. Yeager, M.D., Ph.D. Professor ADJUNCT APPOINTMENTS James Quigley, Ph.D. Professor Shelley Halpain, Ph.D.***** Associate Professor John R. Yates III, Ph.D. Professor Alan McLachlan, Ph.D. †† University of Illinois Chicago, Illinois Velia Fowler, Ph.D.** Professor Larry R. Gerace, Ph.D.* Professor Elizabeth Winzeler, Ph.D. ††† Associate Professor Heidi Stuhlmann, Ph.D. Associate Professor Kevin F. Sullivan, Ph.D.** Associate Professor Peter N.T. Unwin, Ph.D.** Professor Clare Waterman-Storer, Ph.D.** Associate Professor Xiaohua Gong, Ph.D. University of California Berkeley, California Klaus Hahn, Ph.D. University of North Carolina Chapel Hill, North Carolina Eric Peeters, Ph.D. Xerox Palo Alto Research Center Palo Alto, California S TA F F S C I E N T I S T S Michael Bracey, Ph.D. Richard Bruce, Ph.D. Xerox Palo Alto Research Center Palo Alto, California Douglas Curry, B.S. (E.E.C.S.) Xerox Palo Alto Research Center Palo Alto, California Bertil Daneholt, M.D. Karolinska Institutet Stockholm, Sweden Scott Elrod, Ph.D. Xerox Palo Alto Research Center Palo Alto, California Anchi Cheng, Ph.D. Elena Deryugina, Ph.D. Robert Fischer, Ph.D. Thomas Schultz, Ph.D. Elizabeth Wilson, Ph.D. Yuanxin Zhu, Ph.D. †† Fair Issac Corporation San Diego, California SENIOR RESEARCH A S S O C I AT E S Mark Ginsberg, M.D. University of California San Diego, California Brian Adair, Ph.D. David Goldberg, Ph.D. Xerox Palo Alto Research Center Palo Alto, California Sean Conner, Ph.D. †† University of Minnesota, Twin Cities Minneapolis, Minnesota Yuriy Chaban, Ph.D. S E C T I O N C O V E R F O R T H E D E P A R T M E N T O F C E L L B I O L O G Y : Intraocular vascula- ture of a mouse imaged in vivo. Visible in this image are the vessels of the iris, hyaloid, and retina. Vessels of this 16-day-old mouse were visualized by using intravenous injection of fluorescent dye and confocal microscopy. Approximately 150 images over a depth of about 3 mm were collected to Mari Manchester, Ph.D.** Associate Professor construct the image shown. This approach allows visualization of functionally intact ocular blood vessels and serial imaging of the same blood vessels over time. Image by Matthew Ritter, Ph.D., in the laboratory of Martin Friedlander, M.D., Ph.D. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 20 CELL BIOLOGY 2005 Ralf-Peter Czekay, Ph.D. †† Albany Medical College Albany, New York Aurelia Cassany, Ph.D. Mark Daniels, Ph.D. Ihsiung Chen, Ph.D. Emily Chen, Ph.D. Thomas Fath, Ph.D. †† The Children’s Hospital at Westmead Sydney, Australia Claire Kidgell, Ph.D. Katsuhiro Kita, Ph.D. Atanas Koulov, Ph.D. Michael Fitch, Ph.D. Scott E. Franklin, Ph.D. †† Rincon Pharmaceuticals La Jolla, California Yei Hua Chen, Ph.D. Paul LaPointe, Ph.D. Maria Gonzalez, Ph.D. Charmian Cher, Ph.D. Nicole Lazarus, Ph.D. Nicolas Grillet, Ph.D. Jeremiah Joseph, Ph.D. Smita Chitnis, Ph.D. Donmienne Leung, Ph.D. Cemal Gurkan, Ph.D. Edward Korzus, Ph.D. Parag Chowdhury, Ph.D. John Lewis, Ph.D. Frank Harmon, Ph.D. Stephan Miller, Ph.D. Jill Chrencik, Ph.D. Lujian Liao, Ph.D. Samuel Hazen, Ph.D. Matthew Ritter, Ph.D. Jessie Chu, Ph.D. Maria Lillo, Ph.D. Johannes Hewel, Ph.D. Martin Schwander, Ph.D. Michael Churchill, Ph.D. Francesco Conti, Ph.D. Edith Hintermann, Ph.D. †† Institut für Allgemeine Pharmakologie und Toxikologie Frankfurt, Germany Judith Coppinger, Ph.D. Hide Hyracoidea, Ph.D. ††††† Michael Hock, Ph.D. Geza Ambrus-Aikelin, Ph.D. Amy Cullinan, Ph.D. †† Invitrogen Corporation Carlsbad, California Angelique Aschrafi, Ph.D. Leif Dehmelt, Ph.D. Eric Hwang, Ph.D. Hongdong Bai, Ph.D. Claudia Dellas, Ph.D. ††††† Sun Wook Hwang, Ph.D. †† Korea University Seoul, Korea Juliana Conkright, Ph.D. †† Scripps Florida Gina Story, Ph.D. Andries Zijlstra, Ph.D. R E S E A R C H A S S O C I AT E S Jessica Alexander, Ph.D. Claudia Barros, Ph.D. Ph.D. †† Janna Bednenko, University of Rochester Rochester, New York Maria Beligni, Ph.D. Ph.D. †† Johannes De Rooij, Netherlands Cancer Institute Amsterdam, the Netherlands Ajay Dhaka, Ph.D. Meng-Qui Dong, Ph.D. K.E. Hua, Ph.D. Takato Imaizumi, Ph.D. Ph.D. †† Miwako Ishido, Trans-Science Inc. Tokyo, Japan Klara Limback-Stokin, Ph.D.†† University of Ljubljana School of Medicine Ljubljana, Slovenia Jennifer Lin, Ph.D. Ryan Littlefield, Ph.D. Chuan-Yin Liu, Ph.D. Songkai Liu, Ph.D. †† University of California San Francisco, California Dinah Loerke, Ph.D. Matthias Machacek, Ph.D. Mark Madsen, Ph.D. Claudio Masuda, Ph.D. Naoki Matsuo, Ph.D. Richard Belvindrah, Ph.D. Michael Dorrell, Ph.D. Khuloud Jaqaman, Ph.D. Daniel McClatchy, Ph.D. Kari Bradtke, Ph.D. Kelly A. Dryden, Ph.D. Anass Jawhari, Ph.D. Ghislain Breton, Ph.D. Anna Durrans, Ph.D. Nadim Jessani, Ph.D. †† Celera Genomics San Francisco, California Caroline McKeown, Ph.D. Ph.D. †† Anja Bubeck, Ph.D. Andrei Efimov, Vanderbilt University Nashville, Tennessee Barbara Calabrese, Ph.D. Samer Eid, Ph.D. Gregory Cantin, Ph.D. Eva Farre, Ph.D. Florence Brunel, Ph.D. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Lin Ji, Ph.D. Kimon Kanelakis, Ph.D. †† Johnson & Johnson San Diego, California Juncai Meng, Ph.D. †† Merck Pharmaceuticals Whitehorse Station, New Jersey Helena Mira, Ph.D. Jennifer Mitchell, Ph.D. CELL BIOLOGY 2005 21 Jacobus Neels, Ph.D. Alan Saghatelian, Ph.D. Ge Yang, Ph.D. Sherry Niessen, Ph.D. Kumar Saikatendu, Ph.D. Jian Ming Yang, Ph.D. †† Office of Environmental Health Hazard Assessment Oakland, California John Offer, Ph.D. †† University of Oxford Glycobiology Institute Oxford, England Silvia Ortega-Gutierrez, Ph.D. Cleo Salisbury, Ph.D. Trey Sato, Ph.D. †† Scripps Florida Defne Yarar, Ph.D. Eric C. Schirmer, Ph.D. †† University of Edinburgh Edinburgh, Scotland. Masahiro Yasuda, Ph.D. Rie Yasuda, Ph.D. Lesley Page, Ph.D. Ning Pan, Ph.D. †† University of Massachusetts Medical School Boston, Massachusetts Thomas Schroeter, Scripps Florida Ph.D. †† Stephan Sieber, Ph.D. Erquan Zhang, Ph.D. Wenhong Zhu, Ph.D. †† Burnham Institute La Jolla, California * Joint appointment in the Department of Molecular Biology ** Joint appointment in the Center for Integrative Molecular Biosciences *** Joint appointments in the Department of Chemistry, the Skaggs Institute for Chemical Biology, and the Helen L. Dorris Child and Adolescent Neurological and Psychiatric Disorder Institute **** Joint appointment in the Skaggs Institute for Chemical Biology ***** Joint appointment in the Institute for Childhood and Neglected Diseases † Pratik Singh, Ph.D. Zhongmin Zou, Ph.D. Alessia Para, Ph.D. Fabienne Soulet, Ph.D. ††††† Brian Petrich, Ph.D. ††††† Scott Stagg, Ph.D. Sarah Stanton, Ph.D. ††††† Barbie Pornillos, Ph.D. †† Appointment completed; new location shown ††† Joint appointments in the Institute for Childhood and Neglected Diseases and the Genomics Institute of the Novartis Research Foundation S C I E N T I F I C A S S O C I AT E S Aaron Ponti, Ph.D. ††††† Hilda Edith Aguilar de Diaz, M.D. Mark Surka, Ph.D. Faith Barnett, Ph.D. Judith Prieto, Ph.D. †††† Claire Tiraby, Ph.D. Alexei Brooun, Ph.D. Natalie Prigozhina, Ph.D. Ph.D. †† Jose Pruneda-Paz, Ph.D. Florian Topert, Uexkull and Stolberg Hamburg, Germany Emily Burke, Ph.D. Claire Delahunty, Ph.D. Rajesh Ramachandran, Ph.D. Tuija Uusitalo, Ph.D. Tinglu Guan, Ph.D. Vandana Ramachandran, Ph.D. John Venable, Ph.D. Leon Reijmers, Ph.D. Josep Villena, Ph.D. Gang Ren, Ph.D. †† Baylor College of Medicine Houston, Texas Xiaodong Wang, Ph.D. Christopher Reyes, Ph.D. Ann Wheeler, Ph.D. Anna Reynolds, Ph.D. Margaret Wilson, Ph.D.††††† Olga Rodriguez, Ph.D. Torsten Wittmann, Ph.D. Benoit Roger, Ph.D. ††††† James Wohlschlegel, Ph.D. Edwin Romijn, Ph.D. Christine Wu, Ph.D. †† University of Colorado Denver, Colorado Anand Kolatkar, Ph.D. Fakhruddin Palida, Ph.D. Cristian Ruse, Ph.D. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Kathryn Spencer, Ph.D. BinQing Wei, Ph.D. Joint appointments in the Institute for Childhood and Neglected Diseases and the Department of Biochemistry ††††† Joint appointment in the Helen L. Dorris Child and Adolescent Neuro-Psychiatric Disorder Institute Appointment completed 22 CELL BIOLOGY 2005 Sandra Schmid, Ph.D. Chairman’s Overview ou would think that with the complete list of parts provided by the human genome sequence, the task of deciphering the inner workings of cells and how cells interact with each other and with their environment would be easier. But in fact, having ready access to the list of parts and the ability to discover their interconnectedness means that cell biologists must grapple with greater complexity than before and with larger questions of physiology and development. For the past several decades, cell biologists have focused on enumerating the parts of subcellular machines and individually assigning functions to the parts, usually in the context of specific pathways or cellular processes. Now, to have a significant impact, the reductionists among us must establish molecular mechanisms and unravel the structure-function relationships for each cellular machine. Those who study specific cellular processes, such as migration, metastasis, intracellular trafficking, and signal transduction, must unravel the complex regulation and cross talk between cellular processes. For example, signal transduction is controlled and modulated by the spatial localization of signaling molecules and second messengers. This spatial localization in turn is regulated Y Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. by intracellular trafficking, by exchange of signaling molecules between subcellular compartments, and by dynamic association with cytoskeletal elements and/or with structurally distinct membrane microdomains. The goal now is to understand how the integration of each of these cellular processes alters cell physiology in health and disease. Those who study higher-order functions, such as sensory perception, learning and memory, circadian rhythms, and development, encounter even greater complexity and must use the powerful new tools of molecular genetics to probe cause and effect and then to understand how anatomy and the organization of tissues affects the higher-order functions. Finally, those who study specific diseases must identify all of the molecular changes that occur and then determine the relevance of the changes to the disease. How are we tackling these complex biological problems to generate important and valuable new insight? We collaborate! As I read through the reports that follow, I was struck by the extent and nature of ongoing collaborations that are driving our discoveries in the Department of Cell Biology. As the accompanying diagram (Fig. 1) illustrates, to tackle the ever-increasing complexity of questions being addressed, members of the department are taking advantage of the breadth and depth of biomedical research ongoing at Scripps and are weaving an increasingly complex network of collaborations. Bill Balch, for example, is fulfilling his reductionist roots by collaborating with Bridget Carragher and Clint Potter, directors of the National Resource for Automated Molecular Microscopy, to solve the structure of a novel coat complex that drives vesicle formation from the endoplasmic reticulum and with Ian Wilson to solve high-resolution x-ray structures that reveal how individual components of the membrane trafficking machinery carry out the cellular functions. Bill is also collaborating with Jeff Kelly to elucidate how the cell’s biosynthetic machinery distinguishes properly folded and misfolded proteins and what properties of this sorting machinery leave humans susceptible to protein misfolding diseases, collectively known as amyloidoses. Finally, he is collaborating with John Yates to use innovative proteomics methods to identify what components of the cell’s biosynthetic machinery might be limiting for controlling the export of mutant proteins. In combination, these multidisciplinary approaches and collaborations are enabling Dr. Balch and his colleagues to reveal potential new avenues for therapeutic intervention in diseases, such as cystic fibrosis, Gaucher’s disease, emphysema, diabetes, CELL BIOLOGY 2005 23 Other transitions include the well-deserved promotions of Phil Dawson and Elizabeth Winzeler to associate professor; Ardem Patapoutian and Clare Waterman-Storer to associate professor with tenure; and Martin Friedlander to full professor. The success of these scientists attests to the richness of the scientific environment at Scripps Research. F i g . 1 . Department of Cell Biology summary. Intradepartmental and interdepartmental collaborations enrich research in all biology. GNF indicates Genomics Institute of the Novartis Research Foundation; MB, Department of Molecular Biology; PARC, Palo Alto Research Center. and amyloidosis. Please explore the following reports to see how interdisciplinary collaborations between Scripps scientists are ensuring that members of the Department of Cell Biology remain innovative leaders in the members’ respective areas of research. Another report worth reading is Dave Loskutoff ’s. Dave retired this year after nearly 25 years at Scripps, where he served as chairman of the Department of Vascular Biology from 1993 to 2003. During his distinguished career, Dave made numerous seminal contributions to our understanding of fibrinolysis and serum proteases and their regulation. More recently, he focused on the cell signaling role of the serine protease inhibitor plasminogen activator inhibitor-1 in obesity and cardiovascular disease. His productivity and the importance of his research efforts remained high throughout his career. Nine articles describing his research findings were published in excellent journals during the past year. Our daily interactions with Dave will be missed, but we look forward to informally enjoying his collegiality for some time. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 24 CELL BIOLOGY 2005 INVESTIGATORS’ R EPORTS Structural Characterization of Macromolecular Machines F.J. Asturias, Y. Chaban, J.Z. Chadick, A. Jawhari, M. Sedova, D. Wepplo e use state-of-the-art electron microscopy and image analysis to calculate the 3-dimensional structures of macromolecular complexes that carry out a variety of cellular processes, including DNA replication and transcription and fatty acid synthesis. Our ultimate goal is to reveal the mechanism of action of a macromolecular complex by determining the structures of the different conformations the complex adopts as it carries out its function. Images of individual complexes are recorded under physiologically relevant conditions and then are computationally combined to obtain structures of low-to-moderate resolution (10–25 Å). Often, further information is obtained by docking atomicresolution structures of component subunits in the lower resolution map of an entire complex. Macromolecular electron microscopy is an ideal technique for these studies because the technique requires only a small amount of material and the conditions for preparing samples are relatively gentle. DNA transcription and its regulation are major areas of interest. Previously, we characterized a number of complexes that form part of the basal transcription machinery in the yeast Saccharomyces cerevisiae, arguably the most important model organism for studies of eukaryotic transcription. More recently, we extended our studies to the human basal machinery. We are characterizing complexes formed by human RNA polymerase II and general transcription factors to understand how these factors contribute to the initiation of transcription. Also under study are complexes involved in the regulation of transcription during initiation and during previous steps in which the structure of chromatin is altered to make DNA accessible to the transcription machinery. By comparing and contrasting the function and structure of homologous complexes in different eukaryotes, we hope to identify fundamental features of the transcription regulation mechanism. Our interest in the importance of conformational changes for the function of macromolecular machines prompted us to study the structure of mammalian W Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. fatty acid synthase (FAS), the enzyme responsible for the synthesis of long-chain fatty acids. In animal cells, the different enzymes involved in the synthesis of fatty acids have fused into a single polypeptide chain with 7 different enzymatic activities. Moreover, the functional form of FAS is a dimer in which 2 such chains cooperate to create 2 centers of fatty acid synthesis. FAS is therefore a true “molecular assembly line.” Despite the importance of FAS in metabolism, cellular development, and certain types of cancer, information on the structure of the enzyme is limited because of the large number of conformations apparently adopted by FAS. Even the organization of the 2 monomers in the FAS dimer had remained uncertain, thereby precluding an understanding of the functional complementation between active sites required for fatty acid synthesis. In addition, the results from the most recent functional complementation studies conflict with the traditional model for FAS organization in which the 2 component monomers are depicted as fully extended and oriented in an antiparallel arrangement. We used a novel approach in which FAS point mutants were imaged in the presence of substrates. With this approach, the enzyme is effectively paused at a given catalytic step and conformational variability is reduced. Distinct but related FAS conformations appear to be associated with specific catalytic steps (Fig. 1). Calculating the structure of an FAS monomer F i g . 1 . Structure in projection of different FAS paused mutants. The active site compromised in each mutant and the specific mutations are indicated. Imaging mutants in the presence of substrates considerably reduced the normal conformational variability of FAS. CELL BIOLOGY 2005 and labeling the N termini of the 2 monomers in the active form of the enzyme resulted in a revised understanding of FAS organization and in a new model in which 2 coiled FAS monomers are intertwined to facilitate different functional interactions in the active FAS dimer (Fig. 2). This revised model for FAS organization is a starting point for further characterization of the enzyme and the related modular polyketide synthases, the molecules responsible for synthesis of a number of antibiotics and other important physiologically active compounds. F i g . 2 . A 16-Å resolution 3-dimensional structure of mammalian FAS. A, Front view of the enzyme. B, Labeling of the 2 FAS N termini indicates that both are located near the center of the FAS structure. C, Structure of an FAS monomer and the way in which 2 monomers might interact to form the active dimeric form of FAS. Scale bar = 100 Å. PUBLICATIONS Asturias, F.J. Another piece in the transcription initiation puzzle. Nat. Struct. Mol. Biol. 11:1031, 2004. Asturias, F.J., Chadick, J.Z., Cheung, I.K., Stark, H., Witkowski, A., Joshi, A.K., Smith, S. Structure and molecular organization of mammalian fatty acid synthase. Nat. Struct. Mol. Biol. 12:225, 2005. Bourbon, H.M., Aguilera, A., Ansari, A.Z., Asturias, F.J., Berk, A.J., Bjorklund, S., et al. A unified nomenclature for protein subunits of Mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14:553, 2004. Chadick, J.Z., Asturias, F.J. Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 30:264, 2005. Witkowski, A., Ghosal, A., Joshi, A.K., Witkowska, H.E., Asturias, F.J., Smith, S. Head-to-head coiled arrangement of the subunits of the animal fatty acid synthase. Chem. Biol. 11:1667, 2004. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 25 Biological Chemistry and Systems Biology of Conformational Disease W.E. Balch, Y. An, C. Chen, J. Conkright-Johnson, D. Fowler, C. Gurkan, D. Hutt, A. Koulov, J. Matteson, P. LaPointe, L. Page, H. Plutner, A. Pottekat, A. Sawkar, S. Stagg, X. Wang major challenge is to understand and treat the many protein-misfolding diseases that affect human health, including cystic fibrosis, emphysema, type 2 diabetes, and amyloidosis. These abnormalities are classified as membrane-trafficking conformational diseases because a defect in protein folding at some stage of the eukaryotic secretory pathway results in loss of activity or protein aggregation. A key concern is to determine the underlying defect in protein folding and how that defect affects the ability of the protein to function normally within the context of the cell’s intracellular transport machinery or in the extracellular environment of the host. Our broad objective is to define the molecular basis for the trafficking of normal and misfolded proteins through the secretory pathway of eukaryotic cells. We use chemical, structural, biological, and bioinformatics approaches. Eukaryotic cells are highly compartmentalized; each compartment of the exocytic and endocytic pathways provides a unique chemical landscape in which protein function and folding may be modulated. Movement between these compartments involves the activity of both anterograde and retrograde transport tubules and vesicles. Many conformational diseases are a consequence of dysfunction at different stages of this transport pathway or outside of the cell. Transport through the secretory pathway involves a selective mechanism in which cargo molecules are concentrated into carrier vesicles. Vesicle-mediated transport is regulated by a diverse group of small GTPases belonging to the Ras superfamily. Each of these molecules acts as a “molecular sensor” to regulate different steps in the reversible assembly of vesicle coats and targeting-fusion complexes. During export from the first compartment of the secretory pathway, the endoplasmic reticulum, coat recruitment to budding sites involves activation of the GTPase Sar1. After activation, the cytosolic coat components Sec23/24 and Sec13/31 A 26 CELL BIOLOGY 2005 form the coatomer complex II coat that polymerizes to promote budding from the surface of the endoplasmic reticulum. Recently, we showed that Sec 13/31 selfassembles to form a cage that drives vesicle budding. In collaboration with C. Potter and B. Carragher, Department of Cell Biology, using single-particle analysis, we solved the 2-dimensional electron cryomicroscopy structure of the Sec13/31 cage. We also found that cargo selection by the assembling polymer involves “exit codes” found on the cytoplasmic domains of cargo and cargo receptors. These exit codes bind to a pocket in the Sec24 coat component. In studies with J.R. Yates, Department of Cell Biology, we are using shotgun proteomics (multidimensional protein identification technology or MudPIT) to identify unknown components involved in cargo selection. After vesicles separate from the endoplasmic reticulum, targeting and fusion of coatomer complex II transport vesicles to the next step of the secretory pathway, the Golgi apparatus, require a different class of Raslike GTPases that belong to the Rab family. Members of the large Rab family (>70 members) act as molecular switches that assemble complexes involved in vesicle tethering and fusion. Using a bioinformatics approach, in collaboration with J. Hogenesch, Genome Technology, Scripps Florida, we found that each Rab GTPase executes targeting and fusion decisions at a distinct step in the exocytic or endocytic pathway. By coordinating the function of multiple distinct effectors at each step, Rab GTPases act as hubs to define the function and highly distinctive membrane architecture of eukaryotic cells found in different tissues. This systems biology approach provides for the first time a global view of membrane traffic from the top down, integrating form with function. An important common effector for all Rab GTPases is the protein GDP-dissociation inhibitor (GDI). GDI extracts Rabs from membranes after vesicle fusion, thereby forming a cytosolic complex that sequesters the prenyl groups for redelivery to the membrane during budding. Combined biochemical and structural studies revealed the surprising importance of the Hsp90 chaperone system in GDI-dependent Rab recycling. In addition, Rab proteins coordinate the activity of tethers that direct vesicle docking and fusion. Our recent structure of the Rab1 tether p115, determined in collaboration with I.A. Wilson, Department of Molecular Biology, reveals a crucial role for coiled coil interactions in mediating membrane docking. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Although traffic from the endoplasmic reticulum can be disrupted by mutations that prevent proper protein folding during synthesis, other protein conformational diseases have mutations that disrupt function at later steps of the secretory pathway and outside the cell in new chemical environments that can alter the protein fold. In collaboration with J. Kelly, Department of Chemistry, we are studying the link between trafficking defects and the protein-folding energetics (Fig. 1) of a number of conformational diseases, including cystic fibrosis, hereditary childhood emphysema, Gaucher disease, familial amyloidosis of Finnish type, and transthyretin amyloidosis. The latter analyses have led to a new understanding of the function of the endoplasmic reticulum in normal physiology, suggesting that this compartment functions as a capacitor for protein folding. F i g . 1 . Export of cargo (ball-and-stick icons) from the endoplasmic reticulum to the cell surface occurs through cargo-selective budding regions (lower part of image). Cargo export is thought to be restricted to fully folded structures by “quality control” pathways. Surprisingly, analysis of protein-folding energetics and secretion indicated that protein variants destabilized relative to the wild-type fold are secreted with wild-type efficiency, triggering deposition of amyloid fibril aggregates in the extracellular space (upper part of image). Appreciation of the importance of the global energetics of the protein fold in secretion leads to a key shift in understanding the secretory pathway, from quality control to mechanistic pathways. CELL BIOLOGY 2005 Through a multidisciplinary approach that combines the tools of chemistry, biology, systems biology, and structure, we hope to gain critical insight into the basis for a variety of inherited transport diseases. Knowledge of the function of these cargo selection pathways will enable the development of small-molecule chemical chaperones to encourage export and stability of misfolded proteins, leading to restoration of normal cellular function. PUBLICATIONS Bannykh, S.I., Plutner, H., Matteson, J., Balch, W.E. COPI-independent polarization of the Golgi stack. Traffic, in press. Gurkan, C., Alory, C., Su, A.I., Lapp, H., Hogenesch, J., Balch, W.E. Large-scale profiling of Rab GTPase trafficking networks: the membrome. Mol. Biol. Cell, in press. Page, L.J, Huff, M.E., Kelly, J.W., Balch, W.E. Ca2+ binding protects against gelsolin amyloidosis. Biochem. Biophys. Res. Commun. 322:1105, 2004. Page, L.J., Suk, J.Y., Huff, M.E., Lim, H.J., Venable, J., Yates, J.R. III, Balch, W.E., Kelly, J.W. Metalloprotease cleavage within the extracellular matrix triggers the final stages of gelsolin amyloidogenesis. EMBO J., in press. Sekijima, Y., Wiseman, R.L., Matteson, J., Hammarstrom, P., Miller, S.R., Sawkar, A.R., Balch, W.E., Kelly, J.W. The biological and chemical basis for tissue-selective amyloid disease. Cell 121:73, 2005. Wang, X., Matteson, J., An, Y., Moyer, B., Yoo, J.S., Bannykh, S., Wilson, I.A., Riordan, J.R., Balch, W.E. COPII-dependent export of cystic fibrosis transmembrane conductance regulator from the ER uses a di-acidic exit code. J. Cell Biol. 167:65, 2004. Wang, X., Riordan, J.R., Balch, W.E. Rescue of ∆508 CFTR by inducible folding chaperones. J. Biol. Chem., in press. Wiseman, L., Balch, W.E. Druging protein folding pathways through stress: a new pharmacological adventure. Trends Biochem. Sci., in press. Automated Molecular Imaging B. Carragher, C.S. Potter, A. Cheng, D. Fellmann, F. Guerra, G. Lander, S. Mallick, P. Mercurio, J. Pulokas, J. Quispe, 27 holds great promise for routinely and efficiently providing structural information at a resolution sufficient to resolve the secondary structure in these large molecular machines. This method could then be used in conjunction with high-resolution x-ray structures of individual proteins to interpret very large complexes to near-atomic resolution. Unfortunately, the methods generally used in molecular microscopy are both time-consuming and labor intensive. These include the preparation of suitable specimens, the acquisition of the required very large numbers of electron micrographs, and the supervision of the sometimes-complex software needed for analysis and reconstruction of the 3-dimensional electron density maps. The challenge then is to transform structure determination via electron microscopy into a high-throughput method. Success in this endeavor will not only facilitate the process of molecular microscopy but also expand the scope of accessible problems and make possible investigations that currently are deemed too high risk because of the inordinate effort involved. To this end, we are developing technologies to address automation for specimen handling, image acquisition, data processing, and integration of data information. We have created an integrated software system, called Leginon, that automatically collects electron micrographs of macromolecular structures (Fig. 1). This system has been integrated with automated particle-selection algorithms and analysis and processing software. A major focus of our activities is the National Resource for Automated Molecular Microscopy (NRAMM), a biotechnology resource center funded by the National S. Stagg, C. Suloway, C. Yoshioka, Y. Zhu lucidating the structure and mechanism of action of molecular machines is an emerging frontier in understanding how the information in the genome is transformed into cellular activities. Molecular machines are associations of individual components (e.g., proteins, nucleic acids, lipids) in the form of large complexes; examples include ribosomes, transcription complexes, track-motor complexes, and membrane-embedded pumps and channels. These machines are large and may also be conformationally and compositionally dynamic or present in comparatively low numbers, factors that make them extremely challenging (or impossible) objects for study by x-ray crystallography and nuclear magnetic resonance methods. Molecular microscopy, however, E Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. F i g . 1 . Multiscale image collection from a transmission electron microscope is controlled by using Leginon, an automated data collection system. Data are managed by using a relational database and can be visualized by using a Web browser. 28 CELL BIOLOGY 2005 Center for Research Resources, National Institutes of Health. The overall mission is to develop, test, and apply technology to completely automate the processes involved in using electron cryomicroscopy to solve macromolecular structures. The current focus of NRAMM is the development of new approaches for specimen handling, automated acquisition, automated processing, and information handling. The activities of the NRAMM are closely coupled to a number of collaborative and service projects in which fundamental biological goals are incentives for developing the new technology. During 2004, more than 20 of these projects were actively pursued. Specific examples include structural studies of coatomer complex II–coated vesicles, which are responsible for transport of proteins from the endoplasmic reticulum to the Golgi apparatus; characterization of viruslike particles manufactured in recombinant expression systems; structural studies of the crustacean clotting protein; structural studies of coronaviruses; and the structural characterization of the chloroplast ribosome. All of these collaborative projects guided the development of new approaches in the 4 core technologies while simultaneously providing new structural information relevant to specific biological problems. An additional project, sponsored by the National Science Foundation, is the development of automated data collection techniques for imaging serial sections obtained by using an electron microscope. Understanding the fine structure of cells and cellular components contributes to a more profound understanding of cellular function and intracellular or intercellular interactions. In order to visualize these large, complex structures in 3 dimensions at resolutions sufficient to observe structure on the nanoscale, the cells must be cut into sections and then examined by using a transmission electron microscope. Acquiring high-magnification images of a long series of sections is difficult and extremely labor intensive. The region of interest in each section must be tracked across sections and across grids, a process that requires examining the sections at a variety of scales before acquiring high-magnification images of interesting areas. Multiscale imaging of this sort is not straightforward because the image formed by an electron microscope shifts and rotates as the magnification is changed. The overall task of reconstructing a 3-dimensional volume from a set of serial sections is challenging and time-consuming, and the number of large-scale reconstructions has been limited to a few spectacular Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. examples. Our objectives are to design, develop, and implement a software application to automate the task of acquiring high-magnification images of specific regions of the cell across tens to hundreds of serial sections. PUBLICATIONS Dang, T.X., Farah, S.J., Gast, A., Robertson, C., Carragher, B., Egelman, E., WilsonKubalek, E.M. Helical crystallization on lipid nanotubes: streptavidin as a model protein. J. Struct. Biol. 150:90, 2005. Mallick, S.P., Carragher, B., Potter, C.S., Kriegman, D.J. ACE: automated CTF estimation. Ultramicroscopy 104:8, 2005. Mallick, S.P., Zhu, Y., Kriegman, D. Detecting particles in cryo-EM micrographs using learned features. J. Struct. Biol. 145:52, 2004. O’Keefe, M.A., Turner, J.H., Musante, J.A., Hetherington, C.J.D., Cullis, A.G., Carragher, B., Jenkins, R., Milgrim, J., Milligan, R.A., Potter, C.S., Allard, L.F., Blom, D.A., Degenhardt, L., Sides, W.H. Laboratory design for high-performance electron microscopy. Microsc. Today 12:8, 2004. Suloway, C., Pulokas, J., Fellmann, D., Cheng, A., Guerra, F., Quispe, J., Stagg, S., Potter, C.S., Carragher, B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol., in press. Chemical Physiology B.F. Cravatt, J. Alexander, K. Barglow, M.H. Bracey, K. Chiang, M. Evans, H. Hoover, N. Jessani, D. Leung, K. Matsuda, A. Mulder, S. Niessen, M. McKinney, A. Saghatelian, S. Sieber, G. Simon, A. Speers, B. Wei e are interested in understanding complex physiology and behavior at the level of chemistry and molecules. At the center of cross talk between different physiologic processes are endogenous compounds that provide a molecular mode for intersystem communication. However, many of these molecular messages remain unknown, and even in the instances in which the participating molecules have been defined, the mechanisms by which these compounds function are for the most part still a mystery. We are investigating a family of chemical messengers termed the fatty acid amides, which affect many physiologic functions, including sleep and pain. In particular, one member of this family, oleamide, accumulates selectively in the cerebrospinal fluid of tired animals. This finding suggests that oleamide may function as a molecular indicator of an organism’s need for sleep. Another fatty acid amide, anandamide, may be an endogenous ligand for the cannabinoid receptor in the brain. The in vivo levels of chemical messengers such as the fatty acid amides must be tightly regulated to maintain proper control over the influence of the messengers W CELL BIOLOGY 2005 on brain and body physiology. We are characterizing a mechanism by which the level of fatty acid amides can be regulated in vivo. Fatty acid amide hydrolase degrades the fatty acid amides to inactive metabolites. Thus, the hydrolase effectively terminates the signaling messages conveyed by fatty acid amides, possibly ensuring that these molecules do not generate physiologic responses in excess of their intended purpose. We are using transgenic and synthetic chemistry techniques to study the role of the hydrolase in the regulation of fatty acid amide levels in vivo. In collaboration with R.C. Stevens, Department of Molecular Biology, we solved the first 3-dimensional structure of fatty acid amide hydrolase. We are using this information to explore the molecular mechanism of action of the enzyme and to design inhibitors of the hydrolase. We are also interested in proteins responsible for the biosynthesis of fatty acid amides. Another area of interest is the design and use of chemical probes for the global analysis of protein function. The evolving field of proteomics, defined as the simultaneous analysis of the complete protein content of given cell or tissue, encompasses considerable conceptual and technical challenges. We hope to enhance the quality of information obtained from proteomics experiments by using chemical probes that indicate the collective catalytic activities of entire classes of enzymes. These activity-based probes could be used to record variations in protein function independent of alterations in protein abundance and would be a powerful and complimentary set of tools for proteome analysis. To date, we have generated activity-based probes for more than a dozen enzyme classes, including serine hydrolases, metalloproteases, glutathione S-transferases, and several oxidoreductases. We are using the probes to explore the roles that enzymes play in a variety of physiologic and pathologic processes, especially the progression of cancer. We are also developing complementary strategies for profiling the complete content of metabolites in cells and tissues (the “metabolome”) to facilitate the assignment of endogenous substrates to enzymes of uncharacterized function. PUBLICATIONS Azad, S.C., Monory, K., Marsicano, G., Cravatt, B.F., Lutz, B., Zieglgansberger, W., Rammes, G. Circuitry for associative plasticity in the amygdala involves endocannabinoid signaling. J. Neurosci. 24:9953, 2004. Barglow, K.T., Cravatt, B.F. Discovering disease-associated enzymes by proteome reactivity profiling. Chem. Biol. 11:1523, 2004. Chiang, K.P., Gerger, A.L., Sipe, J.C., Cravatt, B.F. Reduced cellular expression and activity of the P129T mutant of human fatty acid amide hydrolase: evidence for a link between defects in the endocannabinoid system and problem drug use. Hum. Mol. Genet. 13:2113, 2004. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 29 Cravatt, B.F., Lichtman, A.H. The endogenous cannabinoid system and its role in nociceptive behavior. J. Neurobiol. 61:149, 2004. Cravatt, B.F., Saghatelian, A., Hawkins, E.G., Clement, A.B., Bracey, M.H., Lichtman, A.H. Functional disassociation of the central and peripheral fatty acid amide signaling systems. Proc. Natl. Acad. Sci. U. S. A. 101:10821, 2004. Jessani, N., Humphrey, M., McDonald, W.H., Niessen, S., Gangadharan, B., Yates, J.R. III, Mueller, B.M., Cravatt, B.F. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc. Natl. Acad. Sci. U. S. A. 101:13756, 2004. Jessani, N., Niessen, S., Mueller, B.M., Cravatt, B.F. Breast cancer cell lines grown in vivo: what goes in isn’t always the same as what comes out. Cell Cycle 4:253, 2005. Jessani, N., Young, J.A., Diaz, S.L., Patricelli, M.P., Varki, A., Cravatt, B.F. Class assignment of sequence-unrelated members of enzyme superfamilies by activitybased protein profiling. Angew. Chem. Int. Ed. 44:2400, 2005. Leung, D., Wu, W., Hardouin, C., Cheng, H., Hwang, I., Cravatt, B.F., Boger, D.L. Discovery of an exceptionally potent and selective class of fatty acid amide hydrolase inhibitors enlisting proteome-wide selectivity screening: concurrent optimization of enzyme inhibitor potency and selectivity. Bioorg. Med. Chem. Lett. 15:1423, 2005. Lichtman, A.H., Leung, D., Shelton, C.C., Saghatelian, A., Hardouin, C., Boger, D.L., Cravatt, B.F. Reversible inhibitors of fatty acid amide hydrolase that promote analgesia: evidence for an unprecedented combination of potency and selectivity. J. Phamacol. Exp. Ther. 311:441, 2004. Saghatelian, A., Cravatt, B.F. Global strategies to integrate the proteome and metabolome. Curr. Opin. Chem. Biol. 9:62, 2005. Saghatelian, A., Jessani, N., Joseph, A., Humphrey, M. Cravatt, B.F. Activitybased probes for the proteomic profiling of metalloproteases. Proc. Natl. Acad. Sci. U. S. A. 101:10000, 2004. Saghatelian, A., Trauger, S.A., Want, E.J., Hawkins, E.G., Siuzdak, G., Cravatt, B.F. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry 43:14332, 2004. Sieber, S.A., Mondala, T., Head, S.R., Cravatt, B.F. Microarray platform for profiling enzyme activities in complex proteomes [published correction appears in J. Am. Chem. Soc. 127:4114, 2005]. J. Am. Chem. Soc. 126:15640, 2004. Mechanics of Cell Migration and Chromosome Segregation G. Danuser, J. Dorn, H. Jaqaman,* K. Jaqaman, L. Ji, A. Kerstens, D. Loerke, M. Machacek, A. Matov, G. Yang, Y. Ying * Bethlehem University, Bethlehem, West Bank ur goal is to understand how the action of thousands of force-generating molecular machines is regulated to achieve complex outputs, such as the directed movement of a cell or of its organelles. Specifically, we investigate how assembly and contraction of the actin cytoskeleton and the structural interactions of actin filaments with other components of the cytoskeleton mediate cell migration. In parallel, we study how assembly and disassembly of microtubules are orchestrated during cell division to symmetrically distribute the replicated DNA from the mother cell into 2 daughter cells. O 30 CELL BIOLOGY 2005 To examine these molecular systems, we develop computational models that predict how the convoluted dynamics of many molecular machines could transform into cell-level responses. Subsequently, we deduce molecular-level mechanisms by fitting the models to measurements of cell dynamics. The challenges arising from such data-driven, multiscale modeling are 2-fold: the precise measurement of single-cell dynamics and the implementation of numerical tools for fitting large sets of cell-level data to models with molecular resolution. During the past year, we made important advances in both directions. In collaboration with C.M. WatermanStorer, Department of Cell Biology, we used quantitative fluorescent speckle microscopy to analyze the architectural dynamics of the actin cytoskeleton during cell migration. We discovered that the initiating events of cell protrusion are mediated by 2 materially distinct, yet spatially overlapping, actin networks. This puzzling find raises several questions that define the focus of our current research: What are the molecular mechanisms by which cells assemble 2 networks of the same polymer? What are the functional differences between the networks? Do signals target the networks differentially, and if so, which signals coordinate the dynamics of the networks? To study chromosome segregation, we have developed automated 3-dimensional light microscopy to probe the changes in microtubule dynamics due to mutations in kinetochore proteins in yeast. We will use this information to model the functional relationships between kinetochore proteins and between the kinetochore and the attached microtubules, which couple the chromosome to the force-generating machinery of the spindle. We made an important step toward this goal by implementing a novel mathematical framework for matching stochastic models of microtubule dynamics to singlecell data. Using this framework not only enables us to test mechanistic models of the kinetochore-microtubule interaction but also provides a generic tool for comparing statistical models of molecular processes with measurements obtained by using light microscopy. We are also using the framework to analyze the functional links between actin cytoskeleton dynamics and clathrinmediated endocytosis. PUBLICATIONS Adams, M.C., Matov, A., Yarar, D., Gupton, S.L., Danuser, G., Waterman-Storer, C.M. Signal analysis of total internal reflection fluorescent speckle microscopy (TIRFSM) and wide-field epi-fluorescence FSM of the actin cytoskeleton and focal adhesions in living cells. J. Microsc. 216(Pt. 2):138, 2004. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Gupton, S., Anderson, K.L.., Kole, T.P., Fischer, R.S., Ponti, A., Hitchcock-DeGregori, S., Danuser, G., Fowler, V.M., Wirtz, D., Hanein, D., Waterman-Storer, C.M. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosins. J. Cell Biol. 168:619, 2005. Künzi, P.A., Lussi, J., Aeschimann, L., Danuser, G., Textor, M., de Rooij, N.F., Staufer, U. Nanofabrication of protein-patterned substrates for future cell adhesion experiments. Microelectron. Eng. 78-79:582, 2005. Ponti, A., Machacek, M., Gupton, S.L., Waterman-Storer, C.M., Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science 305:1782, 2004. Vallotton, P., Danuser, G., Bohnet, S., Meister, J.-J., Verkhovsky, A. Tracking retrograde flow in keratocytes: news from the front. Mol. Biol. Cell 16:1223, 2005. Bioorganic Chemistry of Proteins P.E. Dawson, F. Brunel, M. Churchill, T. Tiefenbrunn, N. Metanis ur focus is the development and use of methods to incorporate unnatural chemical groups into proteins. We developed a chemical approach for producing the large polypeptide chains that make up protein molecules, enabling us to change the structure of a protein in ways impossible by natural means. This chemical ligation approach greatly facilitates the synthesis of proteins of moderate size and has opened the world of proteins to the synthetic tools of organic chemistry. During the past year, we used these tools to study the agouti protein, vitronectin, basic helix-loop-helix DNA-binding domains, and the enzyme 4-oxalocrotonate tautomerase. Our goal is to introduce noncoded amino acids and other chemical groups into proteins to better understand the molecular basis of protein function. O E L E C T R O S TAT I C I N T E R A C T I O N S I N E N Z Y M E C ATA LY S I S A N D D N A B I N D I N G The relative contributions of electrostatic interactions and hydrogen-bonding interactions of arginine residues in proteins have been difficult to evaluate directly because this amino acid side chain has no natural isosteric replacement. To solve this problem, we used the unnatural amino acid citrulline to probe these interactions in the small enzyme 4-oxalocrotonate tautomerase. Citrulline can present 2 neutral hydrogen bonds from the urea side chain in a manner superimposable to the charged guanidinium group of arginine. We found that neutralization of arginine at position 39, which binds to the ketoacid group of the substrate, resulted in a 1000-fold change in the catalytic turnover rate, strongly supporting the key role of electrostatic interactions in this enzyme. Recently, we used CELL BIOLOGY 2005 these citrulline analogs of 4-oxalocrotonate tautomerase to design an enzyme with high specificity for an unnatural ketoamide substrate. These studies complement our earlier work indicating that 4-oxalocrotonate tautomerase, which naturally uses an acid-base mechanism, could be converted, with a change in a single amino acid, into a nucleophilic catalyst for the decarboxylation of oxaloacetate. We also used citrulline analogs to probe interactions between transcription factors and the phosphate backbone of DNA. As was the case with 4-oxalocrotonate tautomerase, the citrulline mutation was destabilizing, although the dynamic range of the assay was limiting. H I V VA C C I N E D E S I G N Eliciting broadly neutralizing antibodies is a major goal of HIV vaccine design. This effort is complicated by the poor accessibility of conserved regions of HIV envelope proteins to antibodies. The membrane proximal region of the HIV envelope protein gp41 has generated marked research because of the discovery of 2 neutralizing antibodies that bind this sequence. We are collaborating with D.R. Burton, Department of Immunology, and I. Wilson, Department of Molecular Biology, to design peptides that mimic the helical conformation of the peptide that binds to the neutralizing antibody 4E10. Immunization of simple peptides obtained from gp41 elicited a nonneutralizing response, presumably because of differences between the neutralizing conformation of the peptide and the most immunogenic conformation of the peptide. We fully characterized the 4E10 epitope and constrained the peptide to adopt the primarily helical conformation recognized by the 4E10 antibody. These studies have yielded a series of peptidomimetics that bind as tightly to the 4E10 antibody as the full-length gp41 glycoprotein does. Our goal is to eliminate all surfaces of the peptide that are not required for 4E10 binding or to mask them with soluble polymers and carbohydrates. We plan to use this negative design approach in combination with binding studies, immunization, and structural analysis by crystallography. PUBLICATIONS Beltran, A.C., Dawson, P.E., Gottesfeld, J.M. Role of DNA sequence in the binding specificity of synthetic basic-helix-loop-helix domains. Chembiochem 6:104, 2004. Brunel, F.M., Dawson, P.E. Synthesis of constrained helical peptides by thioether ligation: application to analogs of gp41. Chem. Commun. (Camb.) 2552, 2005, Issue 20. Deechongkit, S., Dawson, P.E., Kelly, J.W. Toward assessing the position-dependent contributions of backbone hydrogen bonding to β-sheet folding thermodynamics employing amide-to-ester perturbations. J. Am. Chem. Soc. 126:16762, 2004. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 31 Kamikubo, Y., De Guzman, R., Kroon, G., Curriden, S., Neels, J.G., Churchill, M.J., Dawson, P., Oldziej, S., Jagielska, A., Scheraga, H.A., Loskutoff, D.J., Dyson, H.J. Disulfide bonding arrangements in active forms of the somatomedin B domain of human vitronectin. Biochemistry 43:6519, 2004. McNulty, J.C., Jackson, P.J., Thompson, D.A., Chai, B., Gantz, I., Barsh, G.S., Dawson, P.E., Millhauser, G.L. Structures of the agouti signaling protein. J. Mol. Biol. 346:1059, 2005. Metanis, N., Brik, A., Dawson, P.E., Keinan, E. Electrostatic interactions dominate the catalytic contribution of Arg39 in 4-oxalocrotonate tautomerase. J. Am. Chem. Soc. 126:12726, 2004. Metanis, N., Keinan, E., Dawson P.E. A designed synthetic analog of 4-OT is specific for a non-natural substrate. J. Am. Chem. Soc. 127:5862, 2005. Actin Dynamics in Cell Morphogenesis and Function V.M. Fowler, T. Fath, R.S. Fischer, C. McKeown, J. Moyer, R. Nowak, J. Palomique egulation of actin dynamics at the ends of filaments determines the organization and turnover of actin cytoskeletal structures and is critical for cell motility and cell architecture. For example, in striated muscle and red blood cells (RBCs), actin filaments are organized into regular architectural arrays that persist for the lifetime of the cell and are important for maintenance of cell shape, mechanical properties, and physiologic function. In contrast, in motile cells, new actin filaments are rapidly assembled at the barbed ends of the filaments and disassembled at the pointed ends during extension of lamellipodia or filopodia. Our goal is to elucidate the distinct regulatory mechanisms that control polymerization and dynamics of the rapidly turning over actin filaments of motile cells as compared with the stable, long-lived actin filaments of nonmotile cells. Specifically, we focus on regulation by the family of tropomodulin proteins that cap the pointed ends of actin filaments and the roles of these proteins in actin-based morphogenetic processes in development. Tropomodulins are a conserved family of tropomyosin-regulated proteins of about 40 kD that cap the pointed ends of actin filaments. Tropomodulins have 2 domains: an unstructured, flexible N-terminal domain and a compact, folded C-terminal domain consisting of 5 leucine-rich repeats. The N-terminal domain binds tropomyosin and caps tropomyosin-actin pointed ends with nanomolar affinity. The C-terminal domain caps actin pointed ends with submicromolar affinity and is unaffected by tropomyosin. In striated muscle myofibrils, tropomodulin 1 is associated with the pointed ends of thin filaments and controls the length of filaments by transiently capping the pointed ends and regulating R 32 CELL BIOLOGY 2005 actin association and dissociation. The N- and C-terminal domains of tropomodulin 1 may have distinct functions in stabilizing thin filaments and in controlling filament length, respectively, but how the 2 domains regulate dynamics of the pointed ends in vivo to control filament assembly and disassembly is unclear. To investigate the in vivo function of tropomodulin 1 in regulating assembly and disassembly of thin filaments in muscle, we use mice that lack the gene for this tropomodulin. We showed previously that myofibril assembly in the heart is grossly aberrant in the embryos of these mutants, leading to aborted cardiac development and the death of embryos between days 9 and 10 of development. To examine the primary defect in myofibril assembly, we examined nascent myofibrils on myocyte membranes in embryos at 7–8 days of development, before the appearance of gross cardiac abnormalities. In wild-type embryos, the earliest myofibrils contain 1–3 sarcomeres in tandem with regularly spaced Z bodies and continuous F-actin, indicative of unregulated filament lengths. Such sarcomere structures are never observed in the absence of tropomodulin 1; instead, α-actinin and F-actin are present in rodlike, aberrant Z disc structures on myocyte membranes. This finding suggests that tropomodulin 1 has a novel early function in the organization of Z discs into sarcomeres before the stage of regulation of the length of thin filaments, a later event of myofibril formation. In ongoing experiments, we are determining how the absence of tropomodulin 1 affects assembly of tropomyosin and other components of thin or thick filaments and are examining how defective assembly of myofibrils in turn leads to myocyte disorganization and disruption of cellular contacts that occur in the absence of tropomodulin 1. Tropomodulin 1 is also present in RBCs, where it caps the pointed ends of the short actin filaments in the spectrin membrane skeleton. To investigate the consequences in RBCs of deleting the gene for this tropomodulin, we prevented death in embryos of the mutant mice by expressing a tropomodulin 1 transgene solely in the heart. The result was viable mice with no tropomodulin 1 in their RBCs. Western blotting indicated that levels of tropomodulin 3 were increased in tropomodulin 1–deficient RBCs, but the levels were considerably less than the normal amount of tropomodulin 1. Hematologic analyses revealed that these mice had a compensated mild hemolytic anemia, with increased reticulocytosis and RBCs that were abnormally variable in size in blood smears. Measurements of mechanical stability and deformability indicated that tropomodulin 1–deficient RBCs were less deformable and more fragPublished by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. ile than were normal RBCs. Using these mutant RBCs, we can test our hypothesis that tropomodulin 1 regulates the lengths of the short actin filaments in RBCs, thus influencing stability of the membrane skeleton and cell survival and function in vivo. The tropomodulin 3 isoform is expressed ubiquitously, including in motile endothelial cells, where it is enriched in leading lamellipodia and acts as a negative regulator of cell motility. Recently, we found that unlike other members of the tropomodulin family, tropomodulin 3 can also bind to actin monomers in addition to capping pointed ends,. Tropomodulin 3 can be chemically cross-linked to actin in a 1-to-1 complex, creating the expected 82-kD species and providing a powerful tool for identifying the amino acids at the tropomodulin 3–actin binding interface. The results of coimmunoprecipitation and fluorescence staining assays for actin monomers indicate that tropomodulin 3 also binds monomers in endothelial cells in vivo. We propose that binding of the actin monomer to tropomodulin 3 can reduce the amount of tropomodulin 3 available for binding to actin pointed ends, providing a mechanism to regulate tropomodulin 3 activity and preferentially target tropomodulin 3 capping to higher affinity tropomyosin-actin pointed ends. PUBLICATIONS Gupton, S.L., Anderson, K.L., Kole, T.P., Fischer, R.S., Ponti, A., HitchcockDeGregori, S.E., Danuser, G., Fowler, V.M., Wirtz, D., Hanein, D., WatermanStorer, C.M. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 168:619, 2005. Schmid, M., Nanda, I., Hoehn, H., Schartl, M., Haaf, T., Buerstedde, J.M., et al. Second report on chicken genes and chromosomes 2005. Cytogenet. Genome Res. 109:415, 2005. Angiogenesis-Dependent Disease and Membrane Protein Topogenesis M. Friedlander, E. Aguilar, E. Banin, F. Barnett, M. Dorrell, S.F. Friedlander, R. Gariano, S. Moreno, A. Otani, M. Ritter, L. Scheppke, H. Uusitalo-Jarvinen, W. Ruf, P.R. Schimmel, D.A. Cheresh,* S.M. Simon,** K. Philipson*** * University of California, San Diego, California ** Rockefeller University, New York, New York *** University of California, Los Angeles, California ANGIOGENESIS-DEPENDENT DISEASE M ost diseases that cause catastrophic loss of vision do so as a result of abnormal growth of blood vessels. Similarly, tumors depend on CELL BIOLOGY 2005 a blood supply for their growth and use these new vessels as an avenue for metastasis. Blood vessels themselves can generate tumors (e.g., hemangiomas) when the growth and organization of vascular endothelial cells is not properly controlled. Our goal is to understand the mechanisms of ocular neovascularization in normal and pathologic situations. We use a neonatal mouse retina model to identify regulators of developmental angiogenesis and understand endothelial guidance mechanisms. In addition, in a long-standing collaboration with D.A. Cheresh, University of California, San Diego, we are using this system to evaluate the role of integrins in these processes. In collaboration with P.R. Schimmel, Department of Molecular Biology, we found that fragments of tryptophan tRNA synthetase are potent angiostatics, and using a recombinant protein, we showed that a form of cell-based delivery caused significantly reduced retinal vascularization. Most recently, we used combination therapy to show that targeting multiple, distinct angiogenic pathways with fragments of tryptophan tRNA synthetase and antagonists of integrins and vascular endothelial cell growth factor provides highly synergistic, potent angiostatic activity. Although this therapeutic approach should be useful in the treatment of diseases in which complete inhibition of angiogenesis is desirable, it may not be efficacious in the treatment of ischemic retinal disease. In ischemic retinal disease, relief of hypoxia by vascular reconstruction, rather than destruction, may be the desired outcome. To examine possible therapy for diseases of retinal ischemia, we explored the potential usefulness of lineage-negative hematopoietic stem cells derived from the bone marrow of adult mice for cell-based delivery of angiostatic and neurotrophic substances and for the trophic actions of the cells themselves in vascular and neuronal degenerative diseases. We found that the stem cells contain a variety of progenitor (stem) cells, including endothelial precursor cells, and that the precursor cells specifically target activated retinal astrocytes, incorporate into new vessels, and, in a mouse model of retinal degeneration, rescue and stabilize a degenerating retinal vasculature. We also showed that these stem cells derived from the bone marrow of adults have a profound neurotrophic effect when injected into the eyes of mice with inherited retinal degeneration; not only is the vasculature rescued in these mice but photoreceptors and visual function are also preserved. The stem cells also can rescue retinal vasculature subject to hypoxic stress and may be useful for the treatment of Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 33 ischemic retinopathies such as diabetes and retinopathy of prematurity. Glioblastoma multiforme is an incurable brain tumor that is usually fatal within 1 year after diagnosis. We are using gene therapy and a rat model of this disease to study the efficacy of an antiangiogenic approach in treating these tumors. Hemangiomas are endothelial tumors that proliferate rapidly and later involute spontaneously. We are using DNA microarrays to study changes in gene expression as hemangiomas progress. Our goal is to identify (1) new targets for therapy for these tumors and (2) novel regulators of angiogenesis. In collaboration with G.R. Nemerow, Department of Immunology, we used pseudotyped adenovirus to selectively target specific cell types in the retina. By using the appropriate fiber type, we can deliver transgenes to cells, such as photoreceptors, that ordinarily are not targeted by adenovirus. MEMBRANE PROTEIN TOPOGENESIS We are also studying the mechanism whereby proteins are asymmetrically integrated into cell membranes. In addition to studies of membrane protein topogenesis at the molecular level, we have been studying defects in protein processing and insertion that occur in several degenerative diseases of the eye. In collaboration with K. Philipson, University of California, Los Angeles, we are investigating the topology of the cardiac sodium-calcium exchanger. On the basis of hydropathy analysis of the amino acid sequence, the exchanger is proposed to contain 12 hydrophobic segments, the first of which is a cleaved signal sequence. Using a variety of reporter domains (glycosylation sites, epitopes, and proteolytic cleavage sites), we analyzed the topology of the exchanger both in vitro and in oocyte expression systems. Because nearly all other polytopic eucaryotic membrane proteins do not have cleaved signal sequences, we are investigating the putative role of such a sequence in the insertion and targeting of these exchangers. Our results indicate that the native, cleaved N-terminal signal sequence is not necessary for insertion of a functional exchanger into the cell membrane. In contrast, the photoreceptor exchanger does not have a cleaved N-terminal signal sequence. If the N-terminal 65 amino acids are deleted, translocation of the N terminus of the protein is disrupted, but the remainder of the exchanger is integrated into the membrane. We are also using large-scale genomic analysis to study transgenic mice in which a mutated exchanger is expressed and mice that lack the gene for the exchanger. 34 CELL BIOLOGY 2005 PUBLICATIONS Barnett, F.H., Scharer-Schusz, M., Wood, M., Yu, X., Wagner, T.E., Friedlander, M. Intra-arterial delivery of endostatin gene to brain tumors prolongs survival and alters tumor vessel ultrastructure. Gene Ther. 11:1283, 2004. Dorrell, M.I., Friedlander, M. Cell guidance in retinal angiogenesis. Prog. Retin. Eye Res., in press. Henderson, S.A., Goldhaber, J.I., So, J.M., Han, T., Motter, C., Ngo, A., Chantawansri, C., Ritter, M.R., Friedlander, M., Nicoll, D.A., Frank, J.S., Jordan, M.C., Roos, K.P., Ross, R.S., Philipson, K.D. Functional adult myocardium in the absence of Na+Ca2+ exchange: cardiac-specific knockout of NCX1. Circ. Res. 95:604, 2004. Otani, A., Dorrell, M.I., Kinder, K., Moreno, S.K., Nusinowitz, S., Banin, E., Heckenlively, J., Friedlander, M. Rescue of retinal degeneration by intravitreally injected adult bone marrow-derived lineage-negative hematopoietic stem cells. J. Clin. Invest. 114:765, 2004. Otani, A., Friedlander, M. Retinal vascular regeneration. Semin. Ophthalmol. 20:43, 2005. Ritter, M., Aguilar, E., Banin, E., Scheppke, L., Uusitalo-Jarvinen, H., Friedlander, M. Three-dimensional in vivo imaging of the mouse ocular vasculature. Invest. Ophthal. Vis. Sci., in press. Ritter, M., Friedlander, M. Integrins in ocular angiogenesis. In: Ocular Angiogenesis. Tobran-Tink, J., Barnstable, C. (Eds.). Humana Press, Totowa, NJ, in press. Nucleocytoplasmic Transport and Role of the Nuclear Lamina in Higher Level Nuclear Organization L. Gerace, G. Ambrus-Aikelin, A. Aschrafi, J. Bednenko, A. Bubeck, A. Cassany, B. Chen, T. Guan, K. Kanelakis he nuclear envelope is a specialized domain of the endoplasmic reticulum that forms the boundary of the nucleus in eukaryotic cells. The envelope consists of inner and outer nuclear membranes, the nuclear lamina, and nuclear pore complexes (NPCs). The nuclear lamina, a protein meshwork lining the inner nuclear membrane, provides a structural scaffold for the nuclear envelope and an anchoring site at the nuclear periphery for chromatin. NPCs are large supramolecular assemblies that span the nuclear envelope and serve as channels for molecular transport between the nucleus and the cytoplasm. We are using a combination of biochemical, structural, and functional approaches to investigate NPCs and the lamina. T NUCLEOCYTOPLASMIC TRANSPORT MECHANISMS Transport of protein and RNA through NPCs is an energy-dependent process mediated by nucleocytoplasmic shuttling receptors of the karyopherin β family. Karyopherins bind to transport signals on protein or RNA cargo molecules, and the receptor-cargo comPublished by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. plexes are translocated through the NPC by binding of the receptors to a group of NPC proteins (nucleoporins) that contain phenylalanine-glycine amino acid motifs. The directionality of nuclear transport is determined largely by the small GTPase Ran, which directly interacts with karyopherins and thereby regulates cargo binding. Conformational flexibility of karyopherins is thought to be fundamental to their dynamic interactions with cargo, Ran, and nucleoporins. We are using in vitro assays with digitonin-permeabilized cells to analyze the molecular events that specify translocation of cargo-receptor complexes through NPCs. Recently, using site-directed mutagenesis of importin β, the prototypical nuclear import receptor, we characterized 2 distinct binding sites in importin β for nucleoporins containing the phenylalanine-glycine motif and defined mutational hot spots for cargo binding. A major goal is to determine how the conformational dynamics of importin β are linked to discrete transport steps. To this end, we are complementing structure-function studies with analysis involving smallmolecule inhibitors. We are also investigating movement of cargo-receptor complexes though the central channels of NPCs, which form the major permeability barrier of the pore. We are focusing on the transport of large protein complexes, which have more stringent Ran and energy requirements for translocation than small proteins do. In a related project, we are analyzing nuclear import of the adenovirus genome, which consists of a 36-kb double-stranded DNA molecule. Results from our in vitro transport studies indicate that adenovirus DNA transport is driven by import signals on DNA-associated proteins. Our characterization of multiple import signals in adenovirus protein VII and the tight association of the protein with the genome suggest that this viral protein may be the protein adaptor involved in the DNA import. Nuclear import of protein VII involves several of the major cellular importins, suggesting that adenovirus has evolved to use redundant import pathways to ensure efficient nuclear delivery of its genome. We also are analyzing nuclear export of HIV type 1 mRNA mediated by the viral regulatory protein Rev. Rev polymerizes on a cis-acting sequence of viral mRNAs, providing a platform for assembly of nuclear export factors. We are using proteomics combined with a permeabilized cell assay for Rev-dependent HIV mRNA export to functionally characterize the proteins assembled on the Rev platform. This project is part of a larger CELL BIOLOGY 2005 collaboration with a research team at Scripps Research to identify small-molecule inhibitors of Rev function; the goal is to find compounds for developing new drugs to inhibit HIV replication in humans. Organization and Function of the Neuronal Cytoskeleton NUCLEAR LAMINA AND HIGHER LEVEL NUCLEAR S. Halpain, J. Braga, B. Calabrese, L. Dehmelt, E. Hwang, 35 O R G A N I Z AT I O N The nuclear lamina in vertebrates contains a polymer of 2–4 related intermediate filament proteins called lamins, which are associated with a number of transmembrane proteins of the inner nuclear membrane. The lamina plays essential roles in nuclear structure and function, as indicated by the recent findings that more than 15 inherited diseases in humans, including a range of muscular dystrophies, are caused by mutations in lamins or lamina-associated transmembrane proteins. The involvement of the lamina in disease is thought to be linked to its roles in nuclear integrity, chromatin structure, and gene expression. Until recently, only about 12 transmembrane proteins specific to the nuclear envelope had been identified. To survey the complete complement of proteins in the nuclear envelope, we carried out a proteomics analysis of the nuclear envelope of rodent liver in collaboration with J.R. Yates, Department of Cell Biology. We identified 67 novel putative nuclear envelope transmembrane proteins. Analysis of a subset of these proteins indicated that most are authentic components of the nuclear envelope. We are now doing a systems-level analysis of the lamina involved in muscle differentiation. For these studies, we are using transcriptional microarray analysis, quantitative proteomics, and gene silencing by RNA interference in cultured myoblasts. The goals are to identify genes that may have a role in human muscular dystrophies and to further elucidate how the protein network consisting of lamins and associated transmembrane proteins acts in nuclear structure and function. PUBLICATIONS Gerace L. TorsinA and torsion dystonia: unraveling the architecture of the nuclear envelope. Proc. Natl. Acad. Sci. U. S. A. 101:8839, 2004. Ohba, T., Schirmer, E.C., Nishimoto, T., Gerace, L. Energy- and temperaturedependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 167:1051, 2004. Schirmer, E.C., Florens, L., Guan, T., Yates, J.R. III, Gerace, L. Identification of novel integral membrane proteins of the nuclear envelope with potential disease links using subtractive proteomics. Novartis Found. Symp. 264:63, 2005. Schirmer, E.C., Gerace, L. The stability of the nuclear lamina polymer changes with the composition of lamin subtypes according to their individual binding strengths. J. Biol. Chem. 279:42811, 2004. Wiethoff, C.M., Wodrich, H., Gerace, L., Nemerow, G.R. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79:1992, 2005. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. K. Spencer uring the past year, we made significant progress in research on the development and regeneration of neurons. In 2 main projects, we focused on cytoskeletal proteins of nerve cells, key proteins that underlie the structure and morphologic flexibility required by neurons for transmitting, storing, and processing synaptic signals (Fig. 1). We used biochemical, molecular biological, and microscopy-based approaches to understand the function of these molecules. Fluorescence time-lapse imaging of living neurons is an important tool that we use to uncover structure-function relationships for cytoskeletal proteins and the consequences of dysfunction of the proteins. D F i g . 1 . Diagram of a brain cell axon illustrating the dense array of protein fibers that make up the neuronal cytoskeleton. Actin filaments and microtubules interact to regulate the initiation, elongation, branching, and guidance of neuronal axons and dendrites in the developing brain. Illustration courtesy of J. Lim, graduate student, Scripps Research. One project concerns microtubule-associated proteins (MAPs). These proteins are important in regulating the assembly and stability of microtubules and the interactions of microtubules with other components of the cytoskeleton. We found that one microtubule-binding protein, MAP2, also directly binds actin filaments and induces filament bundling. Using fluorescencebased time-lapse imaging and high-resolution confocal microscopy, we tracked the behaviors of microtubules 36 CELL BIOLOGY 2005 and actin filaments in living neuronal cells with normal and mutant forms of MAP2. Recently, we found that the microtubule-based molecular motor dynein plays a key role in transporting microtubule bundles toward the cell periphery. This dynein-dependent activity provides a key force that pushes the cell membrane outward during neurite initiation. Currently, we are identifying other cytoskeletal proteins and signal transduction pathways crucial to the initiation of neurites. A second project concerns the regulation of dendritic spines, specialized cellular protrusions that form the receptive, postsynaptic element at glutamate synapses. Spines become lost or dysmorphic in many types of mental retardation and in psychiatric conditions such as chronic depression and schizophrenia. Furthermore, spines are vulnerable to injury in diseases such as stroke and epilepsy, in which excessive release of glutamate can induce neuronal injury and subsequent cell death (a condition termed excitotoxicity). Understanding how spines form, what regulates their stability, and how they recover from injury is therefore of therapeutic interest for several neurologic conditions. Our most recent results suggest a neuroprotective role for spines, because preventing the collapse of dendritic spines attenuates neuronal cell death induced by a subsequent lethal stimulus. The spine cytoskeleton is composed mainly of actin filaments. We discovered that actin filaments in spines are rapidly broken down within minutes of an injury-inducing stimulus. However, this damage to the spine can be rapidly reversed within minutes under appropriate conditions, indicating for the first time that spines can regrow after they collapse. We identified 2 key molecules involved in regulating the shape and stability of spines. The first is the serine protease cathepsin B, a classical lysosomal protease that functions in other subcellular domains. We also discovered that myristoylated alanine-rich C kinase substrate (MARCKS), a protein that regulates both membrane trafficking and the actin cytoskeleton, is one of the many potential substrates of cathepsin B. MARCKS is a major downstream target of protein kinase C, and our recent studies implicated a key role for protein kinase C and MARCKS in the regulation of spine shape and stability. Together these projects contribute to our understanding of molecular events in normal brain development and in regeneration of neuronal structure after injury and disease. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. PUBLICATIONS Calabrese, B., Halpain, S. Essential role for the PKC target MARCKS in maintaining dendritic spine morphology. Neuron 48:77, 2005. Dehmelt, L., Halpain, S. The MAP2/tau family of microtubule-associated proteins. Genome Biol. 6:204, 2005. Graber, S., Maiti, S., Halpain, S. Cathepsin B-like proteolysis and MARCKS degradation in sub-lethal NMDA-induced collapse of dendritic spines. Neuropharmacology 47:706, 2004. Graber, S., Morrison, M.E., Halpain, S. Dendritic spines in cognition and neurological disease. Cyberounds [on-line review journal]. Available at: http://www.cyberounds.com/ conf/phsychiatryneuroscience/2004. Halpain, S., Spencer, K., Graber, S. Dynamics and pathology of dendritic spines. Prog. Brain Res. 147:29, 2005. Genetics and Genomics of Circadian Clocks S.A. Kay, G. Breton, J. Chu, A. DeSchopke, E. Farre, F.G. Harmon, S.P. Hazen, T. Imaizumi, B. Jenkins, C.Y. Liu, C. Masuda, A. Para, J. Pruneda, T.F. Schultz, H.G. Tran, D.K. Welsh,* E. Zhang * University of California, San Diego, California vast array of cellular processes fluctuate with a 24-hour periodicity, and an endogenous circadian clock is responsible for generating these biological rhythms. Circadian rhythms are found in all kingdoms of life and control diverse events ranging from sleep-wake cycles in mammals to the overall rate of photosynthesis in plants. Many pathologic changes in humans, such as sleep disorders, most likely are due to a defect in circadian rhythms, so understanding how the circadian clock operates within the cell will have significance for human health. To study how circadian clocks are built inside of cells, we use molecular, genetic, and genomic approaches in 2 model systems: mouse and Arabidopsis. In mammals, the circadian clock plays an integral role in timing many physiologic rhythms, such as blood pressure, body temperature, and liver metabolism, in anticipation of dusk and dawn. The master circadian clock resides within a region of the brain that receives light information from the eyes. However, this brain region can keep time even in the absence of light, as occurs in some who are visually blind. Mutations in the genes that encode components of the circadian clock are manifested as abnormal activity rhythms in rodents and as sleeping disorders in humans, although which photoreceptors set the clock is unclear. Thus, although marked advancements have been made in A CELL BIOLOGY 2005 understanding how the mammalian clock itself runs, little is known about how light transduces synchronizing signals to the clock. To address this major question, we are using genetic and genomic approaches to identify new gene functions in circadian biology. We are generating a number of mouse strains with mutations in known and potential photoreceptors and are testing the mice for defects in circadian rhythm. Thus far, we have determined that one photoreceptor, melanopsin, is an important contributor in maintaining synchrony between the clock and environmental light conditions. With the recently completed sequencing of the human and mouse genomes, we now know the sequences of more than 30,000 genes that can be investigated for potential roles in circadian function. We developed large-scale, in vitro, cell-based assays that can be used to rapidly determine if genes control clock activity. Combining this approach with genetic analysis will enable us to further dissect the connection between environmental stimuli, in the form of light, and the behavioral and physiologic events regulated by the circadian clock. In recent years, researchers have found that intrinsic circadian clocks exist in various peripheral tissues and cell types, directly controlling local physiology and behavior. We are studying the circadian oscillators in the liver and in the vasculature. As the first step, we are investigating the heterogeneity and distinct functions of the central and peripheral oscillators. In particular, we are examining the distinct roles of the retinoid-related orphan nuclear receptors in the clock mechanism. These nuclear receptors were recently identified in our functional genomics studies. Second, we are asking how environmental cues, mainly light-dark cycles and feeding time, entrain or synchronize peripheral oscillators. Peripheral oscillators most likely are synchronized by hormonal outputs of the suprachiasmatic nucleus or by physiologic inputs such as feeding-mediated metabolic changes. We are using transgenic mice, mice lacking certain genes, and immortalized hepatocytes and vascular smooth muscle cells in these studies along with real-time bioluminescence imaging and biochemical and genetic approaches. Furthermore, we are using high-resolution bioluminescence imaging to determine whether single neurons in the suprachiasmatic nucleus and peripheral cells are autonomous circadian clocks and to characterize the precise nature of synchronization of the molecular clockwork of individual cells. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 37 Flowering is a major event in the life cycle of higher plants. Many plants use seasonal changes in the length of days as a signal to flower, and higher plants use their circadian clocks to perceive these changes. Recently, we defined a molecular link between the circadian clock and day length–dependent regulation of flowering. A flowering time gene known as CONSTANS was identified a number of years ago and is regulated by the circadian clock. We showed that clock regulation of CONSTANS expression is the key to seasonal control of flowering in Arabidopsis. We are extending these studies by comparing gene expression profiles under conditions of long days and short days to identify other components involved in perception of day length. By combining molecular, genetic, and genomic approaches, we are beginning to define a number of molecular links between the circadian clock and rhythmic regulation of behavior and development. Analysis of circadian rhythms in multiple organisms provides a unique opportunity to define molecular controls for the behavior of whole organisms. These results will provide targets for clinical and agricultural applications to improve the quality of life. PUBLICATIONS Flechner, S.M., Kurian, S.M., Head, S.R., Sharp, S.M., Whisenant, T.C., Zhang, J., Chismar, J.D., Horvath, S., Mondala, T., Gilmartin, T., Cook, D.J., Kay, S.A., Walker, J.R., Salomon, D.R. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am. J. Transplant. 4:1475, 2004. Sato, T.K., Panda, S., Miraglia, L.J., Reyes, T.M., Rudic, R.D., McNamara, P., Naik, K.A., FitzGerald, G.A., Kay, S.A., Hogenesch, J.B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43:527, 2004. Ueda, H.R., Hayashi, S., Matsuyama, S., Yomo, T., Hashimoto, S., Kay, S.A., Hogenesch, J.B., Iino, M. Universality and flexibility in gene expression from bacteria to human. Proc. Natl. Acad. Sci. U. S. A. 101:3765, 2004. Welsh, D.K., Yoo, S.H., Liu, A.C., Takahashi, J.A., Kay, S.A. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 14:2289, 2004. Function of Nuclear Receptors in Stress and Mitochondrial Homeostasis A. Kralli, J. Cardenas, M.B. Hock, C. Tiraby-Nguyen, J. Villena uclear receptors are ligand-regulated transcription factors with important roles in mammalian development and physiology. Although many nuclear receptors are activated by recognized small N 38 CELL BIOLOGY 2005 lipophilic ligands, such as steroids and other lipids, some have no known ligands and are called orphans. We focus on nuclear receptors and receptor cofactors that affect energy metabolism pathways. GLUCOCORTICOIDS AND RESPONSE TO STRESS The ability of organisms to respond and adapt to stressors is fundamental for life. Response to stress involves activation of the neuroendocrine system and the secretion of adrenal glucocorticoids. Glucocorticoids act via the glucocorticoid receptor to enable mobilization of energy resources, recovery from the stress response, and preparation for future stressors. The transcriptional response mediated by receptors for glucocorticoids integrates the hormonal signal with signals indicating the type of stressor, the physiologic state of the organism, and the cellular environment. We are studying the regulatory mechanisms that enable the integration of such diverse signals in the activity of glucocorticoid receptors. In particular, we address the function of transcriptional coactivators as integrators of such responses. Peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α), an inducible coactivator that coordinates transcriptional programs important for energy homeostasis, is a potent, tissue-specific modulator of glucocorticoid receptors. Our studies suggest that PGC-1α, glucocorticoid receptors, and the orphan nuclear receptor estrogen-related receptor α (ERRα) are parts of the transcriptional network that coordinates the stress response. ERRα AND MITOCHONDRIAL FUNCTION ERRα was the first orphan nuclear receptor identified, yet we are just starting to understand its physiologic function. We showed that ERRα is regulated by PGC-1α, which binds ERRα and converts it from a factor with little or no transcriptional activity to a potent activator of gene expression. This activation enables the induction of ERRα target genes, including the ERRα gene itself. As a result, levels of ERRα in vivo are highest in tissues that express PGC-1α, such as heart, kidney, brown adipose tissue, and muscle, and are induced in response to signals that relay metabolic needs and that upregulate PGC -1α, such as exposure to cold, fasting, and exercise. The similar spatial and temporal expression of PGC-1α and ERRα suggests that the complex consisting of these 2 molecules mediates the induction of some of the known PGC-1α–responsive pathways. Indeed, the results of our gain-of- and loss-of-function studies support the notion that ERRα regulates facets of mitochondrial metabolism, including mitochondrial biogenesis, Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. fatty acid oxidation, the tricarboxylic acid cycle, and oxidative phosphorylation. Mitochondrial dysfunction and the reduced expression of genes associated with oxidative phosphorylation have been implicated as underlying factors in the development of insulin resistance. Our findings suggest that strategies aiming at increasing ERRα activity could be beneficial in the treatment of metabolic disease, such as type 2 diabetes. PUBLICATIONS Teyssier, C., Ma, H., Emter, R., Kralli, A., Stallcup, M.R. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes Dev., in press. Detection of Circulating Tumor Cells in Peripheral Blood P. Kuhn, R. Bruce,* K. Bethel, B. Hsieh,* M. Humphrey,* R. Krivacic,* L. Kroener, N. Lazarus,* A. Ladanyi,* D. Marrinucci, J. Nieva * Palo Alto Research Center, Palo Alto, California he spread of cancer from a primary site to secondary sites, known as metastasis, is correlated with a high incidence of mortality. Research indicates that metastasis begins when primary tumor cells disseminate, infiltrate, and invade the circulatory system. Although most disseminated cells are destroyed in the circulatory system via apoptosis, a surviving cell can invade a secondary organ, micrometastasize, and form a secondary tumor. Detection of these circulating tumor cells (CTCs), especially before metastasis occurs, is critical for increasing survival rates. CTCs exist in the peripheral blood in cancer patients at ultralow concentrations, at an estimated rate of about 1 in 10 million leukocytes. Although existing technology can be used to detect CTCs in patients with metastasis and to correlate the cells with overall disease survival, no method has sufficient sensitivity to reliably measure a statistically significant number of cells at early stages of the disease when treatment can be most effective. Generally, optical scan technologies are used to detect immunocytochemically labeled tumor cells, and with some of the technologies, pathologic examination of the cells is possible. Without cell enrichment, scan rates are far too slow for analysis of the sufficiently large population of cells needed to be statistically relevant. Immunomagnetic enrichment relies on consistent and substantial expression of the membrane adhesion recep- T CELL BIOLOGY 2005 tor EpCAM. Although this method has been approved by the Food and Drug Administration for use in patients with metastasis, the phenotypic heterogeneity of CTCs suggests that the sensitivity of this approach is too low to detect all of the CTCs present in peripheral blood. We have developed an instrument that can be used to directly scan and analyze 50 million nucleated cells and detect rare cells in less than 2 minutes without using antigen-dependent enrichment technology. This method is 500 times faster than other cell-based scan approaches because of the instrument’s exceptionally wide field of view made possible by the use of fiberoptic array scanning technology (FAST). With the FAST cytometer (Fig. 1), cells can be detected at a rate of 25 million cells per minute, with a sensitivity of 98% and a specificity of 4 x 10 −6 . In addition, we have developed a sample preparation method, the live-cell attachment protocol, that is optimized for maximal cell retention. F i g . 1 . Major components of the FAST system. Reprinted with permission from Curry, D.N., Krivacic, R.T., Hsieh, H.B., Ladanyi, A., Bergsrud, D.E., Ho, M.Y., Chen, L.B., Kuhn, P., Bruce, R.H. High-speed detection of occult tumor cells in peripheral blood. Proc. IEEE Eng. Med. Biol. Soc. Vol. 26, 2004. © 2004 IEEE. We are using the FAST instrument and the cellpreparation method to investigate the prevalence of CTCs in patients with metastasis and in patients with early-stage disease. Figure 2 shows a CTC in a background of leucocytes. Peripheral blood from patients is processed and labeled for detection of cytokeratin, a suitable biomarker for epithelial cancers. Because cytokeratin is not expressed in normal hematopoietic cells, CTCs can be identified by using a mixture of monoclonal antibodies against 9 different human cytokeratin proteins. The samples are then scanned by using the FAST cytometer. During the scan, the locations of detected objects are collected. These locations are specified by a substrate coordinate system determined by alignment marks on Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 39 F i g . 2 . A circulating tumor cell from a sample of cells obtained from a patient with metastasis and labeled for cytokeratin is easily distinguishable from a background of leukocytes. the substrate. In order to relocate a FAST object with a microscope, these coordinates are transformed to a coordinate system on the microscope stage with the same alignment marks. Via object relocation, the objects detected by using FAST are scanned in the microscope to detect true cancer cells. In addition to determining numbers of CTCs, object relocation will be useful in further characterizing these rare cells. In a preliminary trial with cells from patients with metastatic cancer and cells from healthy donors, CTCs were located in all of the cell samples from cancer patients and in none of the samples from healthy donors. These findings suggest that this method is promising both for diagnosis of cancer and for monitoring responses to therapy. PUBLICATIONS Krivacic, R.T., Ladanyi, A., Curry, D.N., Hsieh, H.B., Kuhn, P., Bergsrud, D.E., Kepros, K.F., Barbera, T., Ho, M.Y., Chen, L.B., Lerner, R.A., Bruce, R.H. A rarecell detector for cancer. Proc. Natl. Acad. Sci. U. S. A. 101:10501, 2004. Novel Mechanisms of Cardiovascular Disease D.J. Loskutoff, R.P. Czekay, C. Dellas, G. Giandomenico, Y. Kamikubo, S. Konstantinides, S. Koschnick, J. Neels P lasminogen activator inhibitor-1 (PAI-1) is a serine protease inhibitor (serpin) that regulates proteases that remove pathologic fibrin deposits 40 CELL BIOLOGY 2005 from the vasculature. PAI-1 differs from most other serpins because it is the product of an immediate-early gene and it binds to the adhesive glycoprotein vitronectin. Leptin is synthesized by adipocytes and acts on hypothalamic receptors to reduce food intake and to increase energy expenditure. Juvenile-onset obesity develops in mice that lack functional leptin (ob/ob mice) or its receptor (db/db mice). Our studies indicate that PAI-1 and leptin promote cardiovascular disease in mice. D E TA C H M E N T O F C E L L S B Y PA I - 1 The binding of PAI-1 to cell-surface urokinase promotes the inactivation and internalization of adhesion receptors (e.g., integrins, the urokinase receptor) and leads to cell detachment from a variety of extracellular matrices. Cell detachment is not a specific property of PAI-1, because protease nexin-1 also detaches cells, and it is not simply the result of the binding of macromolecules to urokinase and/or of the inactivation of urokinase, because neither antibodies to urokinase nor active-site inhibitors of urokinase can detach the cells. The binding of PAI-1 and protease nexin-1 to urokinase leads to the specific inactivation of the matrixengaged integrins, a process that requires a direct interaction between urokinase receptors and those integrins. These inhibitor-mediated changes in the subcellular distribution of the urokinase receptor and integrins may influence a variety of cardiovascular disorders. THROMBOTIC PHENOTYPE OF MICE WITH A C O M B I N E D D E F I C I E N C Y I N PA I - 1 A N D V I T R O N E C T I N The role of vitronectin in thrombosis is not fully understood, primarily because this adhesive glycoprotein not only stabilizes PAI-1 and thus protects fibrin from premature lysis but also binds to platelet integrins and may influence platelet aggregation. Using novel quantitative approaches, we examined the thrombotic phenotype of mice with a combined deficiency in both PAI-1 and vitronectin. Although unstable thrombi developed in all deficient mice, the thrombotic phenotype of mice with the combined deficiency did not differ significantly from the phenotype of mice with deficiencies in only PAI-1 or vitronectin. This observation suggests that vitronectin may influence thrombus stability by regulating PAI-1 and not platelets. L E P T I N A N D C A R D I O VA S C U L A R D I S E A S E Obesity in humans is associated with elevated levels of leptin and an increased risk for thrombosis and for cardiovascular diseases. Endogenous leptin appears to regulate thrombosis in vivo, because administration of leptin promoted arterial thrombosis in mice, and comPublished by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. pared with lean mice, leptin-deficient ob/ob mice had unstable thrombi and an attenuated thrombotic response to injury. Moreover, when lean mice were pretreated with a leptin-neutralizing antibody before injury of the carotid artery, the mice had prolonged times to thrombotic occlusion and the development of unstable, embolizing thrombi. Thus, inhibiting leptin converted the thrombotic phenotype of wild-type mice into one that closely resembled that of ob/ob mice. The antibody also protected mice from experimentally induced venous thrombosis and pulmonary embolism. Thus, inhibition of circulating leptin protects against arterial and venous thrombosis in mice, and possibly in hyperleptinemic humans with obesity. In other studies, compared with mice fed normal chow, wild-type mice fed an atherogenic, high-fat diet had elevated (9-fold) leptin levels and significantly enhanced neointimal thickening after injury of the carotid artery. Unexpectedly, the atherogenic diet had no effect on injured vessels from leptin-deficient ob/ob mice despite aggravating obesity, diabetes, and hyperlipidemia in these animals. Daily administration of leptin to ob/ob mice during the 3-week period after injury dramatically increased neointimal thickness and the severity of luminal stenosis but had no effect on vessels from db/db mice, which lack the receptor for leptin. These results suggest that a direct, leptin receptor–mediated link exists between the hyperleptinemia in human obesity and the increased risk for cardiovascular complications associated with this condition. We also found that administration of leptin promoted platelet activation and thrombosis in mice and potentiated the aggregation of human platelets induced by low concentrations of ADP, collagen, and epinephrine. However, the responses of human platelets varied significantly according to the donor of the cells. Platelets from some donors (~40%) consistently responded to leptin (responders), whereas platelets from other donors (~60%) never responded (nonresponders). Although platelets from both groups expressed only the signaling form of the leptin receptor, the platelets from responders expressed higher levels of this receptor than did those of nonresponders. Ligand-binding assays indicated specific, saturable binding of leptin to platelets from both groups. Thus, the decreased sensitivity to leptin of platelets from nonresponders is not due to the absence of the signaling form of the leptin receptor but may reflect differences in its level of expression and/or affinity CELL BIOLOGY 2005 for leptin. These studies indicate that platelets are a major source of leptin receptors in the circulation and suggest that leptin-responsive persons may have a higher risk for obesity-associated thrombosis than do nonresponsive persons. Preliminary studies indicate that the leptin receptor is upregulated upon platelet activation and that platelets do not store or release significant amounts of leptin. These results suggest that activation of platelets leads to increased binding of circulating leptin, further enhancing platelet activation. PUBLICATIONS Campagnolo, L., Leahy, A., Chitnis, S., Koschnick, S., Fitch, M.J., Fallon, J.T., Loskutoff, D., Taubman, M.B., Stuhlmann, H. EGFL7 is a chemoattractant for endothelial cells and is up-regulated in angiogenesis and arterial injury. Am. J. Pathol. 167:275, 2005. Czekay, R.P., Loskutoff, D.J. Unexpected role of plasminogen activator inhibitor 1 in cell adhesion and detachment. Exp. Biol. Med. (Maywood) 229:1090, 2004. Degryse, B., Resnati, M., Czekay, R.-P., Loskutoff, D.J., Blasi, F. Domain 2 of the urokinase receptor contains an integrin-interacting epitope with intrinsic signaling activity: generation of a new integrin inhibitor. J. Biol. Chem., in press. Dellas, C., Loskutoff, D.J. Historical analysis of PAI-1 from its discovery to its potential role in cell motility and disease. Thromb. Haemost. 93:631, 2005. Giandomenico, G., Dellas, C., Czekay, R.-P., Koschnick, S., Loskutoff, D.J. The leptin receptor system of human platelets. J. Thromb. Haemost. 3:1042, 2005. Konstantinides, S., Schäfer, K., Neels, J.G., Dellas, C., Loskutoff, D.J. Inhibition of endogenous leptin protects mice from arterial and venous thrombosis. Arterioscler. Thromb. Vasc. Biol. 24:2196, 2004. Koschnick, S., Konstantinides, S., Schäfer, K., Crain, K., Loskutoff, D.J. Thrombotic phenotype of mice with a combined deficiency in plasminogen activator inhibitor 1 and vitronectin. J. Thromb. Haemost., in press. Pandey, M., Loskutoff, D.J., Samad, F. Molecular mechanisms of tumor necrosis factor-α-mediated plasminogen activator inhibitor-1 expression in adipocytes. FASEB J., in press. Schäfer, K., Halle, M., Goeschen, C., Dellas, C., Pynn, M., Loskutoff, D.J., Konstantinides, S. Leptin promotes vascular remodeling and neointimal growth in mice. Arterioscler. Thromb. Vasc. Biol. 24:112, 2004. Viral Nanoparticles: Pathogen Inhibitors and Biomolecular Sensors M. Manchester, E. Burke, G. Destito, M. Estrada, 41 Previously, researchers focused primarily on chemically derived materials such as dendrimers or polymers to develop synthetic nanosized particles (<100 nm in diameter) for biological applications such as molecular therapeutics, tumor targeting, and in vivo biomedical imaging. We use cowpea mosaic virus as a nanoparticle platform for antivirals, antitoxins, vaccines, and tumor-targeting agents. The virus is an icosahedral, 31-nm particle that can be produced easily and inexpensively in blackeyed pea plants. In contrast to the structure of most other nanomaterials, the structure of the capsid of cowpea mosaic virus is defined and can be engineered to display peptides or proteins in controlled orientations on particle surfaces via either genetic manipulation of the viral genome or by chemical attachment to the particle surface. V I R A L N A N O PA R T I C L E S A S VA C C I N E S A N D ANTITOXINS We have generated viral nanoparticles that display T-cell epitopes on the surface in a multivalent fashion. In vitro, these nanoparticles can be taken up into several types of antigen-presenting cells, including dendritic cells and macrophages, and in vivo, particles given orally or intravenously localize to antigen-presenting cells. Viral nanoparticles pass through the gut epithelial lining via M cells that sample particulate matter in the gastrointestinal tract. The uptake appears to be via a slow endocytic pathway, with the nanoparticles localizing in perinuclear endosomes. Viral nanoparticles displaying T-cell epitopes can induce pathogen-specific T-lymphocyte responses in vivo that are protective against pathogen challenge, indicating that the particles leave the endosomes to enter the antigen-presentation pathway associated with class I MHC molecules. We are also developing combination inhibitor-vaccine approaches for other viral and bacterial pathogens. Finally, we have shown that soluble pathogen receptors are protective against challenge with anthrax lethal toxin in vivo, and we are studying the efficacy of multivalent arrays of these receptor decoy domains on viral nanoparticles. M.J. Gonzalez, K. Koudelka, E. Powell, C. Rae, P. Singh, V I R A L N A N O PA R T I C L E S F O R VA S C U L A R I M A G I N G D. Thomas A N D T U M O R TA R G E T I N G he goal of nanotechnology in biomedical science is the design of tiny nanomachines with multiple functions that can be used to detect, target, and treat human disease in vivo, thereby eliminating the need for invasive diagnostic or therapeutic procedures. Our goal is to develop viral nanoparticles that can be delivered in a noninvasive manner, home to a tumor in vivo, act as an image-contrast agent for detection by magnetic resonance imaging or other noninvasive imaging techniques, and deliver an antitumor therapeutic T Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 42 CELL BIOLOGY 2005 or tumoricidal gene. These studies are being done in collaboration with M.G. Finn, Department of Chemistry, J. Johnson and A. Schneemann, Department of Molecular Biology, and H. Stuhlmann, Department of Cell Biology. Fluorescently labeled viral nanoparticles are very bright, nontoxic materials that are excellent reagents for imaging the vasculature in live animals. Working with Dr. Stuhlmann and J. Lewis, Department of Cell Biology, we showed that viral nanoparticles can be used to effectively image the complete embryonic vasculature in several species. These particles are also useful for highlighting tumor vasculature in the chick chorioallantoic membrane tumor onplant model. Uptake of particles into endothelial cells occurs, yielding a bright imaging signal that can also be used to differentiate between arterial and venous vessels. We have also investigated the usefulness of viral nanoparticles as biomolecular sensors to detect, image, and treat tumors in vivo. We have now designed several tumor-specific nanoparticles that target various types of tumor cells specifically and are taken up into tumor cells both in vivo and in vitro. These studies will allow us to further the design of antitumor agents that can provide localized, specific imaging and therapy in order to visualize and eliminate cancer in its earliest stages. PUBLICATIONS Naniche, D., Garenne, M., Rae, C., Manchester, M., Buchta, R., Brodine, S.K., Oldstone, M.B. Decrease in measles virus-specific CD4 T cell memory in vaccinated subjects. J. Infect. Dis. 190:1387, 2004. Portney, N.G., Singh, K., Chaudhary, S., Destito, G., Schneemann, A., Manchester, M., Ozkan, M. Organic and inorganic nanoparticle hybrids. Langmuir 21:2098, 2005. Scobie, H.M., Thomas, D., Marlett, J.M., Destito, G., Wigelsworth, D.J., Collier R.J., Young J.A.T., Manchester, M. A soluble receptor decoy protects rats against anthrax lethal toxin challenge. J. Infect. Dis., in press. Singh, P., Gonzalez, M.J., Manchester, M. Viruses and their uses in nanotechnology. Drug Dev. Res., in press. Translational Regulation in Chloroplasts and Expression of Human Monoclonal Antibodies in Eukaryotic Algae S.P. Mayfield, M. Beligni, D. Barnes, E. Brown, A. Coragliotti, K. Espina, S. Franklin, R. Henry, A. Manuell, J. Schultz G ene expression in chloroplasts is primarily controlled during the translation of plastid mRNAs into proteins, and understanding how this pro- Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. cess is regulated is key to understanding plant development and function. Controlling chloroplast translation is also an essential component of optimizing the production of human therapeutic proteins in algae. Using proteomic and bioinformatic analysis, we identified the set of proteins that function in chloroplast translation. These studies indicated that the translational apparatus of chloroplasts is related to that of bacteria but that chloroplasts have incorporated additional proteins that allow more complex regulatory mechanisms. Chloroplasts also contain a number of RNA elements that are not found in bacteria, and even conserved RNA elements, such as ribosome-binding sequences, are positioned in chloroplast mRNAs far upstream compared with their bacterial counterparts. These unique components provide the opportunity for regulation of chloroplast translation that cannot be achieved in simpler bacterial systems. To understand the unique, and conserved, aspects of plant translation, we have begun a structural analysis of both the chloroplast and the cytoplasmic ribosomes of Chlamydomonas reinhardtii. Using electron cryomicroscopy and single-particle reconstruction, we determined the structure of cytoplasmic ribosomes and found that they are almost identical to ribosomes from animals, including ribosomes from mammals. We also determined a preliminary structure of chloroplast ribosomes and found that they differ substantially from ribosomes of bacteria, as predicted. These studies have revealed the structural basis for identifying the molecular and biochemical interactions of mRNA and ribosomes that result in regulated translation of mRNAs. In addition to these basic studies on translational regulation, we have developed a system for the expression of recombinant proteins, including human therapeutic agents, in C reinhardtii. We constructed strains of C reinhardtii that express variants of human monoclonal antibodies against herpes simplex virus. These antibodies assemble in the algal cells to form fully functional antibodies that bind herpes simplex proteins. We also showed that this alga-based system can be used to produce a number of other proteins with potential human therapeutic value. These studies indicate that eukaryotic algae have tremendous potential for the expression of recombinant human therapeutic proteins, because algae can be grown economically on a very large scale. Our continued genetic, biochemical, and structural studies should lead to a greater understanding of the mechanism of chloroplast translation and to higher levels of expression of therapeutic proteins. CELL BIOLOGY 2005 PUBLICATIONS Barnes, D., Cohen, A., Bruick, R.K., Kantardjieff, K., Fowler, S., Efuet, E., Mayfield, S.P. Identification and characterization of a novel RNA binding protein that associates with the 5′-untranslated region of the chloroplast psbA mRNA. Biochemistry 43:8541, 2004. Beligni, M.V., Yamaguchi, K., Mayfield, S.P. Chloroplast elongation factor Ts proprotein is an evolutionarily conserved fusion with the S1 domain-containing plastidspecific ribosomal protein-7. Plant Cell 16:3357, 2004. Beligni, M.V., Yamaguchi, K., Mayfield, S.P. The translational apparatus of Chlamydomonas reinhardtii chloroplast. Photosyn. Res. 82:315, 2004. Franklin, S.E., Mayfield, S.P. Recent developments in the production of human therapeutic proteins in eukaryotic algae. Expert Opin. Biol. Ther. 5:225, 2005. Manuell, A., Beligni, M.V., Yamaguchi, K., Mayfield, S.P. Regulation of chloroplast translation: interactions of RNA elements, RNA-binding proteins, and the plastid ribosome. Biochem. Soc. Trans. 32(Pt. 4):601, 2004. Mayfield, S.P., Franklin, S.E. Expression of human antibodies in eukaryotic microalgae. Vaccine 23:1828, 2005. Somanchi, A., Barnes, D., Mayfield, S.P. A nuclear gene of Chlamydomonas reinhardtii, Tba1, encodes a putative oxidoreductase required for translation of the chloroplast psbA mRNA. Plant J. 42:341, 2005. Molecular Basis of Cognitive Function and Dysfunction M. Mayford, E. Korzus, K. Limbaeck-Stokin, G.J. Reijmers, M. Yasuda, R. Yasuda, S. Miller, N. Matsuo he ability to remember is perhaps the most significant and distinctive feature of our cognitive life. We are who we are in large part because of what we have learned and what we remember. Impairments in learning and memory are a component of disorders that affect human beings throughout life, from childhood forms of mental retardation to psychiatric disorders such as schizophrenia with onsets in late adolescence and early adulthood to diseases of aging such as Alzheimer’s disease. T CALCIUM SIGNALING AND MEMORY We know relatively little at a molecular level about how the brain stores new information. One hypothesis, which we tested, is that calcium-regulated changes in the strength of synaptic connections between nerve cells can store information. The enzyme calcium/calmodulin-dependent protein kinase is abundant at synapses and when activated by calcium can strengthen synaptic connections. We used genetic manipulations in mice to indiscriminately activate this kinase at all synapses in the entorhinal cortex, a part of the brain that is important for memory and is affected in the earliest stages of Alzheimer’s disease in humans. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 43 We found not only that the formation of new memories was impaired but also that previously established memories could be erased. If memories are stored as precise patterns of synaptic weights, then the indiscriminate strengthening of synapses might be expected to erase memories in a manner similar to the way writing all 1’s in computer memory will erase previously stored information. GENETIC MODELS OF DISEASE The recent determination of the complete sequence of the mouse and human genomes indicates that humans are highly similar to mice at the genetic level. One approach to understanding genetic diseases in humans is to introduce the same mutations into mice to produce a model of the disease for better understanding of the molecular pathology and for testing possible treatments. Rubenstein-Taybi syndrome is a developmental and cognitive disorder that results from mutation in the gene CBP. We produced a strain of mice with a defect in CBP and found that the mice were impaired in several learning and memory tasks. More important, we showed that these impairments were not due to problems in development of the brain because they could be reversed by providing a normally functioning CBP gene to adult mice. The protein encoded by CBP chemically modifies histones to allow the expression of a large variety of other genes. We found that the memory deficits in the mice could be reversed by treatment with a drug that targets this histone-modifying function, suggesting a treatment for this and possibly other cognitive disorders. M O L E C U L A R A N AT O M Y O F M E M O R Y When humans learn new information, they use only a tiny fraction of the neurons in brain. One of the difficulties in studying memory is an inability to identify and specifically manipulate those neurons that participate in a particular memory trace. We recently developed a genetic technique for use in mice that enables us to specifically introduce genetic changes into neurons that are activated by behavioral stimuli. We are using this approach to introduce marker proteins that enable us to see the connections between neurons that have been activated during learning. We can follow these neurons over long periods to detect changes in their structure and function as the particular memory trace decays or strengthens with repeated training. PUBLICATIONS Limback-Stokin, K., Korzus, E., Nagaoka-Yasuda, R., Mayford, M. Nuclear calcium/calmodulin regulates memory consolidation. J. Neurosci. 24:10858, 2004. 44 CELL BIOLOGY 2005 Regulation of the Plasminogen Activation System A. Baik, F. Garcia-Bannach,* N. Gingles, J. Mitchell, R.J. Parmer,* L. Teyton,** L.A. Miles * University of California, San Diego, California ** Department of Immunology, Scripps Research ssembly of plasminogen and plasminogen activators on cell surfaces is a key control point for positive regulation of cell-surface proteolytic activity necessary in physiologic and pathologic processes. Plasminogen-binding sites are markedly upregulated when monocytoid cells undergo apoptosis. Monocytes are major mediators of inflammation, and apoptosis is a mechanism for regulating the inflammatory response by eliminating activated macrophages. Therefore, we are investigating the ability of plasminogen to modulate apoptosis in monocytes. We cultured monocytoid cells (freshly isolated human monocytes and U937 cells) in plasminogen-deficient serum and induced the cells to undergo apoptosis by using either TNF-α or cycloheximide. When induced in the presence of increasing concentrations of plasminogen, apoptosis was inhibited in a dose-dependent manner; full inhibition occurred at a concentration of plasminogen equal to its physiologic concentration. Treatment with plasminogen also markedly reduced intranucleosomal DNA fragmentation and the active caspase-3, caspase-8, and caspase-9 induced by TNF-α or by cycloheximide. We also examined the requirement for plasmin proteolytic activity in the cytoprotective function of plasminogen. A plasminogen active-site mutant did not recapitulate the cytoprotective effect of wild-type plasminogen. Furthermore, antibodies against protease-activated receptor 1 blocked the antiapoptotic effects of plasminogen. The cytoprotective effect of plasminogen required plasmin proteolytic activity and was mediated by protease-activated receptor 1. Because apoptosis of monocytes plays a key role in inflammation and atherosclerosis, our studies provide insight into a novel role of plasminogen in these processes. A PUBLICATIONS Garcia-Bannach, F.G., Gutierrez-Fernandes, A., Parmer, R.J., Miles, L.A. Interleukin-6- induced plasminogen gene expression in murine hepatocytes is mediated by transcription factor CCAAT/enhancer binding protein β (C/EBPβ). J. Thromb. Haemost. 2:2205, 2004. Griffin, M.O, Jinno, M., Miles, L.A., Villarreal, F.J. Reduction of myocardial infarct size by doxycycline: a role for plasmin inhibition. Mol. Cell. Biochem. 270:1, 2005. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Miles, L.A., Hawley, S.B., Baik, N., Andronicos, N.M., Castellino, F.J. Parmer, R.J. Plasminogen receptors: the sine qua non of cell surface plasminogen activation. Front. Biosci. 10:1754, 2005. Structure and Action of Molecular Machines R.A. Milligan, J. Chappie, P. Chowdhury, T. Dang, A. Efimov, A. Mulder, G. Orca, M. Reedy,* M.K. Reedy,* C. Reyes, B. Sheehan, K. Thompson, A.B. Ward, E.M. Wilson-Kubalek, C. Yoshioka * Duke University Medical Center, Durham, North Carolina acromolecular assemblies may be composed of from 2 to perhaps scores of proteins and are the functional units—the molecular machines— of the cell. We use electron cryomicroscopy and image analysis to study the structure and mechanism of action of several of these machines. We combine the 3-dimensional maps calculated from electron images of the machines with biochemical data and high-resolution x-ray structures of the individual components to provide insight into the operation of the machines. During the past year, we continued our work on members of the myosin and kinesin superfamilies, microtubule-stabilizing proteins, and membrane proteins. Although the mechanism of plus end–directed, processive motion by conventional kinesins is now well understood, the mechanism by which members of the kinesin 14 class move toward the minus ends of microtubules is not. Likewise, in the myosin superfamily, how nucleotide-mediated conformational changes in the motor domain of class VI myosins result in “backward” motility is not known. We are elucidating the molecular mechanisms of these more unusual members of the myosin and kinesin superfamilies. (Movies showing the motions of conventional myosin and kinesin can be viewed at www.scripps.edu/milligan/projects.html.) The kinesin Ncd belongs to the kinesin 14 class of motor proteins. Compared with the situation with plus end–directed kinesins, the nature and timing of the structural changes that underlie the motility of kinesin 14 motors are poorly understood. We used electron cryomicroscopy and image analysis to calculate 3-dimensional maps of Ncd bound to microtubules in various stages in its mechanochemical cycle. The maps revealed a minus end–directed rotation of approximately 70° of a coiled coil mechanical element of microtubule-bound Ncd upon ATP binding. In parallel M CELL BIOLOGY 2005 with these structural studies, our collaborators, N. Endres and R. Vale at the University of California, San Francisco, showed that extending or shortening this mechanical element respectively increases or decreases movement velocity without affecting ATPase activity. These results indicate that as with other kinesins, the force-producing conformational change of Ncd occurs upon ATP binding but, unlike the situation with other kinesins, involves the swing of a rigid, lever arm–like mechanical element similar to that described for myosins. Whereas most kinesins move along intact microtubules, members of the kinesin 13 class destabilize and depolymerize microtubules and do not appear to have motile properties. We found that a KinI fragment consisting of only the conserved motor core is necessary and sufficient for ATP-dependent depolymerization. The motor core binds along microtubules in all nucleotide states, but in the presence of a nonhydrolyzable ATP analog, depolymerization also occurs. Structural characterization of the analog-induced depolymerization products provided a snapshot of the disassembly machine at the microtubule ends. Our data indicate that whereas conventional kinesins use the energy of ATP binding to execute a power stroke that results in unidirectional motion along the microtubule surface, KinIs at the ends of microtubules use the energy to bend the underlying protofilament, thereby destabilizing the microtubule lattice and leading to microtubule depolymerization. Furthermore, when the motor core is associated with the microtubule wall, the core is stalled in a weakly bound, nucleotide-free state. Progression to the strongly bound, ATP-containing state is possible only when the KinI encounters a microtubule end, where it can catalyze deformation of protofilaments and disassembly of microtubules. The unusual mechanochemical coupling of this kinesin provides an elegant mechanistic basis for its microtubule-depolymerizing activity. The protein doublecortin is expressed in migrating and differentiating neurons. In humans, mutations in this protein disrupt brain development, causing lissencephaly. Although doublecortin is associated with and stabilizes the microtubule cytoskeleton, it has no homology with other microtubule-binding proteins such as MAP2 or tau. We found that doublecortin preferentially nucleates and binds to 13-protofilament microtubules. This specificity was explained when we discovered that the protein binds in the valleys between the protofilaments of the microtubule wall. This binding site is Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 45 unique and appears to be ideally located for microtubule stabilization. In this location, doublecortin most likely contributes to both the longitudinal and the lateral interactions that stabilize the microtubule wall. In collaboration with G. Chang, Department of Molecular Biology, we have grown well-ordered arrays of several membrane proteins that are involved in multidrug resistance. These arrays, helical tubes and 2-dimensional crystals of membrane-embedded proteins, are suitable for structural studies via electron microscopy. In one instance, we trapped a drug transporter in various stages of its mechanistic cycle and with substrates bound. We anticipate that 3-dimensional electron microscopy maps of membrane-embedded transporters in various states, together with the high-resolution x-ray structures of the detergent-solubilized protein, will provide insights into the mechanisms used to transport metabolites and drugs across membranes. In other studies, we developed a general method for helical crystallization of proteins on lipid tubules that we are using to study the virulence factor PFO from Clostridium perfringens. PFO is a cytolysin, an important class of proteins that oligomerize and embed within membranes as part of their lytic function. We obtained helical crystals of wild-type and several mutant forms of PFO on nickel-lipid tubules. Three-dimensional maps of these proteins derived from images of the helical crystals will be used to complement our studies of PFO pore formation on lipid layers. These studies will provide a better understanding of the pathogenic function of cytolysins. Additional studies involving tubular crystallization of membrane proteins and other bacterial toxins are opening up promising new areas for future research. PUBLICATIONS Dang, T.X., Farah, S.J., Gast, A., Robertson, C., Carragher, B., Egelman, E., Wilson-Kubalek, E.M. Helical crystallization on lipid nanotubes: streptavidin as a model protein. J. Struct. Biol. 150:90, 2005. Dang, T.X., Hotze, E.M., Rouiller, I., Tweten, R.K., Wilson-Kubalek, E.M. Prepore to pore transition of a cholesterol-dependent cytolysin visualized by electron microscopy. J. Struct. Biol. 150:100, 2005. Neuman, B., Adair, B.D., Burns, J.W., Milligan, R.A., Buchmeier, M.J., Yeager, M. Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J. Virol. 79:3822, 2005. O’Keefe, M.A., Turner, J.H., Musante, J.A., Hetherington, C.J.D., Cullis, A.G., Carragher, B., Junkins, R., Milgrim, J., Milligan, R.A., Potter, C.S., Allard, L.F., Blom, D.A., Degenhardt, L., Sides, W.H.. Laboratory design for high-performance electron microscopy. Microsc. Today 12:8, 2004. 46 CELL BIOLOGY 2005 Molecular Mechanisms of CNS Development and Mechanosensory Perception U. Müller, R. Banan, C. Barros, R. Belvindrah, F. Conti, S. Hankel, E. Hintermann, P. Kazmierczak, R. Radakovits, C. Ramos, A. Reynolds, A. Sczaniecka, M. Schwander, M. Senften, S. Webb disproportionately large number of genes in the genomes of vertebrates encode cell recognition molecules that mediate cell-cell interactions and interactions between cells and the extracellular matrix. This finding most likely reflects an evolutionary trend toward increasingly more complex cellular interactions in higher metazoans. The highest diversity of such interactions occurs in the CNS, where thousands of different neuronal subtypes are connected into defined neuronal circuits. We use mouse genetics, genomics, cell biology, biochemistry, and imaging technology to analyze the function of cell recognition molecules during the development of neuronal circuits in the CNS. In another project, we are elucidating the mechanisms by which cell recognition molecules contribute to mechanosensory perception. A F O R M AT I O N O F C O R T I C A L S T R U C T U R E S I N T H E C N S The establishment of the 3-dimensional cytoarchitecture of the nervous system depends on interactions of receptors on neuronal cells with molecules presented within the extracellular matrix and by neighboring cells. Integrins are a class of neuronal receptors that mediate interactions with glycoproteins secreted by the extracellular matrix and with membrane-anchored counterreceptors. Recently, we found that integrins cooperate with secreted signaling molecules such as sonic hedgehog and Reelin to regulate important steps during CNS development, such as cell proliferation and formation of neuronal layers during the development of the cerebral and cerebellar cortex. We are identifying the downstream signaling pathways activated by integrins during cortical development. We are also studying signaling interactions between integrins and other receptors such as receptor tyrosine kinases. Finally, we have extended our studies to the analysis of integrin functions in the CNS in adults. CELL RECOGNITION MOLECULES, MECHANOSENSORY PERCEPTION, AND DEAFNESS Mechanosensation, the transduction of mechanical force into an electrochemical signal, allows living organisms to detect touch, hear, register movement and gravity, Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. and sense changes in cell volume and shape. In mammals, the hair cells of the inner ear are the principle mechanosensors for the detection of sound and movement. Hair cells elaborate stereocilia that contain mechanosensitive ion channels. The stereocilia of a hair cell are interconnected by extracellular bridges into a bundle and are situated next to specialized extracellular matrix assemblies. Sound waves or head movements lead to deflection of the stereocilia bundle, changes in the ion permeability of the mechanosensitive channels, and depolarization of the hair cells. The molecules that regulate development and function of hair cells are poorly defined. Because defects in hair cells cause inherited forms of deafness, we use human and mouse genetics as a guideline to identify and study molecules that regulate the development and function of mechanosensory hair cells. Currently, about 70 genes have been identified in which mutations lead to deafness. Many of these genes encode membrane-anchored cell adhesion molecules and molecules secreted by the extracellular matrix. Mutations in the gene for the cell adhesion molecule cadherin 23 cause deafness in mice and humans. Our findings provide strong evidence that cadherin 23 is a component of the so-called tip-link, which has been predicted to transmit force onto mechanically gated ion channels in the stereocilia of hair cells. We are analyzing the function of cadherin 23, proteins that interact with this cell adhesion molecule, and proteins encoded by additional “deafness” genes for mechanotransduction. We are also doing genetic screens in mice to identify novel recessive deafness traits. Using this strategy, we have already identified several novel genes that may be associated with deafness. PUBLICATIONS Barros, C., Müller, U. Cell adhesion in nervous system development. In: Integrins in Development. Danen, E. (Ed.). Landes Bioscience, Georgetown, TX, in press. Belvindrah, R., Müller, U. Integrin signaling and central nervous system development. In: Extracellular Matrix and Disease. Miner, J. (Ed.). Elsevier, St. Louis. Advances in Developmental Biology and Biochemistry, Vol. 15, in press. Escher, P., Lacazette, E., Courtet, M., Blindenbacher, A., Landmann, L., Bezakova, G., Lloyd, K., Müller, U., Brenner H.R. Synapse formation in skeletal muscle lacking neuregulin receptors. Science, in press. Li, N., Zhang, Y., Naylor, M.J., Schatzmann, F., Maurer, F., Wintermantel, T., Schuetz, G., Müller, U., Streuli, C.H., Hynes, N.E. β1 Integrins regulate mammary gland proliferation and maintain the integrity of mammary alveoli. EMBO J. 24:1942, 2005. Müller, U. Integrins and extracellular matrix in animal models. In: Cell Adhesion. Behrens, J., Nelson, W.J. (Eds.). Springer Verlag, New York, 2004, p. 217. Handbook of Experimental Pharmacology, Vol. 165. White, D.E., Kurpios, N.A., Zuo, D., Hassell, J.A., Blaess, S., Müller, U., Muller, W.J. Targeted disruption of β1 integrins in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell 6:159, 2004. CELL BIOLOGY 2005 Molecular Mechanisms of Thermosensation A. Patapoutian, A. Dhaka, T. Earley, S. Eid, S.W. Hwang, L. Macpherson, T. Miyamoto, A. Murray, G. Story e are interested in the molecular description of the function of sensory neurons. Of the 5 popularly characterized senses—sight, hearing, taste, smell, and touch—touch is among the most varied and least understood. Within this sense is the ability to sense mechanical forces, chemical stimuli, and temperature, and the molecules that mediate this ability have been a long-standing mystery. Temperature sensation in particular has received relatively little attention from biologists and yet is critical for interactions with the environment. We recently discovered proteins that may enable sensory neurons to convey information about temperature. These proteins are ion channels activated by specific changes in temperature; thus they act as the molecular thermometers of the body. Specifically, our results have led to the identification and characterization of a novel warm-activated transient-receptor-potential (TRP) channel, TRPV3 (33°C threshold) and 2 novel cold-activated TRP channels, TRPM8 (25°C threshold) and TRPA1 (ANKTM1, 17°C threshold). We found that TRPM8 is also the receptor for the compound menthol, providing a molecular explanation of why mint flavors are typically perceived as cooling. Furthermore, we discovered that TRPA1 is activated by cinnamaldehyde and other compounds with a burning sensory quality, consistent with a role of TRPA1 in the detection of noxious cold sensations. Together these temperature-activated channels represent a new subfamily of TRP channels that we have dubbed thermoTRPs. In agreement with a role in initiating temperature sensation, most of the thermoTRPs are normally found in subsets of neurons in dorsal root ganglia. A surprisingly distinct expression pattern was observed for TRPV3, the warm receptor. High levels of TRPV3 are observed solely in skin keratinocytes in mice, suggesting that skin cells might be able to “sense” temperature and then communicate this information to dorsal root ganglia neurons. How temperature information is coded from the skin to the spinal cord is not well understood, and we are using a variety of approaches to answer this question. For example, recent data from mice lacking the gene for TRPV3 suggest that TRPV3 is indeed required W Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 47 for proper heat sensation in vivo, reinforcing a role of skin in thermosensation. All organisms have a need for thermosensation. Because some invertebrate species are more amenable to genetic studies than mammals are, we asked whether nonvertebrates also use thermoTRPs to sense temperature. We recently showed that the Drosophila ortholog of TRPA1 is an ion channel activated by warm temperatures, suggesting an evolutionary conserved role of TRP channels in temperature sensing. In collaborative efforts with P. Garrity, Massachusetts Institute of Technology, Cambridge, Massachusetts, and W. Shafer, University of California, San Diego, we are using genetic studies to examine the role of TRPA family members in invertebrate species. Another key question is what makes thermoTRPs temperature sensitive whereas other TRPs are not? Answering this question requires insight into the fundamental biophysical mechanism of how temperature activates ion channels. Our ongoing structure-function experiments, including mutagenesis and chimeric protein analysis of the thermoTRPs, will provide us with important clues about how cold or heat activates these ion channels. Our long-term goal is to synthesize an integrated picture of sensory neuron function. By identifying the proteins that most likely initiate the molecular cascade leading to temperature perception, we have provided the basis for probing the foundation of the sense of temperature. We now have the opportunity to extend these insights into important areas of human health, such as pain pathophysiology. For example, TRPA1 is a potential target for treating pain, and we are identifying small-molecule inhibitors of TRPA1 in collaboration with scientists at the Genomics Institute of the Novartis Research Foundation, San Diego, California. Therefore, the approaches we are using will yield insights into the basic biology of the peripheral nervous system and may also have an effect on novel treatments for pain. PUBLICATIONS Moqrich, A., Earley, T., Watson, J., Andahazy, M., Backus, C., Martin-Zanca, D., Wright, D.E., Reichardt, L.F., Patapoutian, A. Expressing TrkC from the TrkA locus causes a subset of dorsal root ganglia neurons to switch fate. Nat. Neurosci. 7:812, 2004. Moqrich, A., Hwang, S.W., Earley, T.J., Petrus, M.J., Murray, A.N., Spencer, K.S.R., Andahazy, M., Story, G., Patapoutian, A. Impaired thermosensation in mice lacking TRPV3, a heat and camphor sensor in skin. Science 307:1468, 2005. Patapoutian A., Wood, J.N. Mechanisms of nociception: molecules to behaviour. J. Neurobiol. 61:1, 2004. Rosenzweig, M., Brennan, K.M., Tayler, T.D., Phelps, P.O., Patapoutian, A., Garrity, P.A. The Drosophila ortholog of vertebrate TRPA1 regulates thermotaxis. Genes Dev. 19:419, 2005. 48 CELL BIOLOGY 2005 Functional Proteins in Tumor Metastasis and Angiogenesis A. Zijlstra, J.P. Partridge, T. Kupriyanova, M. Madsen, T. Papagiannakopoulas, M.C. Subauste, E.I. Deryugina, J.P. Quigley e have established a number of in vivo model systems that can recapitulate the major cellular and tissue events that occur during tumor metastasis and angiogenesis. The model systems allow quantitative measurements, microscopic analysis in real time, biochemical and immunologic probing, and direct molecular and therapeutic interventions. Recently, use of short interfering RNA molecules directed against specific expressed genes provided insights into the contributory role of the gene products in tumor dissemination and neovascularization. In addition, use of subtractive immunization, which is used to generate unique neutralizing monoclonal antibodies, in combination with immunoproteomics enables us to identify specific antigenic molecules that are functionally active in metastasis and angiogenesis. W M E TA S TA S I S Selected human tumor cells inoculated onto the chorioallantoic membrane of developing chick embryos form primary tumors on the membrane in 4–7 days. A small percentage of the cells in the primary tumor disseminate through the vasculature and within 3–4 days, arrest and proliferate in secondary organs of the embryo. Measuring a small number of early-arriving metastatic cells (<200) growing and expanding in the secondary organ has always been technically difficult. We now use an approach in which unique regions of human DNA, known as Alu repeat sequences, are amplified by polymerase chain reaction from the total DNA extracted from various organs of the tumor-bearing chick embryo. Chicken DNA contains no Alu sequences, so any product generated by the polymerase chain reaction indicates that human tumor cells are present in the chick embryo organ and would have arrived there via the known sequential steps in metastasis. We can now detect as few as 25–50 human tumor cells present in the entire chick embryo lung, liver, or brain and can measure the expansion of these metastatic cells by using the real-time polymerase chain reaction. We are using various screening procedures in this model system to identify molecules that enhance, or conversely inhibit, the appearance of metastatic human Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. tumor cells in organs of chick embryos. The screening procedures include direct inoculation of primary tumor cells that have been transfected with various short interfering RNA constructs to delete specific genes that might contribute to metastatic dissemination. Inoculating monoclonal antibodies directly into the tumor-bearing embryos and monitoring the influence of the antibodies on metastasis are also part of our screening procedures. We are also using a more conventional method of monitoring human tumor metastasis in specific immunodeficient mice. However, compared with our chick embryo metastasis assay, this method is less quantitative, requires more time (3–5 weeks), and is more difficult to use for inhibitor screening and molecular intervention. We are using the mouse metastasis assay to take advantage of mouse genetics and to confirm the efficacy of various effector molecules and inhibitors that initially are identified in the chick embryo metastasis assay. ANGIOGENESIS One of the most commonly used in vivo assays for angiogenesis is the chick embryo chorioallantoic membrane assay. We developed a quantitative variation of this assay that allows detection and measurement of the newly sprouting blood vessels responding to an angiogenic stimulus such as a specific growth factor or a growing tumor (Fig. 1). A highly specific metalloproteinase, MMP-13, has been implicated in the tissue remodeling that occurs during the formation of the new blood vessels. We characterized this proteolytic event and found that collagen-cleaving metalloproteinases are implicated directly in the outgrowth of new vessels. We also discovered that another metalloproteinase, MMP-9 (gelatinase B), most likely is involved in angiogenic tissue remodeling. The proteolytic activity of this enzyme, which is quite distinct from that of MMP-13, also appears to be necessary for a full angiogenic response. Interestingly, these 2 critical enzymes are actively imported into the vascular/stromal tissue by distinct inflammatory cells responding to the angiogenic stimulation. Neutrophil-like heterophils rapidly and almost immediately import MMP-9 into the tissue, whereas monocyte/macrophages actively deliver MMP-13 1–2 days later, possibly in response to specific secreted products of the early arriving heterophils. Thus, normal angiogenesis and tumor angiogenesis are closely linked to an accompanying host inflammatory response that contributes critical functional molecules to the angiogenic process. We are dissecting out and identifying the molecules and cells that link the inflammatory response to the CELL BIOLOGY 2005 49 PUBLICATIONS Blancafort, P., Chen, E.I., Gonzalez, B., Bergquist, S., Zijlstra, A., Guthy, D., Brachat, A., Brakenhoff, R.H., Quigley, J.P., Erdmann, D., Barbas, C.F. III. Genetic reprogramming of tumor cells by zinc finger transcription factors. Proc. Natl. Acad. Sci. U. S. A. 102:11716, 2005. Deryugina, E.I., Zijlstra, A., Partridge, J.J., Kupriyanova, T.A., Madsen, M., Papagiannakopoulos, T., Quigley, J.P. Unexpected effect of matrix metalloproteinase downregulation on vascular intravasation and metastasis of human fibrosarcoma cells selected in vivo for high rates of dissemination. Cancer Res., in press. Wilson, S., Greer, B., Hooper, J., Zijlstra, A., Walker, B., Quigley, J.P., Hawthorne S. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem. J. 388(Pt. 3):967, 2005. Zijlstra, A., Seandel, M., Kupriyanova, T.A., Partridge, J.J., Madsen, M., HahnDantona, E.A., Quigley, J.P., Deryugina, E.I. Pro-angiogenic role of neutrophil-like inflammatory heterophils during neovascularization induced by growth factors and human tumor cells Blood, in press. Regulators of Clathrin-Mediated Endocytosis S.L. Schmid J. Chappie, S.D. Conner, M. Ishido, M. Leonard, R. Ramachandran, F. Soulet, B.D. Song, M.C. Surka, D. Yarar lathrin-mediated endocytosis is essential for the efficient uptake of nutrients and other macromolecules into cells and for the regulation of signaling by cell-surface receptors. The process occurs at clathrin-coated pits, which concentrate receptor-ligand complexes, deform the membrane, invaginate, and eventually pinch off, forming clathrin-coated vesicles (CCVs). The major components involved in the formation of CCVs are clathrin, adaptor proteins, and dynamin. Clathrin self-assembles into a polygonal lattice and serves as a scaffold for the formation of coated pits. Adaptor protein-2 (AP2) is a heterotetrameric protein that triggers clathrin assembly at the plasma membrane and interacts directly with the cytoplasmic tails of surface receptors to concentrate the receptors into the assembling coated pit. Dynamin is a multidomain GTPase that regulates endocytosis. Each of these proteins also interacts with a myriad of accessory proteins, whose function in the formation of CCVs is poorly understood. In the past year, we focused on factors that regulate clathrin-mediated endocytosis. Dynamin has an unusually low affinity for GTP and a relatively high basal rate of GTP hydrolysis. Moreover, dynamin self-assembles into rings and helical stacks of rings that are localized to the necks of deeply invaginated coated pits. Dynamin self-assembly stimulates its GTPase activity approximately 100-fold; this stimulation is mediated by dynamin’s GTPase effector C F i g . 1 . In vivo model to quantitate angiogenesis induced by growth factors and tumor cells. Collagen onplants are placed on the chorioallantoic membrane (CAM) of day 10 chick embryos developing ex ovo (white arrows, left panel, top row). In 3 days, new vessels sprouting from preexisting vessels in the membrane grow up through the collagen into the gridded nylon mesh (right panel, top row) and with a dissecting microscope are clearly visible within the grids (black arrowheads, middle row) and easily distinguished from the background preexisting vessels (open circles, middle row). The number of new vessels in the grids can be easily scored. Differential angiogenesis can be quantitated as indicated in the bar graph. Aggressive human tumor cells, such as HT1080 cells, placed into the onplants induce elevated levels of angiogenesis similar to those induced by purified growth factors (VEGF/FGF) and blocked by specific metalloproteinase inhibitors (MMP inhibitor and TIMP-2). Nonaggressive human tumor cells, such as HeLa cells, do not induce significant angiogenesis. If onplants containing HT1080 cells are allowed to incubate on the embryo for 6 days, large vascularized primary tumors develop (right panel, bottom row), resulting in dissemination of metastatic cells into secondary organs of the embryo. angiogenic process and to the progression of malignant neoplasms. We are also trying to decipher whether the relevant functional molecules are derived from host cells or from tumor cells. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 50 CELL BIOLOGY 2005 domain. Although dynamin clearly is essential for clathrin-mediated endocytosis, the role of GTP binding and/or hydrolysis in endocytosis remains unclear. Insight into the function of dynamin in endocytosis was derived from collaborative studies with M. Ramaswami, University of Arizona, Tucson, Arizona. Point mutations in shibire, the gene for dynamin in Drosophila, cause temperature-sensitive defects in endocytosis. We showed that the ts2 mutation, which occurs in the switch 2 region of the dynamin GTPase domain, compromises GTP-binding affinity. Three second-site suppressor mutations, 1 in the switch 1 region of the GTPase domain and 2 in the GTPase effector domain, fully rescued the ts2 defects in synaptic vesicle recycling. The functional rescue in vivo correlated with a reduction in both the basal and assembly-stimulated GTPase activity in vitro. These findings indicated that the GTPase effector domain is indeed an internal dynamin GTPase-activating protein and establish that, as for other GTPases, the function of dynamin in vivo is negatively regulated by its activity as a GTPase-activating protein. On the basis of these findings, we proposed a 2-step model (Fig. 1) for dynamin during the formation of vesicles. In this model, an early regulatory GTPase-like function precedes late, assembly-dependent steps during which GTP hydrolysis is required for vesicle release. A 2-step model for dynamin function in clathrin-mediated endocytosis. In stage 1, dynamin (ovals) plays a regulatory role and functions as a kinetic timer controlling the recruitment and/or activity of SH3 domain–containing molecules. In stage 2, dynamin assembly and assembly-stimulated GTPase activity are required as Fig. 1. a fission apparatus, a curvature sensor, and/or a structural scaffold. In this model, dynamin self-assembly marks the transition between early and late functions of dynamin. What might control this transition? We identified a new protein partner of dynamin, sorting nexin 9 (SNX9). SNX9 binds directly to dynamin and potentiates dynamin assembly and assembly-stimulated GTPase activity on liposomes and in solution. Using total internal reflection fluorescence microscopy in living cells, we detected a transient burst of recruitment of SNX9 labeled with green fluorescent protein to clathrin-coated pits that occurs during the Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. late stages of vesicle formation and coincides spatially and temporally with a burst of fluorescence of dynamin labeled with red fluorescent protein. These data suggest that dynamin effectors, like SNX9, might coordinate dynamin self-assembly with coat assembly, cargo recruitment, or membrane invagination. Clathrin-mediated endocytosis is also regulated by phosphorylation events, but the kinases responsible, some of which cofractionate with coat proteins, have not been identified. Through mass spectroscopic analysis of preparations of adaptor proteins, we identified a novel serine/threonine kinase, the coated vesicle–associated kinase of 104 kD (CVAK104), that belongs to the SCY1-like family of protein kinases, previously thought to be catalytically inactive. CVAK104 cofractionates with adaptor proteins extracted from CCVs and directly binds to both clathrin and the plasma membrane adaptor AP2. CVAK104 binds ATP, and kinase assays indicate that it functions as a poly-L-lysine–stimulated kinase that can autophosphorylate and phosphorylate the β 2 -adaptin subunit of AP2. Further studies are needed to determine the functional consequences of this phosphorylation reaction. Actin assembly is spatially and temporally coordinated with endocytosis, and numerous endocytic accessory proteins directly or indirectly regulate actin dynamics. However, functional evidence for a role of actin during the formation of CCVs has been lacking. In collaboration with C. Waterman-Storer, Department of Cell Biology, using parallel biochemical and microscopic approaches, we reexamined the role of cortical actin dynamics of clathrin-mediated endocytosis. Using total internal reflection fluorescence microscopy, we found that disruption of the F-actin assembly and disassembly cycle with latrunculin A or jasplakinolide resulted in nearly complete cessation of all aspects of the dynamics of clathrin-coated structures labeled with red fluorescent protein. Stage-specific biochemical assays and quantitative fluorescence and electron microscopic analyses established that F-actin dynamics are required for multiple distinct stages of the formation of CCVs, including the formation, constriction, and internalization of coated pits. In future studies, we will address the molecular mechanisms that link actin dynamics to clathrin-mediated endocytosis. PUBLICATIONS Conner, S.D., Schmid, S.L. CVAK104 is a novel poly-L-lysine-stimulated kinase that targets the β2 subunit of AP2. J. Biol. Chem. 280:21539, 2005. Leonard, M., Song, B.D., Ramachandran, R., Schmid, S.L. Robust colorimetric assays for dynamin’s basal and stimulated GTPase activities. Methods Enzymol., in press. CELL BIOLOGY 2005 Miwako, I., Schmid, S.L. Coated vesicle formation from isolated plasma membranes. Methods Enzymol., in press. Narayanan, R., Leonard, M., Song, B.D., Schmid, S.L. Ramiswami, M. An internal GAP domain negatively regulates presynaptic dynamin in vivo: a two-step model for dynamin function. J. Cell Biol. 169:117, 2005. Song, B.D., Leonard, M., Schmid, S.L. Dynamin GTPase domain mutants that differentially affect GTP binding, GTP hydrolysis, and clathrin-mediated endocytosis. J. Biol. Chem. 279:40431, 2004. Soulet, F., Yarar, D., Leonard, M., Schmid, S.L. SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol. Biol. Cell 16:2058, 2005. Yarar, D., Waterman-Storer, C., Schmid, S.L. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol. Biol. Cell 16:964, 2005. Molecular Biology of Olfaction L. Stowers, K. Flanagan, M. Gubernator, J. Lin, T. Marton, C. Ramos, J. Riceberg very breath samples the environment for olfactory chemical information, determining the quality of food, warning of danger, and confirming safety. The sense of olfaction is composed of 2 types of neurons: those that mediate an evocative perception that varies with each individual’s experience and those that regulate stereotyped innate social behaviors such as aggression and mating. Neurons that elicit odorant perception reside in the olfactory epithelium and relay chemical information through activation of cAMP-responsive channels. Recently, we showed that behavior-generating neurons are located in the vomeronasal organ and respond to pheromones through a cascade that ultimately activates C-type transient-receptor-potential 2 (TrpC2) channels. We are using a molecular genetic approach to characterize the function of these pheromoneresponsive neurons. Through electrophysiologic recordings, we showed that neurons in mutant mice lacking TrpC2 do not depolarize in response to natural sources of pheromones. Behavioral assays with these animals revealed that this pheromone response is necessary for both intermale aggression and gender recognition. We are identifying other unique molecular subpopulations of pheromoneresponsive neurons, and through genetic ablation, biochemistry, and electrophysiology, we are assigning a biological function to each neuron type. A full characterization of the repertoire of chemosensory neurons will be essential in understanding the logic of olfactory information coding. To this end, we are investigating a novel class of olfactory neurons that lack both the cAMP and TrpC2 signaling components. E Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 51 Analysis of these neurons by transcriptional profiling and then molecular genetics and biochemistry is being used to identify their role in olfactory function. Elucidation of the function of specific neural circuits that regulate mammalian behavior has been hindered because the pheromone compounds that signal each behavior have not been identified from the complex natural sources of the compounds. To obtain these important molecules, we are establishing a high-throughput screening assay of chemical compounds that we expect will provide identification of both agonists and antagonists specific to each of the 400 pheromone receptors. These data will enable us both to activate specific neural circuits and analyze the natural production and regulation of the signaling ligands. In total, we expect to define the pheromone response pathway of mice and to reveal general principles of neurons that govern complex social behavior. PUBLICATIONS Stowers, L., Marton, T.F. What is a pheromone? Mammalian pheromones reconsidered. Neuron 46:699, 2005. Molecular Regulation of Vascular System Development in Mammals M.J. Fitch, Z. Zou, S. Chitnis, J.D. Lewis, A. Durrans, M. Schroeter,* L. Campagnolo,** W. LeVine, H. Stuhlmann * University of Göttingen, Göttingen, Germany ** Università degli Studi di Roma Tor Vergata, Rome, Italy he cardiovascular system is the first organ system to develop in mammalian embryos. Establishment of a functional circulatory system is crucial for delivery of nutrients and oxygen to the embryos, and defects result in death before birth or in congenital cardiovascular abnormalities. Our major goal is to dissect the molecular pathways that regulate the 2 principal processes of vascular development: vasculogenesis and angiogenesis. We focus on the mouse model because of the ready availability of genetic information on mice and experimental tools and because of similarities between mice and humans. Previously, we used a “gene trap” screen in mouse embryonic stem cells and embryos to detect novel genes involved in these processes. T A ZINC FINGER GENE ESSENTIAL FOR NORMAL VA S C U L A R A N D LY M P H AT I C D E V E L O P M E N T One endothelial-expressed gene identified in the screen, Vezf1, encodes a 56-kD nuclear transcription 52 CELL BIOLOGY 2005 factor with 6 putative zinc finger domains. The gene and its family members are highly conserved in vertebrate species. Using transgenic mice and mice deficient in Vezf1, we determined that this gene plays an essential and dosage-dependent role in the proliferation, remodeling, and integrity of the developing vasculature. Homozygous and a fraction of heterozygous mutant embryos had vascular and lymphatic endothelial abnormalities and died during midgestation. Electron and confocal microscopy indicated that the primary phenotype involves defects in endothelial cell junctions and basal membrane. These findings are supported by recent studies in the embryoid body in vitro differentiation model and in teratocarcinomas derived from mutant embryonic stem cells. Of interest, the lethal phenotype in mice lacking Vezf1 can be at least partially rescued by endothelial overexpression of the gene. How does Vezf1 control vascular development? We are beginning to explore the molecular pathways of Vezf1 function. We have identified the regulatory regions in the Vezf1 promoter in vitro and in vivo. To identify target genes of Vezf1, we are using subtractive hybridization and cDNA/expressed sequence tag microarray analysis. In addition, we are analyzing the properties of primary cells from Vezf1 mutant embryos. A N E A R LY M A R K E R F O R E N D O T H E L I A L C E L L S A N D thelial progenitor cells in mouse embryos and during differentiation of embryonic stem cells and to study the potential of the cells to contribute to or regenerate vascular tissues in embryos and adults. D E V E L O P M E N T O F M U LT I VA L E N T V I R A L N A N O PA R T I C L E S F O R I N V I V O VA S C U L A R TA R G E T I N G A N D I M A G I N G We are developing, in collaboration with M. Manchester, Department of Cell Biology, viral nanoparticles for noninvasive imaging and targeting of the cardiovascular system in mammals. For these studies, fluorescent dyes and peptides are chemically attached to lysine residues of the capsid of cowpea mosaic virus (CPMV). In initial studies, we showed that fluorescent CPMV produces a bright and stable signal that allows visualization of blood flow in developing mouse and chick embryos and in tumor angiogenesis models. We are extending these studies to target CPMV with multivalent attached peptides to the vasculature both during development and in disease models such as tumor angiogenesis. PUBLICATIONS Campagnolo, L., Leahy, A., Chitnis, S., Koschnick, S., Fitch, M.J., Fallon, J.T., Loskutoff, D., Taubman, M.B., Stuhlmann, H. EGFL7 is a chemoattractant for endothelial cells and is up-regulated in angiogenesis and arterial injury. Am. J. Pathol. 167:275, 2005. Feral, C.C., Nishiya, N., Fenczik, C.A., Stuhlmann, H., Slepak, M., Ginsberg, M.H. CD98hc (SLC3A2) mediates integrin signaling. Proc. Natl. Acad. Sci. U. S. A. 102:355, 2005. THEIR PROGENITORS A second endothelial gene identified in our screen, Egfl7, encodes a 30-kD secreted protein with 2 internal epidermal growth factor–like domains. Expression of Egfl7 is restricted to the vascular endothelium and its progenitors in the yolk sac mesoderm. Expression is downregulated in most of the quiescent vasculature in adults but is transiently upregulated during vascular injury and endothelial regeneration and during physiologic angiogenesis in the uterus during pregnancy. Clues about a possible role of Egfl7 come from our studies in which we found that EGFL7, the protein encoded by the gene, acts as a chemoattractant for endothelial cells. What is the function of Egfl7 during development and adult angiogenesis? We are using gain- and lossof-function approaches in the mouse model to test our hypothesis that Egfl7 plays a crucial role during vascular development in embryos and in angiogenesis in adults. We are also determining if EGFL7 acts as a growth factor or a cytokine through direct binding to receptors or through interaction with proteins in the extracellular matrix. Finally, we are using Egfl7 as a molecular marker to isolate early populations of endoPublished by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. Kuhnert, F., Campagnolo, L., Xiong, J.-W., Lemons, D., Fitch, M.J., Zou, Z., Kiosses, W.B., Gardner, H., Stuhlmann, H. Dosage-dependent requirement for mouse Vezf1 in vascular system develpment. Dev. Biol. 283:140, 2005. Kuhnert, F., Stuhlmann, H. Identifying early vascular genes through gene trapping in mouse embryonic stem cells. Curr. Top. Dev. Biol. 62:261, 2004. Ion Channels and Fast Synaptic Transmission N. Unwin on channels play a central role in the rapid transmission of electrical signals throughout the nervous system. To determine how these membrane proteins work, my colleagues and I are using electron microscopy to analyze their structures trapped in different physiologic states. Current studies center on the nicotinic acetylcholine receptor at the nerve-muscle synapse. We wish to find out how this ion channel achieves its ion selectivity and high transport rate and how it opens and desensitizes in response to acetylcholine released into the synaptic cleft. For our studies, we use post- I CELL BIOLOGY 2005 synaptic membranes isolated from the (muscle-derived) electric organ of a Torpedo ray, which form tubular crystals of acetylcholine receptors. The acetylcholine receptor is a member of a superfamily of transmitter-gated ion channels, which includes the serotonin 5-HT3, γ-aminobutyric acids A and C, and glycine receptors. It has a cation-selective pore, delineated by a ring of 5 similar subunits, that opens upon binding of acetylcholine to the 2 ligand-binding (α) subunits at the subunit interfaces. Recently, we obtained a refined atomic model of the acetylcholine receptor in the closed-channel form. We found that the individual subunits in the N-terminal ligand-binding domain are organized around 2 sets of β-sheets packed in a curled β-sandwich, as in the related soluble pentameric acetylcholine-binding protein. Each of the subunits in the membrane- spanning domain is made from 4 α-helical segments. The helical segments arrange symmetrically, forming an inner ring of helices that shape a water-filled pore and an outer shell of helices that coil around each other and shield the inner ring from the lipids. In the closed channel, the helices in the inner ring come together near the middle of the membrane and make a constricting hydrophobic girdle. This girdle, which is about 50 Å from the acetylcholine-binding sites, constitutes an energetic barrier to ion permeation and functions as the gate of the channel. These details, together with those obtained earlier from studies of the receptor trapped in the open-channel form, have enabled us to understand in outline the allosteric mechanism by which acetylcholine opens the pore. In the absence of acetylcholine, the pore is normally closed. When acetylcholine enters the binding sites, localized rearrangements in the α-subunits occur that stabilize an alternative extended conformation of the channel in which the inner sets of β-sheets are rotated by about 10° about axes perpendicular to the membrane plane, relative to their orientations in the closed channel. These rotations are communicated through the inner membrane-spanning helices and open the pore by breaking the hydrophobic girdle apart. Improvements in resolution of the 3-dimensional structure, in both the closed- and the open-channel forms, are now being attempted so that the structural mechanism of gating of the channel can be described in greater detail. The knowledge gained from the refined structure of the locations of amino acid residues, in relation to the ion pathway, is also being used to develop Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 53 quantitative explanations of how the high cation selectivity and high conduction rates of this channel are achieved. These studies are yielding crucial insight into the nature of a number of neuromuscular disorders, including several well-characterized congenital myasthenic syndromes. They are also providing important 3-dimensional information about the binding sites for drugs that affect the brain by modulating the function of the related γ-aminobutyric acid, serotonin, glycine, and neuronal acetylcholine receptors. PUBLICATIONS Unwin, N. Refined structure of the nicotinic acetylcholine receptor at 4Å resolution. J. Mol. Biol. 346:967, 2005. Microscopes and Motility: Systems Integration in Cell Migration C.M. Waterman-Storer, T. Wittmann, N. Prigozhina, O. Rodriguez, S.L. Gupton, K. Kita, R. Littlefield, K. Hu, A. Wheeler, W. Shin, M.L. Gardel ell migration is critical to development, the immune response, and wound healing. In cancer cells, loss of regulation of cell motility results in deadly metastasis. The locomotion of vertebrate tissue cells is thought to require complex and dynamic interactions between the microtubules and actin cytoskeletal polymers, the endomembrane trafficking system, and focal adhesions to the substrate. We develop quantitative light microscopy methods to analyze the dynamic interactions between these complex macromolecular systems in living cells to understand how the systems are spatiotemporally coordinated to drive directed cell movement. We then use these microscopic assays to analyze cells with specific perturbations of cytoskeletal, membrane, or adhesive proteins to dissect the molecular mechanisms of the regulation of the proteins and their contribution to cell morphogenesis and migration. We pioneered fluorescent speckle microscopy, a powerful method that allows quantitative analysis of the dynamics of macromolecular assemblies in living cells. In the past year, we enhanced the sensitivity of this technique by extending the technology to multispectral total internal fluorescence reflection fluorescence microscopy, allowing the first-ever analysis of C 54 CELL BIOLOGY 2005 the integration of proteins within focal adhesion complexes with the actin cytoskeleton during cell migration. The microtubule and actin cytoskeletons may interact in cells via coregulation by the same signaling cascades to promote a coordinated effort that drives polarized cell migration. In support of this notion, we showed that a signaling cascade downstream of Rac1 GTPase coordinates the regulation of both actin and microtubules at the protruding edge of migrating cells. We found that Rac1 interacts directly with and activates p21-activated kinase 1, whose activity is required for the assembly of both microtubules and actin filaments in cell protrusion. Microtubule growth is promoted when the kinase directly phosphorylates and inactivates the microtubule-destabilizing oncoprotein Op18/stathmin. We also identified a second Rac1 effector, CLASP, that regulates polarized microtubule behavior in migrating cells. CLASP interaction with microtubules specifically in the leading edge of migrating cells is promoted by Rac1 activity via glycogen synthase kinase 3, the wellknown regulator of cell polarity. It has been thought for some time that localized, microtubule-dependent delivery of endomembrane components to the leading-edge plasma membrane is required for continued cell motility. We used PKD, a mutant kinase of the trans-Golgi network, to specifically inhibit the transport of membrane from the trans-Golgi network to the plasma membrane in cells and monitored cell motile functions by using time-lapse microscopy. The results indicated that PKD-mediated transport across the trans-Golgi network to the plasma membrane along microtubules is required for fibroblast locomotion and localized Rac1-dependent leading-edge activity. The actin cytoskeleton is locally regulated for functional specializations for cell motility. In collaboration with G. Danuser, Department of Cell Biology, using computational analysis of quantitative fluorescent speckle microscopy of the actin cytoskeleton, we defined 2 spatially colocalized but functionally distinct actinbased machines in migrating epithelial cells. The lamellipodium consists of a treadmilling F-actin array with rapid retrograde flow in which Arp2/3, a complex containing the actin-related proteins 2 and 3, and the actin depolymerization factor cofilin concentrate, whereas the lamella has spatially random punctae of F-actin assembly and disassembly with slow retrograde flow and contains myosin II and tropomyosin. We sought to specifically inhibit the formation of the lamellipodium to test its requirement in cell migraPublished by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. tion. We increased cellular tropomyosin levels in epithelial cells, and using quantitative fluorescent speckle microscopy, electron microscopy, and immunolocalization, showed that this increase blocked formation of functional lamellipodia. Cells lacking a lamellipodium had more persistent leading-edge protrusion and rapid cell migration than did cells with a lamellipodium. Inhibition of endogenous tropomyosin reduced the persistence of lamellipodial protrusion. Thus, in stark contrast to the dogma in cell migration, cells can migrate in the absence of a lamellipodium and tropomyosin is a major regulator of functional specialization of the F-actin cytoskeleton in migrating cells. PUBLICATIONS Adams, M.C., Matov, A., Yarar, D., Gupton, S.L., Danuser, G., Waterman-Storer, C.M. Signal analysis of total internal reflection fluorescent speckle microscopy (TIRFSM) and wide-field epi-fluorescence FSM of the actin cytoskeleton and focal adhesions in living cells. J. Microsc. 216(Pt. 2):138, 2004. Gupton, S.L., Anderson, K.L., Kole, T.P., Fischer, R.S., Ponti, A., HitchcockDeGregori, S.E., Danuser, G., Fowler, V.M., Wirtz, D. Hanein, D., WatermanStorer, C.M. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 168:619, 2005. Gupton, S.L., Waterman-Storer, C.M. Live-cell fluorescent speckle microscopy (FSM) of actin cytoskeletal dynamics and their perturbation by drug perfusion. In: Cell Biology: A Laboratory Handbook, 3rd ed. Celis, J., Small, J.V. (Eds.). Elsevier, St. Louis, in press. Ponti, A., Machachek, M., Gupton, S.L., Waterman-Storer, C.M., Danuser, G. Two distinct actin networks drive the protrusion of migrating cells. Science 305:1782, 2004. Ponti, A., Matov, A., Waterman-Storer, C.M., Danuser, G. Computational fluorescent speckle microscopy, II: high-resolution comapping of F-actin flow and turnover in migrating cells. Biophys. J., in press. Torreano, P.A., Waterman-Storer, C.M., Cohan, C.S. The effects of collapsing factors on F-actin content and microtubule distribution of Helisoma growth cones. Cell Motil. Cytoskeleton 60:166, 2005. Wittmann, T., Littlefield, R., Waterman-Storer, C.M. Fluorescent speckle microscopy of cytoskeletal dynamics living cells. In: Live Cell Imaging: A Laboratory Manual. Spector, D.L., Goldman, R.D. (Eds.). Cold Spring Harbor Press, Cold Spring Harbor, NY, 2004, p. 187. Wittmann, T., Waterman-Storer, C.M. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3β in migrating epithelial cells. J. Cell Biol. 169:929, 2005. Yarar, D., Waterman-Storer, C.M., Schmid, S.L. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol. Biol. Cell 16:964, 2005. Systems Biology of Malaria E. Winzeler, C. Kidgell, J. Young, J. Johnson s the causative agent of human malaria, parasites of the genus Plasmodium are major contributors to global morbidity and mortality. Approximately 300 million to 500 million cases of malaria occur each year. Individuals who are repeatedly exposed to the par- A CELL BIOLOGY 2005 asite become immune but not completely. The immunity is incomplete because either the parasites alter the complement of antigens on the surface of the red blood cells or on the surface of the merozoites during the merozoites’ brief extracellular phase in the blood or because a great deal of genetic diversity exists in the parasite population. Because no licensed malaria vaccine is available, drugs are the best therapy for the disease. However, resistance to inexpensive drugs such as chloroquine has emerged and spread quickly. Multidrug-resistant strains are now common. Global warming and human migration may bring the disease back to areas where it was once eliminated. The economic cost of malaria to the developing world is enormous. Despite the impact of malaria parasites on global health and productivity, little is known about the proteins encoded by the Plasmodium genome. Many proteins do not have recognizable homologs in other species, either because the proteins are involved in parasite-specific processes or because of the parasite’s preference for amino acids encoded by codons rich in adenine and thymine. Thus, we also do not understand how protein expression is regulated or know the identity of any key regulators of development. Because traditional methods for discovering gene function are laborious, we are using a systematic approach to begin to predict the function of proteins encoded by the genome of Plasmodium falciparum and to discover networks and protein-protein interactions. We are interested in describing how the transcript and protein levels change for all the genes in the genome under as many natural life-stage and environmental conditions as possible. Some of our current studies involve globally comparing transcript and protein levels to systematically identify proteins whose levels are controlled posttranscriptionally and extending our transcriptional analysis of the parasite’s genome to the sexual development phase, which is responsible for the transmission of malaria. This work has led to the identification of sequence motifs in the parasite genome that most likely are responsible for controlling transcription during development. We are also interested in developing novel algorithms that can be used to create probabilistic models of gene function based on gene expression and proteomic and homology data. PUBLICATIONS Daily, J.P., Le Roch, K.G., Sarr, O., Ndiaye, D., Lukens, A., Zhou, Y., Ndir, O., Mboup, S., Sultan, A., Winzeler, E.A., Wirth, D.F. In vivo transcriptome of Plasmodium falciparum reveals overexpression of transcripts that encode surface proteins. J. Infect. Dis. 191:1196, 2005. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 55 Kidgell, C., Winzeler, E.A. Elucidating genetic diversity with oligonucleotide arrays. Chromosome Res. 13:225, 2005. Le Roch, K., Johnson, J.R., Florens, L., Zhou, Y., Santrosyan, A., Grainger, M., Yan, S.F., Williamson, K.C., Holder, A.A., Carucci, D.J., Yates, J.R. III, Winzeler, E.A. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 14:2308, 2004. O’Neill, B.M., Hanway, D., Winzeler, E.A. Coordinated functions of WSS1, PSY2 and TOF1 in the DNA damage response. Nucleic Acids Res. 32:6519, 2004. Simpson, K.M., Baum, J., Good, R.T., Winzeler, E.A., Cowman, A.F., Speed, T.P. A comparison of match-only algorithms for the analysis of Plasmodium falciparum oligonucleotide arrays. Int. J. Parasitol. 35:523, 2005. Stubbs, J., Simpson, K.M., Triglia, T., Plouffe, D., Tonkin, C.J., Duraisingh, M.T., Maier, A.G., Winzeler, E.A., Cowman, A.F. Molecular mechanism for switching of P falciparum invasion pathways into human erythrocytes. Science 309:1384, 2005. Young, J.A., Fivelman, Q.L., Blair, P.L., de la Vega, P., Le Roch, K.G., Zhou, Y., Carucci, D.J., Baker, D.A., Winzeler, E.A. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol. Biochem. Parasitol. 143:67, 2005. Young, J.A., Winzeler, E.A. Using expression information to discover new drug and vaccine targets in the malaria parasite Plasmodium falciparum. Pharmacogenomics 6:17, 2005. Zhou, Y., Young, J.A., Santrosyan, A., Chen, K., Yan, F., Winzeler, E.A. In silico gene function prediction using ontology-based pattern identification. Bioinformatics 21:1237, 2005. Advancing Applications in Mass Spectrometry–Based Proteomics J.R. Yates III, A.O. Bailey, G.T. Cantin, E. Chen, D. Cociorva, J. Coppinger, C. Delahunty, M.Q. Dong, J. Hewel, J.R. Johnson, L. Liao, I. MacLeod, D. McClatchy, J. Meng, S. Niessen, R. Park, J.H. Prieto, E. Romijn, C.I. Ruse, R. Sadygov, J. Venable, J. Wohlschlegel, C. Wu, T. Xu, W.H. Zhu ass spectrometry has emerged as a powerful technique for cellular proteomics, complementing traditional gene-by-gene approaches with a comprehensive description of the molecular factors that contribute to a biologically relevant system. We remain at the forefront of this field, developing new strategies to address more sophisticated scientific questions through proteomics, such as how to quantify global changes in protein abundance for mammalian systems, how to characterize complex posttranslational modifications, and what information can be gained by comparing the proteome with the transcriptome. Proteomics methods based on quantitative mass spectrometry rely on strategies that introduce an internal isotopic standard for every protein to be characterized. The preferred method for introducing these standards is metabolic labeling (i.e., providing an isotope-labeled M 56 CELL BIOLOGY 2005 amino acid in culture medium); however, this approach has been limited to microorganisms and simple model organisms. Recently, we developed a method to generate proteins and peptides labeled with nitrogen 15 in Rattus norvegicus tissues; the method resulted in an atomic enrichment greater than 90% in liver and plasma. This labeling strategy has countless possibilities for use in comparative proteomic analyses of mammalian tissues, particularly studies of animal models of disease. In addition, we developed a novel technique for comparing the abundance of peptides with internal isotopic standards from tandem mass spectrometry spectra. Whereas in most quantitative proteomics approaches, these differences are measured at the level of the mass spectra of intact peptides, by quantifying the fragmented peptide instead, we dramatically improved the signalto-noise ratio and dynamic range of these measurements. The characterization of posttranslational modifications is also an emerging application of mass spectrometry–based proteomics. The modification of cellular factors by small ubiquitin-like modifiers (SUMOs) is an essential process in budding yeast, but the identities of the substrates remain largely unknown. Using proteomics techniques, we identified 271 new SUMO targets, illustrating for the first time the diverse roles that SUMO plays in regulating eukaryotic cells. This research also revealed coordinated SUMO modification of multiple proteins in well-defined macromolecular complexes. This intriguing result suggests that sumoylation may target protein complexes rather than individual proteins. In other studies, we are characterizing the mechanism that underlies this observation. Finally, in collaboration with E.A. Winzeler, Department of Cell Biology, we did a genome-wide comparison of the abundance of global protein and mRNA transcripts for several stages throughout the life cycle of Plasmodium falciparum, the parasite that causes malaria. Previous work in other laboratories suggested that little correlation exists between mRNA and protein abundance. However, our comparison showed a significant correlation in P falciparum, particularly when a delay between the synthesis of mRNA transcripts and proteins was considered. Interestingly, correlating mRNA and protein expression profiles for individual genes revealed particular families of functionally related genes that appeared to have similar patterns of mRNA and protein accumulation. Using this gene-by-gene correlation analysis, we were able to identify those genes that most likely are regulated by posttranscriptional controls Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. and will be useful for future studies to unravel the molecular mechanisms that underlie this type of regulation. PUBLICATIONS Le Roch, K.G., Johnson, J.R., Florens, L., Zhou, Y., Santrosyan, A., Grainger, M., Yan, S.F., Williamson, K.C., Holder, A.A., Carucci, D.J., Yates, J.R. III, Winzeler, E.A. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 14:2308, 2004. Venable, J.D., Dong, M.Q., Wohlschlegel, J., Dillin, A., Yates, J.R. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1:39-45, 2004. Wohlschlegel, J.A., Johnson, E.S., Reed, S.I., Yates, J.R. III. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279:45662, 2004. Wu, C.C., MacCoss, M.J., Howell, K.E., Matthews, D.E., Yates, J.R. III. Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal. Chem. 76:4951, 2004. Macromolecular Assemblies Visualized by Electron Cryomicroscopy and Image Analysis: Membrane Proteins and Viruses M. Yeager, B.D. Adair, K. Altieri, A. Cheng, M.J. Daniels, K.A. Dryden, B. Ganser, J. Harless, Y. Hua, R. Nunn, F.A. Palida, M.A. Arnaout,* A.R. Bellamy,** N. Ben-Tal,*** M.J. Buchmeier,**** F.V. Chisari,**** K. Coombs,***** H.B. Greenberg,† J.E. Johnson,**** S. Matsui,† L.H. Philipson,†† T.D. Pollard,††† A. Rein†††† A. Schneeman,**** J.A. Tainer,**** J.A. Taylor,** V.M. Unger††† * Harvard Medical School, Boston, Massachusetts ** University of Auckland, Auckland, New Zealand *** Tel-Aviv University, Tel-Aviv, Israel **** Scripps Research ***** University of Manitoba, Winnipeg, Manitoba † Stanford University, Stanford, California †† University of Chicago, Chicago, Illinois ††† Yale University, New Haven, Connecticut †††† National Cancer Institute, Frederick, Maryland he ultimate goal of our studies is to gain a deeper understanding of the molecular basis of important human diseases, such as sudden death, heart attacks, and HIV infection, that cause substantial mortality and suffering. The structural details revealed by our research may provide clues for the design of more effective and safer medicines. At the basic science level, we are intrigued by questions at the interface between cell biology and struc- T CELL BIOLOGY 2005 tural biology: How do membrane proteins fold? How do membrane channels open and close? How are signals transmitted across a cellular membrane when an extracellular ligand binds to a membrane receptor? How do viruses attach to and enter host cells, replicate, and assemble infectious particles? To explore such problems, we use high-resolution electron cryomicroscopy and computer image processing. With this approach, we can examine the molecular architecture of supramolecular assemblies such as membrane proteins and viruses. In electron cryomicroscopy, biological specimens are quick frozen in a physiologic state to preserve their native structure and functional properties. A special advantage of this method is that we can capture dynamic states of functioning macromolecular assemblies, such as open and closed states of membrane channels and viruses actively transcribing RNA. Three-dimensional density maps are obtained by digital image processing of the high-resolution electron micrographs. The rich detail in the density maps indicates the power of this approach to reveal the structural organization of complex biological systems that can be related to the functional properties of such assemblies. Research projects under way include the structure analysis of (1) membrane proteins involved in cell-tocell communication (gap junctions), water transport (aquaporins), ion transport (potassium channels), transmembrane signaling (integrins), and viral recognition (rotavirus NSP4); (2) viruses responsible for significant human diseases (retroviruses, hepatitis B virus, rotavirus, astrovirus); and (3) viruses used as model systems to understand mechanisms of pathogenesis (arenaviruses, reoviruses, nodaviruses, tetraviruses, and sobemoviruses). The following sections summarize selected projects that exemplify the themes of our research program. 57 We have now extended this analysis to 5.7-Å in-plane and 19.8-Å vertical resolution, a step that enables us to identify the positions and tilt angles for the 24 α-helices within each hemichannel (Fig. 1). The 4 hydrophobic segments in connexin sequences were assigned to the α-helices in the map on the basis of biochemical and phylogenetic data. Evolutionary conservation and an analysis of compensatory mutations in connexin evolution were used to identify the packing interfaces between the helices. The final model, which specifies the coordinates of Cα atoms in the transmembrane domain, provides a structural basis for understanding the different physiologic effects of almost 30 mutations and polymorphisms in terms of structural deformations at the interfaces between helices, revealing an intimate connection between molecular structure and disease. INTEGRINS Integrins are a large family of heterodimeric transmembrane receptor proteins that modulate important GAP JUNCTION MEMBRANE CHANNELS Gap junction channels connect the cytoplasms of adjacent cells by means of an intercellular conduit formed by the end-to-end docking of 2 hexameric hemichannels called connexons. Gap junctions play an essential functional role by mediating metabolic and electrical communication within tissues. For instance, in the heart, gap junction channels organize the pattern of current flow to allow a coordinated contraction of the muscle. We expressed a recombinant cardiac gap junction protein, termed connexin 43, and produced 2-dimensional crystals suitable for electron cryocrystallography. Our previous findings indicated that each hexameric connexon is formed by 24 closely packed α-helices. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. F i g . 1 . Intercellular gap junction channels have a diameter of about 65 Å and are formed by the end-to-end docking of 2 hemichannels, each composed of a hexamer of connexin subunits. A Cα model (ribbons) for the membrane-spanning domain of the hemichannels was derived by combining the information from a computational analysis of connexin sequences, the results of more than a decade of biochemical studies, and the constraints provided by a 3-dimensional map derived by electron cryocrystallography. Although individually none of these approaches provided high-resolution information, their sum yielded an atomic model that predicts how connexin mutations (spheres) may interfere with formation of functional channels by disrupting helix-helix packing. 58 CELL BIOLOGY 2005 biological processes such as development, cell adhesion, angiogenesis, wound healing, and neoplastic transformation. The ectodomain of the integrin α v β 3 crystallizes in a bent, genuflexed conformation, which is considered to be inactive (i.e., unable to bind physiologic ligands in solution) unless it is fully extended by activating stimuli. To assess whether the bent integrin can bind physiologic ligands, we collaborated with M.A. Arnaout, Harvard Medical School, Boston, Massachusetts, to generate a stable, soluble complex of the manganese-bound αvβ3 ectodomain with a fragment of fibronectin containing type III domains 7–10 and the EDB domain. Electron microscopy and single-particle image analysis were used to determine the 3-dimensional structure of this complex (Fig. 2). F i g . 2 . The 3-dimensional density map (gray-scale transparency) of the integrin αvβ3 in a complex with fibronectin was determined by using electron microscopy and image analysis. The x-ray structures of the αv and β3 chains have been docked into the electron microscopy density envelope. Additional density (lower right) can accommodate fibronectin domain 10 adjacent to the ligand-binding site as well as domain 9 at the synergy site. The complex is shown adjacent to the white box, which represents the 30-Å-thick hydrophobic part of the cellular membrane across which signals are transmitted. Most αvβ3 particles, whether unliganded or bound to fibronectin, had compact, triangular shapes. A difference map comparing ligand-free and fibronectinbound integrin revealed density that could accommodate the fibronectin type III domain 10 containing arginine– glycine–aspartic acid in proximity to the ligand-binding site of β3, with domain 9 just adjacent to the synergy site binding region of αv. We conclude that the ectodomain of αvβ3 has a bent conformation that can stably bind a physiologic Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. ligand in solution. These results are relevant for understanding how binding of ligands to the extracellular domain leads to conformational changes that transmit signals across the plasma membranes of cells, culminating in changes in gene transcription in the nucleus. N O D AV I R U S E S The nodavirus Flock House virus (FHV) has a genome consisting of 2 strands of RNA that are packaged in an icosahedral capsid formed by 180 protein subunits. FHV is an exceedingly useful system for understanding mechanisms of viral assembly. We used electron cryomicroscopy and image reconstruction to determine the structure of 4 types of FHV particles that differed in RNA and protein content (Fig. 3). 3 . Electron cryomicroscopy and image reconstruction of 4 types of FHV particles that differed in RNA and protein content revealed almost identical capsid shells. Despite differences in the encapsidated RNA (electrophoretic profiles, a–d), a substantial fraction of the packaged nucleic acid, either viral or heterologous, was organized as a dodecahedral cage of duplex RNA. A, Native FHV particles purified from infected Drosophila melanogaster cells contain both RNA1 and RNA2. B, Particles obtained by expression of Fig. the FHV coat protein in Sf21 cells contain primarily cellular RNAs. C, FHV particles assembled in Drosophila cells from coat protein subunits lacking basic residues at the N terminus primarily encapsidate RNA1. D, FHV particles assembled in Sf21 cells from coat protein subunits lacking the N terminus contain primarily cellular RNAs. RNA-capsid interactions were primarily mediated via the N and C termini, which are essential for RNA recognition and particle assembly. A substantial fraction of the packaged nucleic acid, either viral or heterologous, was organized as a dodecahedral cage of duplex RNA. The similarity in RNA tertiary structure suggests CELL BIOLOGY 2005 that RNA folding is independent of sequence and length. Computational modeling indicated that RNA duplex formation involves both short- and long-range interactions. These studies suggest that the capsid protein can exploit the plasticity of the RNA secondary structures, capturing those that are compatible with the geometry of the dodecahedral cage. Further analysis of capsid protein mutants and designed RNA molecules is under way. PUBLICATIONS Adair, B.D., Xiong, J.-P., Maddock, C., Goodman, S.L., Arnaout, M.A., Yeager, M. Three-dimensional EM structure of the ectodomain of integrin αvβ3 in complex with fibronectin. J. Cell Biol. 168:1109, 2005. Becker, C.F.W., Strop, P., Bass, R.B., Hansen, K.C., Locher, K.P., Ren, G., Yeager, M., Rees, D.C., Kochendoerfer, G.G. Conversion of a mechanosensitive channel to a water-soluble form by covalent modification with amphiphiles. J. Mol. Biol. 343:747, 2004. Fleishman, S.J., Unger, V.M., Yeager, M., Ben-Tal, N. A Cα model for the transmembrane α-helices of gap-junction intercellular channels. Mol. Cell 15: 879, 2004. Gollapudi, R.R., Yeager, M., Johnson, A.D. Left ventricular cardiac tamponade in the setting of cor pulmonale and circumferential pericardial effusion: case report and review of the literature. Cardiol. Rev. 13:214, 2005. Neuman, B.W., Adair, B.D., Burns, J.W., Milligan, R.A., Buchmeier, M.J., Yeager, M. Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J. Virol. 79:3822, 2005. Turbedsky, K.T., Pollard, T.D., Yeager, M. Assembly of Acanthamoeba myosin-II minifilaments: model of anti-parallel dimers based on EM and x-ray diffraction of 2D and 3D crystals. J. Mol. Biol. 345:363, 2005. Published by TSRI Press®. © Copyright 2005, The Scripps Research Institute. All rights reserved. 59