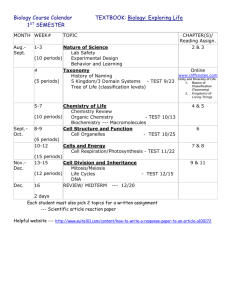
T S I C
advertisement

THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY S c i e n t i f i c R e p o r t 2 0 0 3 O n t h e f r o n t c o v e r : Acetylcholinesterase, a crucial enzyme in the cholinergic signal transduction pathways in muscles and the brain and a well-established therapeutic target. In a novel approach termed in situ click chemistry, the enzyme preferentially assembles 1 pair of the reactants, each of which bears a group that binds to adjacent positions on the protein structure (the choline-binding site of the active center and the peripheral site), into a triazole adduct that is the most potent noncovalent inhibitor of the enzyme yet developed. This approach is more than a novel way to design drugs with exquisite selectivity for the native biological target and hence fewer unwanted side effects; it also provides unique insights into protein flexibility and how enzymes, as dynamic catalysts, truly function as fluctuating, breathing structures and not as rigid templates. This work was carried out in the laboratories of K. Barry Sharpless, Ph.D. Image by Flavio Grynszpan, Ph.D., and Steven Lustig, BioDesign Communications. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY In 1996, The Scripps Research Institute established The Skaggs Institute for Chemical Biology, made possible by a gift of more than $100 million to The Skaggs Institute for Research from Aline W. and L.S. Skaggs. Scientific members of the Skaggs Institute hold dual appointments in various departments at TSRI. These scientists have broad expertise in areas including the structure of biological macromolecules, chemical and antibody catalysis, synthetic and combinatorial chemistry, molecular recognition, and molecular modeling methods. With the achievements of its staff, the Skaggs Institute has assumed its research identity in the United States and throughout the world at the interface of biology and chemistry. 2 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 MEMBERS OF THE SKAGGS INSTITUTE Back row, left to right Seated, Middle row, left to right Front row, left to right Not present Martha J. Fedor, Ph.D. Ernest Beutler, M.D. Elizabeth D. Getzoff, Ph.D. Geoffrey Chang, Ph.D. Ian A. Wilson, D.Phil. Peter E. Wright, Ph.D. Dale L. Boger, Ph.D. Ehud Keinan, Ph.D. Jeffery W. Kelly, Ph.D. Kim D. Janda, Ph.D. Erik Sorensen, Ph.D.* Richard A. Lerner, M.D. Carlos F. Barbas III, Ph.D. Gerald M. Edelman, M.D., Ph.D. Benjamin F. Cravatt, Ph.D. Peter Schultz, Ph.D. Stephen P. Mayfield, Ph.D. Gerald F. Joyce, M.D., Ph.D. K. Barry Sharpless, Ph.D. Paul R. Schimmel, Ph.D. Julius Rebek, Jr., Ph.D. Chi-Huey Wong, Ph.D. Subhash C. Sinha, Ph.D. K.C. Nicolaou, Ph.D. M. Reza Ghadiri, Ph.D. Kurt Wüthrich, Ph.D. James R. Williamson, Ph.D. Albert Eschenmoser, Ph.D. John A. Tainer, Ph.D. Philip Dawson, Ph.D. M.G. Finn, Ph.D. * Appointment completed THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 With a contribution from the Skaggs family, TSRI established the Skaggs Clinical Scholars Program in an effort to more closely integrate clinical and basic research within the Scripps organization. The program identifies research-oriented clinicians and funds meritorious collaborative research projects between the clinical scholar and appropriate TSRI scientists. A broader goal is to expand the body of knowledge related to human disease and to develop effective therapeutic interventions. THE SKAGGS SCHOLARS Standing, Back row, left to right Seated, Front row, left to right Not present Huan B. Giap, M.D. Alan Saven, M.D. Faith H. Barnett, M.D., Ph.D. George E. Dailey, M.D. James R. Mason, M.D. John F. Bastian, M.D. Daniel A. Nachtsheim, M.D. Prabhakar Tripuraneni, M.D. Sheila F. Friedlander, M.D. Donald D. Stevenson, M.D. Michael P. Kosty, M.D. Merlin L. Hamer, M.D. Robert J. Russo, M.D., Ph.D. Katharine M. Woessner, M.D. Mary A. Kalafut, M.D. Darlene J. Elias, M.D. Williamson B. Strum, M.D. David A. Mathison, M.D. Jack C. Sipe, M.D. Richard A. Smith, M.D. Jorge J. Nieva, M.D. Ken D. Pischel, M.D. Clifford W. Colwell, Jr., M.D. Paul S. Phillips, M.D. Steven J. Poceta, M.D. Paul J. Pockros, M.D. Ronald A. Simon, M.D. 3 VOLUME 8 PRESIDENT’S INTRODUCTION 7 DIRECTOR’S OVERVIEW 9 I N T R A C E L L U L A R R N A A S S E M B LY A N D C ATA LY S I S 24 M.J. Fedor, K.F. Baban, C.P. Da Costa, Y.I. Kuzmin, E.M. Mahen, S.B. Voytek, R.S. Yadava S TA F F 11 VIRUSES AS MOLECULAR BUILDING BLOCKS I N V E S T I G AT O R S ’ R E P O R T S 25 M.G. Finn, D.D. Díaz, W.G. Lewis, J.-C. Meng, S. Meunier, C A N C E R , A I D S , C ATA LY S I S , A N D T H E R E G U L AT I O N D. Prasuhn, S. Punna, K.S. Raja, Q. Wang OF GENES: INVENTING MOLECULES WITH DEFINED FUNCTIONS 13 C.F. Barbas III, R. Lerner, P. Blancafort, N.S. Chowdari, R. Fuller, T. Gräslund, N. Jendreyko, C. Lund, L. Magnenat, N. Mase, M. Popkov, C. Rader, D.B. Ramachary, S. Sinha, H. Shim, F. Tanaka, R. Thayumanavan, J. Turner T R A I N I N G I N M O L E C U L A R A N D E X P E R I M E N TA L MEDICINE INSIGHTS INTO PROTEIN CHEMISTRY AND BIOLOGY FROM PROTEIN STRUCTURE 27 E.D. Getzoff, M. Aoyagi, A.S. Arvai, D.P. Barondeau, R.M. Brudler, J.M. Castagnetto, D. Cerutti, T. Cross, M. DiDonato, E.D. Garcin, U.K. Genick, S.W. Hennessy, C. Hitomi, K. Hitomi, C.J. Kassmann, I. Li, S.J. Lloyd, M.E. Pique, R.J. Rosenfeld, M.E. Stroupe, M.M. Thayer, M.J. Thompson, J.L. Tubbs, T.I. Wood 16 SUPRAMOLECULAR APPROACHES TO DRUG DESIGN, E. Beutler CELL DIAGNOSTICS, AND COMPLEX SYSTEM ENGINEERING SYNTHETIC, MEDICINAL, AND BIOORGANIC CHEMISTRY 18 D.L. Boger, Y. Ambroise, B. Blagg, S. Buck, K. Capps, H. Cheng, Y. Choi, B. Crowley, P. Desai, J. Desharnais, R. Dominique, W. Du, R. Ducray, G. Elliott, R. Fecik, Y. Ham, N. Haq, C. Hardouin, I. Hwang, S. Ichikawa, D. Kastrinsky, G. Kim, P. Krenitsky, M. Lall, C. McComas, Y. Mori, J. Parrish, S. Pfeiffer, A. Pichota, Y. Rew, D. Shin, D. Soenen, J. Trzupek, W. Tse, J. Wanner, G. Wilkie, S. Wolkenberg, Z. Yuan, B. Yeung, J. Zimpleman STRUCTURAL BIOLOGY OF MEMBRANE-BOUND TRANSPORTERS AND RECEPTORS 20 A. Ma, T. Nguyen, P. Szewczyk, Y. Yin, C. Reyes, S. Wada, O. Pornillos, M. Grant, G. Chang 29 M.R. Ghadiri, M. Al-Sayah, G. Ashkenasy, A. Chavochi, J. Fletcher, V. Haridas, W.S. Horne, R. Jagasia, L. Leman, S. Rahimipour, M. Yadav N E W V I S TA S I N I M M U N O P H A R M A C O T H E R A P Y 30 K.D. Janda, J.-M. Ahn, J. Ashley, G. Boldt, R. Carrera, Y. Chen, B. Clapham, A. Coyle, T. Dickerson, Y. Ding, L. Eubanks, T. Fujimori, R. Galve, C. Gambs, C. Gao, T. Hoffman, L. Hom, G. Kaufmann, Y.-S. Kim, E. Laxman, B.-S. Lee, S.-H. Lee, S.-J. Lee, M. Lillo, S. Mahajan, H. Matsushita, M. Matsushita, J. McDunn, G. McElhaney, J. Mee, M. Meijler, J. Moss, J.-I. Park, N. Reed, A. Shafton, C. Sun, R. Troseth, A.D. Wentworth, P. Wentworth, Jr., P. Wirsching, Y. Xu, N. Yamamoto, K. Yoshida, B. Zhou C ATA LY T I C N U C L E I C A C I D S F O R T R E AT I N G T H E CHEMICAL PHYSIOLOGY 20 MOLECULAR BASIS OF DISEASE B.F. Cravatt, G.C. Adam, K.T. Barglow, M.H. Bracey, K. Chiang, M. Evans, G. Hawkin, M. Humphrey, N. Jessani, A. Joseph, D. Leung, K. Masuda, M. McKinney, A. Saghatelian, A. Speers 33 G.F. Joyce, J.T. Hillman, R.M. Kumar, N. Paul, W.M. Shih B I O M O L E C U L A R C O M P U T I N G , C ATA LY T I C SYNTHETIC PROTEIN CHEMISTRY 21 P. Dawson, R. Balambika, J. Blankenship, M. Churchill, J. Offer, C. Neidre, F. Topert SYNTHETIC ENZYMES 22 M E TA B O L I T E - I N I T I AT E D P R O T E I N M I S F O L D I N G G.M. Edelman, S. Aschrafi, A. Atkins, S. Chappell, G.W. Rogers, F. Smart, T. Stevens, M. Tsatmali, W. Zhou 23 A. Eschenmoser, R. Krishnamurthy, Z. Wang, G.R. Vavilala, C. Mang, V. Rajwanshi, J. Nandy, T. George Published by TSRI Press ®. All rights reserved. © Copyright 2003, The Scripps Research Institute. 36 J.W. Kelly, S. Deechongkit, M.A. Dendle, T. Foss, D. Fowler, K. Frankenfeld, N. Green, A. Hurshman, M.B. Huff, S. Johnson, H.-J. Lim, E. Powers, A. Sawkar, Y. Sekijima, S. Werner, L. Wiseman, S.-L. You, Q. Zhang CHEMICAL ETIOLOGY OF THE STRUCTURE OF NUCLEIC ACIDS 34 E. Keinan, H.C. Lo, H. Han, S. Sasmal, M. Arifuddin, A. Brik, S. Saphier, G. Sklute, N. Metanis, M. Soreni, D. Vebenov, L. Kosoy, M.K. Sinha, A. Alt, I. Ben-Shir, R. Piran, P.E. Dawson F U N D A M E N TA L P R O C E S S E S I N N E U R A L DEVELOPMENT ANTIBODIES, ORGANIC SYNTHESIS, AND THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 EXPRESSION OF THERAPEUTIC PROTEINS IN EUKARYOTIC ALGAE MACROMOLECULAR MACHINES AS MASTER KEYS 38 S.P. Mayfield, E. Brown, M. Beligni, A. Manuell, K. Yamaguchi, S.E. Franklin I N G E N O M E I N T E G R I T Y, T H E C E L L C Y C L E , PCONTROL OF REACTIVE OXYGEN SPECIES, A N D PAT H O G E N E S I S 48 J.A. Tainer, A.S. Arvai, D.P. Barondeau, M. Bjoras, R. Brudler, CHEMICAL SYNTHESIS AND CHEMICAL BIOLOGY R.M. Cardoso, J.M. Castagnetto, B.R. Chapados, C. Chahwan, L. Craig, 38 K.C. Nicolaou, R. Baati, T. Bando, M. Bella, F. Bernal, W. Brenzovich, S. Bulat, D. Chen, J. Chen, E. Couladouros, P. Dagneau, P. Diamandis, A. Estrada, M. Follmann, T. Francis, M. Frederick, L. Gomez Paloma, M. Govindasamy, D. Gray, P. Guntupalli, S. Harrison, J. Hao, X. Huang, R. Hughes, M. Jennings, F. Kaiser, D. Kim, T. Koftis, S. Lee, Y. Li, T. Ling, D. Longbottom, F. Marr, C. Mathison, T. Montagnon, M. Nevalainen, A. Nalbandian, R. Noronha, . Paraselli, B. Pratt, W. Qian, G. Rassias, M. Reddy, M. Rodriguez, A. Roecker, B. Safina, P. Sasmal, D. Seigel, S. Snyder, X. Sun, G. Vasilikogiannakis, S. Vyskocil, H. Xu, Y. Yamada, M. Zak D.S. Daniels, M. DiDonato, G. Divita, L. Fan, S. Han, C. Hitomi, K. Hitomi, J.L. Huffman, K.-P. Hopfner, D. Hosfield, C.D. Mol, G. Moncalian, S.S. Parikh, C.D. Putnam, D.S. Shin, M.E. Stroupe, O. Sundheim, T.I. Wood, A. Yamagata TA R G E T I N G H I V R N A 50 J.R. Williamson, A. Bunner, J. Chao, S. Edgcomb, P.M. Funke, M. Hennig, E. Johnson, K.A. Lehmann, P.K. Radha, M. Recht, S.P. Ryder, L.G. Scott, E. Sperling, M.W. Trevathan C R Y S TA L L O G R A P H I C S T U D I E S O F I M M U N E M O L E C U L A R R E C O G N I T I O N A N D E N C A P S U L AT I O N 40 J. Rebek, Jr., T. Amaya, P. Ballester,* S. Biros, W.-D. Cho, T. Dale, J. Friese, A. Gissot, S. Gu, F. Hof, D.W. Johnson, L. Kröck, Y. Kim, H. Onagi, L. Palmer, B. Purse, D. Rachavi-Robinson, A. Scarso, A. Shivanyuk, L. Trembleau, E. Ullrich, M. Yamanaka 42 K. Ewalt, J. Liu, M. Lovato, D. Metzgar, C. Myers, L. Nangle, B. Nordin, F. Otero, J. Reader, L. Ribas de Pouplana, B. Slike, M. Swairjo, K. Tamura, W. Waas, X. Yang CLICK CHEMISTRY AND THE SEARCH FOR 45 K.B. Sharpless, Y. Baba, S. Boral, J. Cappiello, T. Chan, A. Converso, M. David, A. Feldman, V. Fokin, R. Hilgraf, P. Holzer, C. Kappe, T. Kitayama, A. Krasinski, I. Lewis, V. Litosh, R. Manetsch, A. McPherson, J. Muldoon, S. Narayan, V. Rostovtsev, S. Silverman, N. Stuhr-Hansen, E. Van der Eycken, P. Wu, Y. Xie A N T I B O D Y C ATA LY S I S , O R G A N I C S Y N T H E S I S , A N D P R O D R U G A N D TA R G E T I N G T H E R A P I E S S.C. Sinha, R.A. Lerner, L.-S. Li, S. Das, S. Dutta 55 44 P.G. Schultz, J.C. Anderson, M. Bose, C. Cho, J. Gildersleeve, J. Hong, Q. Huang, J. Liu, K.H. Min, M. Mukherji, S. Santoro, F. Tian, L. Wang, P. Yang, J. Yin, C. Yu, Z. Zhang, L.X. Zheng BIOLOGICAL ACTIVITY DRUG DISCOVERY C.-H. Wong, F. Agnelli, C. Behrens, M. Best, A. Brik, M.C. Bryan, A. Buchnyskyy, G. Cain, A. Chang, W.C. Cheng, A. Datta, S. Duron, F. Fazio, D. Franke, J.I. Furukawa, M.C. Galan, S. Hanson, Z.-Y. Hong, J. Hsu, C.-Y. Huang, H.K. Lee, L. Lee, F.-S. Liang, D. Lin, H. Liu, J. Liu, M. Numa, P. Nyffeler, T. Polat, D. Thayer, S.K. Wang, G. Watt, D. Wu, Y.Y. Yang, N. Yu, G.-W. Xing, R. Xu P. Schimmel, J. Bacher, A. Bates, K. Beebe, V. de Crécy-Lagard, EXPANDING THE GENETIC CODE 51 I.A. Wilson, D.A. Calarese, R.M.F. Cardoso, C.-G. Cheong, A.L. Corper, M.D.M. Crispin, M.-A. Elsliger, S. Ferguson, A. Gakhal, P.A. Horton, S. Ito, N.A. Larsen, J.G. Luz, E. Ollmann Saphire, J.B. Reiser, R. Sanders, D.A. Shore, R.L. Stanfield, R.S. Stefanko, J. Stevens, P. Verdino, D.W. Wolan, D. Wu, L. Xu, D.M. Zajonc, Y. Zhang, X. Zhu BIOORGANIC CHEMISTRY AND POTENTIAL MEDICAL CONSEQUENCES OF AMBIGUITY IN THE GENETIC CODE R E C O G N I T I O N A N D T H E R A P E U T I C TA R G E T S 46 STUDIES OF MACROMOLECULAR RECOGNITION B Y M U LT I D I M E N S I O N A L N U C L E A R M A G N E T I C RESONANCE 55 P.E. Wright, H.J. Dyson, M. Martinez-Yamout, J. Covalt, R. De Guzman, T. Dunzendorfer-Matt, N. Goto, B. Hudson, J. Lansing, T. Nishikawa, G. Perez-Alvarado, J. Wojciak, T. Zor, J. Hosea, M. Landes S TA F F AWA R D S A N D A C T I V I T E S 58 SUBJECT INDEX 60 5 6 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 THE SKAGGS INSTITUTE THE SCRIPPS THE SCRIPPS FOR RESEARCH RESEARCH INSTITUTE RESEARCH INSTITUTE BOARD OF TRUSTEES BOARD OF TRUSTEES OFFICERS Susie Balukoff The Hon. Alice D. Sullivan (Ret.) Chair Paul L. Herrling, Ph.D. Vincent E. Benstead Thomas H. Insley Mrs. William McCormick Blair, Jr. Janet R. Kellogg Ernest Beutler, M.D. The Most Reverend Robert H. Brom Hubert T. Greenway, Jr., M.D. Lawrence C. Horowitz, M.D. Arnold LaGuardia Herbert Boyer, Ph.D. Richard A. Lerner, M.D. Richard A. Lerner, M.D. Thomas E.K. Cerruti, Esq. Claudia S. Luttrell Gary N. Coburn Don L. Skaggs Gerald Cohn Frank Lowy AC Claudia S. Luttrell Mark S. Skaggs Thomas E.K. Cerruti Richard A. Lerner, M.D. President James R. Mellor George H. Conrades John J. Moores Rod Dammeyer John G. Davies, Esq. The Hon. Paul G. Rogers, Esq. Thomas E. Dewey, Jr. John Safer John D. Diekman, Ph.D. The Hon. Lynn Schenk Charles C. Edwards, M.D. John R. Seffrin, Ph.D. Richard J. Elkus, Jr. Ralph J. Shapiro Jane E. Henney, M.D. David Tappan Charles C. Edwards, M.D. Arnold LaGuardia Chris D. Van Gorder Arnold LaGuardia Executive Vice President and Treasurer Douglas Bingham Executive Vice President and General Counsel and Secretary Donna Weston Senior Vice President and Chief Financial Officer THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Richard A. Lerner, M.D. President’s Introduction s is the norm, members of The Skaggs Institute for Chemical Biology at The Scripps Research Institute this year contributed a prodigious volume of work to the body of scientific knowledge in a broad range of disciplines, work that changes the way we think about biological mechanisms and the course of human disease. The following merely skims the surface of the knowledge they created and the importance of their discoveries. A RESEARCH ACTIVITIES Work in the laboratory of Peter Schultz effectively removed a billion-year constraint on the ability to manipulate the structure and function of proteins. Dr. Schultz and his research group completed the synthesis of a form of the bacterium Escherichia coli with a genetic code that uses 21 basic amino acid building blocks to synthesize proteins, instead of the 20 that occur naturally. This creation was the first one of an autonomous organism that uses 21 amino acids and has the metabolic machinery to build those amino acids. Further, the group introduced revolutionary changes into the genetic code of organisms such as yeast that allow the mass production of proteins with unnatural amino acids. By so doing, Dr. Schultz and his team set the stage for an entirely new approach to applying the same technology to other eukaryotic cells, and even to multicellular organisms. Simply stated, these scientists have opened up the whole pathway to higher organisms. Researchers in the laboratory of Stephen Mayfield and in my laboratory used algae to express an antibody that targets herpesvirus. The usefulness of the antibody lies not only in the potential production of an antiherpes topical cream or treatment but also in the development of technology that could facilitate the production of multiple human antibodies and other proteins on a massive scale. This technology enables the generation of antibodies, soluble receptors, and other proteins so much more cheaply than previous technology that an entire new class of therapeutic agents may become available. Jeffery Kelly and his colleagues discovered a new approach for treating amyloid diseases, particularly transthyretin amyloid diseases, which are similar to Parkinson’s and Alzheimer’s diseases. Amyloid diseases are caused by misfolding of proteins into a structure that leads the proteins to cluster, forming microscopic fibrillar plaques that are deposited in internal organs and interfere with normal function. Dr. Kelly and his team showed the efficacy of using small molecules to stabilize the normal folding of transthyretin, preventing this protein from misfolding. By so doing, they were able to inhibit the formation of fibrils by a mechanism that can ameliorate disease. In what was a first for biology, researchers in my laboratory reported that the human body makes ozone. Ozone appears to be produced in a process involving human immune cells known as neutrophils and human antibodies. The presence of ozone in the body may be linked to inflammation, and the research may have important ramifications for treating inflammatory diseases. In addition to killing bacteria, the neutrophils feed singlet oxygen to the antibodies, which convert it into ozone. Carlos F. Barbas and his group designed a hybrid anticancer compound that combines the efficacy of a cancer cell–targeting agent with the long-lasting dose of an antibody. This potent combination has a profound effect on the size of tumors in animal models; in preclinical studies, the compound shrank both Kaposi sarcomas and colon cancers. The approach is general enough to be used to design hybrids against numerous different cancers; a single antibody can be mixed with multiple small molecules, resulting in a multiplicity of therapeutic agents. 7 8 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Scientists led by Kim D. Janda designed a new way to make a vaccine against nicotine that could become a valuable tool for treating nicotine addiction by helping the body clear the drug from the bloodstream. The vaccine, which eventually would be given to patients in smoking cessation programs, greatly suppresses the reinforcing aspect of nicotine. The researchers used an “immunopharmacotherapy” approach, designing a drug that stimulates the immune system to clear the nicotine from the body. In a related study, Dr. Janda and his group discovered that a chemical called nornicotine, a major metabolite of nicotine, modifies proteins that misfold and form the fibril plaques found in abundance in the brains of patients who have Alzheimer’s disease. Simply stated, this process physically inhibits the formation of the fibrils. The research is promising—not because nornicotine most likely would be an effective therapeutic agent, but because it shows how a single molecule can cause a chemical interaction that may alter a mechanism important in Alzheimer’s disease. This research could lead to the development of small molecules similar to nornicotine that are not toxic but that could interact in a similar fashion, preventing the aggregation of amyloid-β protein and perhaps Alzheimer’s disease. A group of scientists including John Tainer of the Skaggs Institute and Lisa Craig, Mark Yeager, and Mike Pique at The Scripps Research Institute solved 2 key structures of a bacterial protein called pilin, which is required for infection by pathogens that cause diseases such as meningitis, gonorrhea, pneumonia, and cholera. The members of the group think that the research provides essential knowledge to help scientists develop novel antibiotics and vaccines against these sometimes deadly bacterial diseases. Because the structures are too large and flexible to be solved by using the traditional techniques of structural biology, the team used both x-ray crystallography and electron microscopy to build a model of the pili that would have otherwise been impossible at that level of molecular detail. In another structural achievement, a multi-institutional group of researchers led by Ian Wilson of the Skaggs Institute and Dennis Burton of The Scripps Research Institute solved the structure of an antibody that effectively neutralizes HIV, an important step toward the goal of designing an effective vaccine against HIV and a new means by which scientists may design antibodies in general. The structure of the antibody had never been seen before, prompting the scientists to speculate on whether they can use this knowledge to engineer antibodies with higher affinity against other antigens. FA C U LT Y H O N O R S A N D AWA R D S Many of our scientists, at various stages of their careers, were honored by their peers this past year with awards for achievement in numerous areas of scientific endeavor. Members of the Skaggs Institute who received recognition include Ernest Beutler, who was awarded the E. Donnall Thomas Lecture and Prize of the American Society of Hematology, and Ben Cravatt, who won the Eli Lilly Award in Biological Chemistry from the American Chemical Society. A L O O K AT T H E F U T U R E The Skaggs Institute for Chemical Biology is an organization that exceeds expectations on multiple levels; its faculty and staff remain at the leading edge of science in an era in which the pace of discovery accelerates on a continual basis. Its new developments and discoveries, all of which were made possible by the vision and remarkable generosity of the Skaggs family and The Skaggs Institute for Research, will provide greater impetus for the Skaggs Institute for Chemical Biology to play an even larger role on the international stage of scientific discovery. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Julius Rebek, Jr., Ph.D. Director’s Overview he Skaggs Institute for Chemical Biology was established in 1996, and as it enters its 8th year, we congratulate our benefactor, L. Sam Skaggs, on his 80th birthday. His generosity, and that of the entire Skaggs family, supports the work of the Institute through its 31 principal investigators, 95 postdoctoral fellows, and some 50 graduate students. The aim of the Institute is to relieve human suffering by moving basic research to applications that lead to cures for diseases. Our researchers authored nearly 300 publications during the past year, loosely clustered in the areas of organic synthesis, antibody science, nucleic acid chemistry, and protein structure. The details of the scientific discoveries appear in the individual reports of the principal investigators, but I present a few of the highlights here. The faculty members involved with nucleic acids, Albert Eschenmoser, Martha Fedor, Gerald Joyce, Ehud Keinan, Peter Schultz, and Jamie Williamson, showed that DNA isn’t just for genes anymore. DNA can fold into a spectacular octahedral shape, visible by electron microscopy; it can do biomolecular computing; and its sequences can be selected and amplified to cat- T alyze the cleavage of RNA. The use of RNA as a target for small-molecule medicinal agents is also growing. Synthetic alterations in either the base pairs or the sugar backbones indicate that nucleic acid structure is enormously versatile and that the information encoded can even be expanded. In the past 50 years, the seed represented by the discovery of the double helix has grown into an orchard. The synthetic team, K.C. Nicolaou, Phil Baran, Dale Boger, Barry Sharpless, and Chi-Huey Wong, created purely synthetic molecules to regulate protein-protein interactions. These interactions emerged as targets for intervention in diseases but have not, could not, be addressed by conventional medicinal chemistry. Specific agents for blocking angiogenesis and stimulating production of blood cells were synthesized, and small molecules that arrest microtubular structures are now in clinical trials. Both plant- and marine-derived natural products were used to produce antitumor agents, and efforts to discover carbohydrate-based drugs led to the development of new HIV protease inhibitors. As a result of methods developed here last year, environmentally benign reagents are now available as building blocks for combinatorial medicinal chemistry. Research led by Richard Lerner indicated that ozone is one of the key reactive species that antibodies can generate to destroy antigens. Steve Mayfield developed methods for large-scale, inexpensive production of antibodies in algae—good news at a time when antibodies are a sizable fraction of the new drugs approved by the Food and Drug Administration. Using x-ray crystallography, Ian Wilson and collaborators determined the structures of antibodies responsible for resistance to HIV; these antibodies, specifically, and structural biology, generally, will take greater roles in the ongoing efforts to develop an AIDS vaccine. Peter Wright used nuclear magnetic resonance to solve protein structures as they exist in solution. Several of these proteins are transcription factors, and their interactions with other proteins are responsible for specific gene expression of tumor viruses. Elsewhere, structures of enzymes that produce nitric oxide, a ubiquitous cellsignaling molecule, were determined, and the behavior of superoxide dismutase, an enzyme involved in amyotrophic lateral sclerosis, was analyzed. The structural details offer a starting point for the development phase of small-molecule medicinal agents. We are proud of these discoveries, for which a number of the Skaggs investigators received national and 9 10 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 international awards, including the Paul Ehrlich Prize and, in the past 2 years, a share of the Nobel Prize in Chemistry. The Skaggs family has enabled the recruitment of more than 15 principal investigators during the Institute’s short lifetime. These new faculty members have propelled our graduate programs to the top in national surveys. At the international level, recognition came through an unprecedented joint Ph.D. program with Oxford University in England. Graduate students in the program will receive Skaggs Scholarships. All of us at Scripps are grateful for the continued support of the Skaggs Institute for Research. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY S TA F F Julius Rebek, Jr., Ph.D.* Director and Professor Carlos F. Barbas III, Ph.D.** Professor Janet and W. Keith Kellogg II Chair in Molecular Biology Ernest Beutler, M.D.*** Professor Chairman, Department of Molecular and Experimental Medicine, TSRI Dale L. Boger, Ph.D.* Richard and Alice Cramer Professor of Chemistry Geoffrey Chang, Ph.D.** Assistant Professor Benjamin F. Cravatt, Ph.D.**** Associate Professor Philip Dawson, Ph.D.***** Assistant Professor Gerald M. Edelman, M.D., Ph.D. † Professor Chairman, Department of Neurobiology, TSRI Albert Eschenmoser, Ph.D.* Professor Martha J. Fedor, Ph.D.** Associate Professor M.G. Finn, Ph.D.* Associate Professor Elizabeth D. Getzoff, Ph.D.†† Professor M. Reza Ghadiri, Ph.D.* Professor Kim D. Janda, Ph.D.* Professor Ely R. Callaway, Jr., Chair in Chemistry Gerald F. Joyce, M.D., Ph.D. ††† Professor Ehud Keinan, Ph.D.** Adjunct Professor Jeffery W. Kelly, Ph.D.* Vice President, Academic Affairs, TSRI Dean, Graduate Program, TSRI Lita Annenberg Hazen Professor of Chemistry Hartmuth Kolb* Associate Professor Richard A. Lerner, M.D.††† President, TSRI Lita Annenberg Hazen Professor of Immunochemistry Cecil H. and Ida M. Green Chair in Chemistry Stephen P. Mayfield, Ph.D.***** Associate Professor Associate Dean, Graduate Program, TSRI K.C. Nicolaou, Ph.D.* Aline W. and L.S. Skaggs Professor of Chemical Biology Darlene Shiley Chair in Chemistry Chairman, Department of Chemistry, TSRI John A. Tainer, Ph.D.** Professor James R. Williamson, Ph.D.** Professor Associate Dean, Graduate Program, TSRI Ian A. Wilson, D.Phil.** Professor Chi-Huey Wong, Ph.D.* Ernest W. Hahn Professor and Chair in Chemistry Peter E. Wright, Ph.D.** Professor Cecil H. and Ida M. Green Investigator in Biomedical Research Chairman, Department of Molecular Biology, TSRI Kurt Wüthrich, Ph.D. ††† Cecil H. and Ida M. Green Visiting Professor of Structural Biology Eidgenössische Technische Hochschule Zürich Zürich, Switzerland RESEARCH A S S O C I A T E S ††††† Toru Amaya, Ph.D. Wook Dong Cho, Ph.D. Paul R. Schimmel, Ph.D. ††† Ernest and Jean Hahn Professor and Chair of Molecular Biology andChemistry Jan Friese, Ph.D. Peter Schultz, Ph.D.* Professor Scripps Family Chair Yoonkyung Kim, Ph.D. Arnaud Gissot, Ph.D. Shen Gu, Ph.D. Darren W. Johnson, Ph.D. Hideki Onagi, Ph.D. Alessandro Scarso, Ph.D. K. Barry Sharpless, Ph.D.* W.M. Keck Professor of Chemistry Subhash C. Sinha, Ph.D.** Associate Professor Erik J. Sorensen, Ph.D. †††† Assistant Professor Alexander Shivanyuk, Ph.D. Laurent Trembleau, Ph.D. Elke Ullrich, Ph.D. Masamichi Yamanaka, Ph.D. 11 * Joint appointment in the Department of Chemistry ** Joint appointment in the Department of Molecular Biology *** Joint appointment in the Department of Molecular and Experimental Medicine **** Joint appointments in the Departments of Cell Biology and Chemistry ***** Joint appointment in the Department of Cell Biology † †† ††† †††† ††††† Joint appointment in the Department of Neurobiology Joint appointments in the Departments of Molecular Biology and Immunology Joint appointments in the Departments of Chemistry and Molecular Biology Appointment completed Research associates in the laboratories of staff other than Dr. Rebek are included in the lists of the respective departments in which the associates hold joint appointments. 12 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Shannon Biros, Graduate Student in Chemistry, The Skaggs Institute for Chemical Biology THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 INVESTIGATORS ’ R EPORTS Cancer, AIDS, Catalysis, and the Regulation of Genes: Inventing Molecules With Defined Functions C.F. Barbas III, R. Lerner, P. Blancafort, N.S. Chowdari, R. Fuller, T. Gräslund, N. Jendreyko, C. Lund, L. Magnenat, N. Mase, M. Popkov, C. Rader, D.B. Ramachary, S. Sinha, H. Shim, F. Tanaka, R. Thayumanavan, J. Turner e are concerned with problems at the interface of molecular biology, chemistry, and medicine. Many of our studies involve learning or improving on Nature’s strategies to prepare novel molecules that perform specific functional tasks, such as regulating a gene, destroying cancer, or catalyzing a reaction with small molecules in an enzymelike manner. We hope to apply these novel insights, methods, and products to provide solutions to human diseases including cancer, HIV disease, and genetic diseases. W C ATA LY T I C A N T I B O D I E S We are extending and refining approaches to catalytic antibodies by using novel recombinant strategies coupled with reactive immunization, chemical-event selections, and the design of unique multiturnover selection chemistries. We are developing in vitro selection and evolutionary strategies as routes for obtaining antibodies of defined biological and chemical activity. These strategies involve the directed evolution of human, rodent, and synthetic antibodies. Essentially, we are evolving proteins to function as efficient catalysts, a task performed naturally over eons, and one that we aim to complete in weeks. The approach is a blend of chemistry, enzymology, and molecular biology. A major focus of our work is the development of strategies to produce antibodies that efficiently form and break carbon-carbon bonds. Much of this work centers on the chemistry of imines and enamines and the development of antibodies that use covalent catalysis. We are examining the aldol, Michael, and Diels-Alder reactions and related reactions such as the Mannich reaction. Many of these catalysts may someday be important in the synthesis of enantiomerically pure drugs. Using novel catalytic antibodies, we showed the efficient asymmetric synthesis and resolution of a variety of molecules, including tertiary and fluorinated aldols, carbo- 13 hydrates, and insect pheromones. In our collaboration with S.C. Sinha, the Skaggs Institute, large-scale antibody-catalyzed reactions have been key to the synthesis of epothilone anticancer drugs. In a new approach to catalytic antibodies, we combined transition-state theory with reactive immunization and produced antibodies with catalytic proficiencies of almost 1014. More recently, we focused on learning how we can use a laboratory-based evolutionary strategy to improve these catalysts even further. A practical goal of this study is the creation of human catalytic antibodies that can be applied therapeutically in the treatment of metabolic diseases, cancer and HIV disease. We showed that we can selectively release insulin from an inactive or captured state in animal models. This result suggests that our catalytic antibody approach can be used in vivo to prepare and selectively deliver proteins and peptides. O R G A N O C ATA LY S I S : E N Z Y M E L I K E C H E M I S T R Y WITH SMALL ORGANIC MOLECULES In studying how proteins catalyze reactions, we often examine how the constituent components react. These studies led to a new green approach to catalytic asymmetric synthesis that can be applied to the synthesis of drugs and druglike molecules. Using insights garnered from our studies of aldolase antibodies, we prepared simple chiral amino acids and amines to catalyze aldol and related imine and enamine chemical actions such as Michael and Mannich reactions. We also studied small amine-bearing peptides that are catalytic. Although aldolase antibodies are superior catalysts, simple chiral amino acids and amines are enabling us to measure the importance of pocket sequestration in catalysis. In addition, we achieved direct asymmetric catalytic aldol reactions by using aldehydes and unmodified ketones together with commercially available chiral cyclic secondary amines as catalysts. This approach can also be applied to Diels-Alder reactions wherein dienes are catalytically generated in situ. We extended the scope of catalytic asymmetric reactions that form carboncarbon bonds to embrace the use of aldehyde donors. We produced catalytic asymmetric aldol, Michael, and Mannich reactions, the first examples of their kind, that use aldehyde donors to create highly desirable aldehydebearing synthons. Using this method, we directly synthesized a wide variety of α- and β-amino acids, carbohydrates, and lactams. Stereochemically complex molecules can now be assembled by using small mol- 14 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 ecules in a manner analogous to that of natural enzymes (Fig. 1). range of drugs to dramatically increase their efficacy. In cancer studies, we showed that we can use this approach to turn weakly active anticancer drugs into potent anticancer therapeutic agents (Fig. 2). F i g . 2 . Chemically programmed antibodies. A, Many small mol- F i g . 1 . Enzymelike asymmetric assembly of simple molecules can be used to create a wide variety of chiral synthons. TA R G E T I N G C A N C E R In targeting cancer, we take a multidisciplinary approach that involves gene regulation, catalytic antibodies, drug design, and combinatorial antibody libraries. Using a combinatorial antibody strategy, we prepared a high-affinity human antibody to an integrin. Our goal is to use the antibody in dual antitumor-antivascular therapy in cancer. Our most recent studies involved Kaposi sarcoma, the most common AIDS-associated malignant neoplasm. We showed that a laboratoryevolved antibody can substantially inhibit growth of Kaposi tumors in mice. Using catalytic antibodies, we are developing a strategy to activate drugs in a highly specific fashion at the site of cancer. Typically, the use of highly potent chemotherapeutic agents is limited by nonspecific toxic effects associated with the inability to direct these agents to the appropriate targets. We showed that a wide variety of clinically relevant anticancer drugs such as doxorubicin, camptothecin, and etoposide can be modified to less toxic prodrugs that can be specifically activated by catalytic antibody 38C2 to kill cancer cells in vivo. This past year, we invented a new approach to therapeutic antibodies in which small molecules are used to chemically program the specificity of a generic catalytic antibody. This approach can be applied to a wide ecules can be endowed with the unique pharmacokinetic properties of antibodies as well as the ability to fix complement and direct cellular cytotoxic reactions. Attachment of a β-diketone functional group allows the targeting molecule to be attached to a aldolase catalytic antibody. B, An integrin-targeting molecule. When this molecule was attached to the catalytic antibody, the combination had potent antiangiogenic and antitumor properties. CONTROLLING GENES AND EXPLORING GENOMES In all organisms, from the simplest to the most complex, proteins that bind nucleic acids control the expression of genes. The nucleic acids DNA and RNA are the molecules that store the recipes of all life forms. The fertilized egg of a human contains the genetic recipe for the development and differentiation of a single cell into 2 cells, 4 cells, and so on, finally yielding a complete individual. The coordinated expression or reading of the recipes for life allows cells containing the same genetic information to perform different functions and to have distinctly different physical characteristics. Proteins that bind nucleic acids enable this coordinated control of the genetic code; lack of coordination due to genetic defects or to viral seizure of control of the cell results in disease. In one project, we are developing methods to produce proteins that bind to specific DNA sequences to control specified genes. As we showed earlier, these proteins can be used as specific genetic switches to turn on or turn off genes on demand, creating an operating system for genomes. To this end, we selected and designed specific zinc finger domains that will constitute an alphabet of 64 domains that will allow any THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 DNA sequence to be bound selectively. The prospects for this “second genetic code” are fascinating and should have a major impact on basic and applied biology. We showed that these domains are functionally modular and can be recombined with one another to create polydactyl proteins capable of binding 18-bp sequences with subnanomolar affinity. Our goal is to develop a new class of therapeutic proteins that inhibit or enhance the synthesis of proteins, providing a new strategy for fighting diseases of either somatic or viral origin. Recently, we created gene switches that can be controlled by small molecules and showed the effectiveness of our zinc finger technology in tobacco and Arabidopsis plants. Using a novel library of transcription factors, we developed a strategy that effectively allows us to turn on and turn off every gene in the genome. With this powerful new strategy, we can quickly regulate a target gene or discover other genes key in disease (Fig. 3). We are targeting a variety of other genes involved not only in cancer and HIV disease but also in genetic diseases such as sickle cell anemia. We hope to take this genetic strategy and our other molecular approaches all the way to clinic trials. 15 Blancafort, P., Magnenat, L., Barbas, C.F. Scanning the human genome with combinatorial transcription factor libraries. Nat. Biotechnol. 21:269, 2003. Chowdari, N.S., Ramachary, D.B., Barbas, C.F. III. Organocatalysis in ionic liquids: highly efficient l-proline-catalyzed direct asymmetric Mannich reactions involving ketone and aldehyde nucleophiles. Synlett, in press. Chowdari, N.S., Ramachary, D.B., Barbas, C.F. III. Organocatalytic asymmetric assembly reactions: one-pot synthesis of functionalized β-amino alcohols from aldehydes, ketones, and azodicarboxylates. Org. Lett. 5:1685, 2003. Chung, J., Rader, C., Popkov, M., Hur, Y.-M., Kim, H.-K., Lee, Y.-J., Barbas, C.F. III. Integrin αIIbβ3-specific synthetic human monoclonal antibodies and HCDR3 peptides that potently inhibit platelet aggregation. FASEB J., in press. Córdova, A., Barbas, C.F. III. Direct organocatalytic asymmetric Mannich-type reactions in aqueous media: one-pot Mannich-allylation reactions. Tetrahedron Lett. 44:1923, 2002. Córdova, A., Notz, W., Barbas, C.F. III. Direct organocatalytic aldol reactions in buffered aqueous media. Chem. Commun. (Camb.) 3024, 2002, Issue 24. Gottesfeld, J.M., Barbas, C.F. III. RNA as a transcriptional activator. Chem. Biol. 10:584, 2003. Holbro, T., Beerli, R.R., Maurer, F., Koziczak, M., Barbas, C.F. III, Hynes, N.E. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 100:8933, 2003. Hyland, S., Beerli, R.R., Barbas, C.F., Hynes, N.E., Wels, W. Generation and functional characterization of intracellular antibodies interacting with the kinase domain of human EGF receptor. Oncogene 22:1557, 2003. Jendreyko, N., Popkov, M., Beerli, R.R., Chung, J., McGavern, D.B., Rader, C., Barbas, C.F. III. Intradiabodies: bispecific, tetravalent antibodies for the simultaneous functional knockout of two cell surface receptors. J. Biol. Chem. 278:47812, 2003. Lin, Q., Barbas, C.F. III, Schultz, P.G. Small-molecule switches for zinc finger transcription factors. J. Am. Chem. Soc. 125:612, 2003. Mase, N., Tanaka, F., Barbas, C.F. III. Rapid fluorescent screening for bifunctional amine-acid catalysts: efficient syntheses of quaternary carbon-containing aldols under organocatalysis. Org. Lett. 5:4369, 2003. Notz, W., Tanaka, F., Watanabe, S.-I., Chowdari, N.S., Turner, J.M., Thayumanavan, R., Barbas, C.F. III. The direct organocatalytic asymmetric Mannich reaction: unmodified aldehydes as nucleophiles. J. Org Chem. 68:9624, 2003. Rader, C., Sinha, S.C., Popkov, M., Lerner, R.A., Barbas, C.F. III. Chemically programmed monoclonal antibodies for cancer therapy: adaptor immunotherapy based on a covalent antibody catalyst. Proc. Natl. Acad. Sci. U. S. A. 100:5396, 2003. Rader, C., Turner, J.M., Heine, A., Shabat, D., Sinha, S.C., Wilson, I.A., Lerner, R.A., Barbas, C.F. III. A humanized aldolase antibody for selective chemotherapy and adaptor immunotherapy. J. Mol. Biol. 332:889, 2003. Ramachary, D.B., Chowdari, N.S., Barbas, C.F. III. The first organocatalytic heterodomino Knoevenagel-Diels-Alder-epimerization reactions: diastereoselective synthesis of highly substituted spiro[cyclohexane-1,2′-indan]-1,3′,4-triones. Synlett, in press. F i g . 3 . Genome-wide transcriptional regulation strategies. Libraries of transcription factors can be rapidly constructed from predefined recognition domains. With this approach, a wide range of regulators can be studied to quickly determine the regulator most optimal for gene regulation. This approach can also be applied to gene discovery. PUBLICATIONS Berry, J.D., Licea, A., Popkov, M., Cortez, X., Fuller, R., Elia, M., Kerwin, L., Kubitz, D., Barbas, C.F. III. Rapid monoclonal antibody generation via dendritic cell targeting in vivo. Hybrid. Hybridomics 22:23, 2003. Berry, J.D., Rutherford, J., Silverman, G.J., Kaul, R., Elia, M., Gobuty, S., Fuller, R., Plummer, F.A., Barbas, C.F. III. Development of functional human monoclonal single-chain variable fragment antibody against HIV-1 from human cervical B cells. Hybrid. Hybridomics 22:97, 2003. Ramachary, D.B., Chowdari, N.S., Barbas, C.F. III. Organocatalytic asymmetric domino Knoevenagel/Diels-Alder reactions: a bioorganic approach to the diastereospecific and enantioselective construction of highly substituted spiro[5,5]undecane1,5,9-triones. Angew. Chem. Int. Ed. 42:4233, 2003. Segal, D.J., Beerli, R.R., Blancafort, P., Dreier, B., Effertz, K., Huber, A., Koksch, B., Lund, C.V., Magnenat, L., Valente, D., Barbas, C.F. III. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry 42:2137, 2003. Segal, D.J., Stege, J., Barbas, C.F. III. Zinc fingers and a green thumb: manipulating gene expression in plants. Curr. Opin. Plant Biol. 6:163, 2003. Tanaka, F., Fuller, R., Shim, H., Lerner, R.A., Barbas, C.F. III. Evolution of aldolase antibodies in vitro: correlation of catalytic activity and reaction-based selection. J. Mol. Biol. 335:1007, 2004. 16 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Tanaka, F., Thayumanavan, R., Barbas, C.F. III. Fluorescent detection of carboncarbon bond formation. J. Am. Chem. Soc. 125:8523, 2003. Tanaka, F., Thayumanavan, R., Mase, N., Barbas, C.F. III. Rapid analysis of solvent effects on enamine formation by fluorescence: how might enzymes facilitate enamine chemistry with primary amines? Tetrahedron Lett., in press. Wantanabe, S., Córdova, A., Tanaka, F., Barbas, C.F. III. One-pot asymmetric synthesis of β-cyanohydroxymethyl α-amino acid derivatives: formation of three contiguous stereogenic centers. Org. Lett. 4:4519, 2002. Training in Molecular and Experimental Medicine E. Beutler he Skaggs Institute for Chemical Biology is currently supporting the training of 5 young scientists in the Department of Molecular and Experimental Medicine at The Scripps Research Institute. Each of the 5 is studying a problem that is important for human health under the direct supervision of a mentor who is an established expert in the field. ISG15 is an ubiquitin-like protein that is strongly induced and forms protein conjugates in mammalian cells after viral infection. Kenneth Ritchie and his mentor, Dong-Er Zhang, have identified UBP43 as an ISG15 protease and have produced UBP43-deficient mice. They have been investigating the response of UBP43–/– mice to infection with lymphocytic choriomeningitis virus (LCMV). In previous studies, UBP43–/– mice were found to have increased levels of ISG15 in ependymal cells and the choroid plexus of the brain. Recently, Dr. Ritchie extended these observations to include the meninges. After intracerebral inoculation with LCMV, viral replication primarily occurs in the ependyma, choroid plexus, and meninges. He found that such infection induces ISGylation with these cell types and the brain microvasculature. In order to investigate if UBP43–/– mice have altered susceptibility to intracerebral viral infection, LCMV was injected intracerebrally and the mice were monitored daily. Confirming successful infection, 100% of wildtype mice died before day 7 postinfection. Contrastingly, no UBP43 –/– mice died up to day 11 postinfection. Investigations into the presence of LCMV in UBP43 –/– brain tissue and serum revealed no detectable virus by plaque assay. Supporting in vitro studies with mouse embryo fibroblasts cells revealed that UBP43 deficiency also confers resistance to infection by Sindbus virus. Dr. Ritchie has therefore provided the first direct evi- T dence that ISGylation has an important role in innate immune defense against viral infection. In doing so, he also extended the repertoire of functions performed by ubiquitin-like molecules to viral defense. Xianghong Li, under the direction of Xiaohua Wu, has participated in the study of DNA replication initiation control and cell-cycle checkpoint control. She has mainly focused on studying a replication licensing factor, Cdt1, in the control of DNA replication initiation. She has shown that Cdt1 degrades when cells enter S phase. On the basis of the observation that overexpression of Cdt1 leads to re-replication, cell cycle–dependent Cdt1 degradation is likely to play a crucial role in restricting DNA replication to once per cell cycle. Dr. Li investigated the mechanisms of human Cdt1 degradation and found that human Cdt1 degrades through the SCF Skp2 -mediated ubiquitination pathway. She also made the observation that Cdt1 is phosphorylated by cyclin-dependent kinase (Cdk) at multiple sites on Cdt1. This Cdk-mediated phosphorylation stimulates Skp2 binding and triggers human Cdt1 for degradation. The biological significance of the Cdk-mediated phosphorylation events is currently under investigation. In addition, Dr. Li investigated Cdt1 function in DNA damage responses and performed experiments to test the association of Cdt1 with other proteins involved in DNA replication initiation. Her findings have contributed significantly to the understanding of cellular mechanisms that control DNA replication and checkpoints and will help to reveal how genome stability, which is essential for preventing cancer, is maintained. The production of reactive oxygen species represents the major microbicidal mechanism of professional phagocytes. The aim of the research of Natalia Sigal, mentored by Bernard Babior, is to study the recruitment of cytosolic components of NADPH oxidase (proteins Rac2, p67 phox, p47 phox, and p40 phox) to the phagosomal membrane that forms around ingested particles. To do this, she has used deconvolution microscopy instead of confocal microscopy, thus making it possible to take images of 3-dimensional objects, allowing her to see proteins associated with the phagosome. Neutrophils were settled on serum-coated coverslips; opsonized particles were then incubated with adhered neutrophils for various periods (0–15 minutes). After fixation with 10% formalin and permeabilization with acetone, cells were blocked with mouse or rabbit serum and incubated with primary antibody and then with secondary fluorescent antibody. Dr. Sigal was able to THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 determine the time course of association of p47 phox , p67 phox, p40 phox, and Rac2 assembly with the phagosomal membrane. Cytosolic proteins p47 phox and p40 phox were found to interact with phosphoinositides that are under control of phosphatidylinositol-3′-kinase (PI3K). PI3K has been known to be required for oxidase function, and PI3K products are believed to be responsible for translocation of cytosolic proteins to the membranes. Dr. Sigal studied the involvement of PI3K in the targeting of cytosolic proteins to the phagosomal membrane and the ability of PI3K inhibitors to inhibit superoxide production in assays with whole cells. She found that wortmannin and LY294002 have a strong inhibitory effect on neutrophils activated by N-formylmethionine– leucine–phenylalanine, but not on cells activated by phorbol myristate acetate. To better characterize the ability of opsonized zymosan particles to activate the NADPH oxidase in the forming phagosome, Dr. Sigal activated neutrophils with opsonized zymosan particles and measured superoxide production. PI3K inhibitors were found to inhibit the zymosan-activated superoxide production in a dose-response manner. Previous studies have shown a major role of protein kinase C in NADPH oxidase activation. To investigate the involvement of protein kinase C in the translocation of cytosolic protein to the phagosomal membrane and to compare the results with the inhibition of translocation by PI3K inhibitors, Dr. Sigal has begun to study the ability of protein kinase C inhibitors to affect translocation of p47phox and p40 phox. Human platelets are the smallest cellular component of blood, but they play a very important role in the prevention of bleeding. At sites of injury, they do this by sticking to exposed substances in the blood vessel wall, such as collagens. Platelets have specific binding sites or receptors on their surface that mediate the attachment to collagens and other substances in the blood vessel wall. Kazunobu Kato, under the supervision of Jerry Ware and Thomas Kunicki, is currently investigating the genetic differences between platelet receptors that can modify the ability of platelets to function properly. One of these receptors for collagen, glycoprotein VI, is unique to platelets. It is important to identify and study natural products that bind to this receptor so that they can be used to develop therapeutic or diagnostic reagents. One such natural product is a substance called convulxin, produced in the venom of a certain South American rattlesnake. 17 Convulxin normally binds to platelets and induces platelet clumping or clot formation. By carefully studying the structure of convulxin, Dr. Kato hopes to learn ways to modify its effect and to turn it into an inhibitor. Such an inhibitor may prove useful for the control of unwanted and dangerous clot formation, as seen in stroke or heart disease. This research involves the isolation of DNA that encodes convulxin, the mutation of this DNA, and the expression of the convulxin protein by using recombinant DNA technology. Sharookh Kapadia, under the guidance of Frank Chisari, is studying hepatitis C virus (HCV), a member of the Flaviviridae family of viruses and a major cause of chronic hepatitis and hepatocellular carcinoma. Viral clearance during acute HCV infection is usually associated with a multispecific CD4+ and CD8 + T-cell response that is weak or undetectable in subjects who do not control the infection. Importantly, most chronically infected patients do not resolve HCV infection after combination therapy with IFN and ribavirin. Recent data have shown that a number of genes involved in lipid metabolism are transcriptionally regulated preferentially in acutely HCV-infected chimpanzees that had the highest acute titers of virus. This finding suggests that these genes may, theoretically, be important under baseline conditions for efficient viral replication to occur. Although there is considerable evidence suggesting that the lipid metabolism pathways may play a role in HCV replication and infection, it is not known whether changes in cholesterol and/or fatty acid biosynthesis directly regulate viral replication or whether these changes merely reflect a pathologic host response to HCV infection. Furthermore, drugs that are known to inhibit key steps in these biosynthetic pathways were also shown to regulate replication of the HCV subgenomic replicon in Huh-7 cells. However, the specificity of these drugs was not assessed. Therefore, Dr. Kapadia wanted to specifically determine whether the cholesterol and/or fatty acid biosynthetic pathways regulated HCV replication by using the subgenomic HCV replicon cell system. The liver X receptors (LXRs) belong to a family of nuclear hormone receptors that play a key role in regulating cholesterol and fatty acid metabolism. Dr. Kapadia demonstrated that activation of the LXRs by a nonsteroidal LXR agonist, T0901317, dramatically regulates HCV replication. Further characterization of the expression of genes encoding enzymes involved in cholesterol and fatty acid biosynthetic pathways are in progress. She is currently 18 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 using short interfering RNAs to specifically inhibit expression of key genes that encode enzymes involved in these pathways. PUBLICATIONS Kapadia, S.B., Brideau-Andersen, A., Chisari, F.V. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 100:2014, 2003. Li, X., Zhao, O., Liao, R., Sun, P., Wu, X. The SCFSkp2 ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 278:30854, 2003. Synthetic, Medicinal, and Bioorganic Chemistry by using combinatorial libraries to target protein-protein interactions. Each example defined or validated new therapeutic targets and provided the first small-molecule leads for each target. We identified an effective inhibitor of Myc-Max that inhibits protein dimerization, subsequent DNA binding, and gene transcription (Fig. 1). In addition to being the first small-molecule inhibitor of a transcription factor dimerization, the lead compound also inhibited Myc-induced cell transformation in a functional cellular assay (the transformation from a normal to a tumor phenotype), validating the target for antineoplastic intervention via small molecules. D.L. Boger, Y. Ambroise, B. Blagg, S. Buck, K. Capps, H. Cheng, Y. Choi, B. Crowley, P. Desai, J. Desharnais, R. Dominique, W. Du, R. Ducray, G. Elliott, R. Fecik, Y. Ham, N. Haq, C. Hardouin, I. Hwang, S. Ichikawa, D. Kastrinsky, G. Kim, P. Krenitsky, M. Lall, C. McComas, Y. Mori, J. Parrish, S. Pfeiffer, A. Pichota, Y. Rew, D. Shin, D. Soenen, J. Trzupek, W. Tse, J. Wanner, G. Wilkie, S. Wolkenberg, Z. Yuan, B. Yeung, J. Zimpleman he research interests of our group include the total synthesis of biologically active natural products, the development of new synthetic methods, heterocyclic chemistry, bioorganic and medicinal chemistry, combinatorial chemistry, the study of DNA-agent interactions, and the chemistry of antitumor antibiotics. We place a special emphasis on investigations to define the structure-function relationships of natural or designed agents in efforts to understand the origin of the biological properties of the agents. T TA R G E T I N G P R O T E I N - P R O T E I N I N T E R A C T I O N S W I T H C O M B I N AT O R I A L L I B R A R I E S Receptor activation and signal transduction by protein-protein homodimerization, heterodimerization, and higher order homo- and hetero-oligomerization have emerged as general mechanisms of initiating and transmitting intracellular signals. We are investigating the fundamental principles and structural features embodied in such receptor activation and signal transduction events with the receptor for erythropoietin. Additional targets under examination include ErbB2; the transcription factor Myc-Max; LEF-1/β-catenin; the androgen receptor; and angiogenesis inhibitors (αvβ3 and αvβ5), including those that act by inhibiting binding of matrix metalloproteinase 2 to αvβ3. This past year, we reported 3 important instances in which we successfully modulated cellular signaling F i g . 1 . Inhibition of transcription factor heterodimerization and aberrant gene transcription: antitumor target validation. ELISA indicates enzyme-linked immunosorbent assay; FRET, fluorescent resonance energy transfer. Similarly, we discovered a potent inhibitor of paxillin binding to the intracellular tail of the cell-surface α4 by screening our combinatorial library of compounds. This discovery provided the first inhibitor of paxillin-α4 protein-protein binding and allowed a critical assessment of small-molecule positional scanning libraries. The most potent lead compound inhibited cell migration, validating the protein-protein interaction as one useful for treatment of chronic inflammatory and autoimmune diseases. Finally, we discovered a series of small-molecule erythropoietin agonists that act by promoting (not inhibiting) the dimerization of the cell-surface erythropoietin receptor, providing a general model for the synthesis, screening, and design of such molecules (agonists from antagonists) (Fig. 2). STRUCTURE-BASED DRUG DESIGN In collaboration with I.A. Wilson, the Skaggs Institute, we are using x-ray crystallographic structures of (1) the apo forms of glycinamide ribonucleotide transformylase and aminoimidazole carboxamide ribonucleotide transformylase inosine monophosphate cyclohydrolase and (2) complexes of the 2 enzymes with their substrates, folate cofactors, and inhibitors to design and THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 19 antitumor agent, involves a hydrogen-bonding triplexlike interaction in the minor groove of duplex DNA. We are extending and exploiting these developments. PUBLICATIONS Ambroise, Y., Boger, D.L. The DNA phosphate backbone is not involved in catalysis of the duocarmycin and CC-1065 DNA alkylation reaction. Bioorg. Med. Chem. Lett. 12:303, 2002. Ambroise, Y., Yaspan, B., Ginsberg, M.H., Boger, D.L. Inhibitors of cell migration that inhibit intracellular paxillin/α4 binding: a well-documented use of positional scanning libraries. Chem. Biol. 9:1219, 2002. F i g . 2 . An erythropoietin (EPO) agonist that functions by pro- moting dimerization of the erythropoietin receptor (EPOR). examine potent inhibitors of the enzymes as antineoplastic agents. One such inhibitor is undergoing extensive in vivo and preclinical evaluation. After the discovery of oleamide, a fatty acid primary amide with sleep-inducing properties, and anandamide, the endogenous ligand for the cannabinoid receptor, the study of these prototypical members of a new class of endogenous chemical messengers led to the identification of fatty acid amide hydrolase, an enzyme responsible for their degradation and regulation. In the past year we produced a series of exceptionally potent α-ketoheterocycle inhibitors of the hydrolase that may be useful in the treatment of sleep disorders or as novel analgesics. We also investigated the substrate specificity of pure recombinant fatty acid amide hydrolase against a complete series of fatty acid primary amides. We identified required structural features and additional potential endogenous members of this class of chemical messengers subject to regulation by the hydrolase. With the determination of the x-ray structure of the enzyme, conducted by R. Stevens, The Scripps Research Institute, and B.F. Cravatt, the Skaggs Institute, retrospective active-site modeling of the inhibitors and substrates has provided new inhibitor classes. DNA-BINDING ANTITUMOR ANTIBIOTICS We are continuing our synthesis and evaluation of naturally occurring antitumor antibiotics, including CC-1065/duocarmycins, yatakemycin, bleomycin A 2 , sandramycin/luzopeptins, and isochrysohermidin, and are studying their sequence-selective interaction with duplex DNA. In the past year, we identified the source of catalysis for the CC-1065/duocarmycin DNA alkylation reaction based on a DNA binding–induced conformational change in the agent, a process we termed “shape-dependent catalysis.” Similarly, we established that the source of the polynucleotide recognition of bleomycin A2, an Berg, T., Cohen, S.B., Desharnais, J., Sonderegger, C., Maslyar, D.J., Goldberg, J., Boger, D.L., Vogt, P.K. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 99:3830, 2002. Blagg, B.S.J., Boger, D.L. Total synthesis of (+)-camptothecin. Tetrahedron 58:6343, 2002. Boger, D.L., Castle, S.L. Synthesis of cycloisodityrosine peptides. In: Synthesis of Peptides and Peptidomimetics, Vol. E22c. Goodman, M., et al. (Eds.). Thieme Medical, New York, 2002, p. 194. Houben-Weyl: Methods of Organic Chemistry, 4th ed. Chen, Y., Bilban, M., Foster, C.A., Boger, D.L. Solution-phase parallel synthesis of a pharmacophore library of HUN-7293 analogues: a general chemical mutagenesis approach to defining structure-function properties of naturally occurring cyclic (depsi)peptides. J. Am. Chem. Soc. 124:5431, 2002. Goldberg, J., Jin, Q., Ambroise, Y., Satoh, S., Desharnais, J., Capps, K., Boger. D.L. Erythropoietin mimetics derived from solution phase combinatorial libraries. J. Am. Chem. Soc. 124:544, 2002. Jiang, W., Wanner, J., Lee, R.J., Bounaud, P.-Y., Boger, D.L. Total synthesis of the ramoplanin A2 and ramoplanose aglycon. J. Am. Chem. Soc. 124:5288, 2002. Krenitsky, P.J., Boger, D.L. Preparation of the 14-membered L,L-cycloisodityrosine subunit of RP 66453. Tetrahedron Lett. 43:407, 2002. Lewy, D., Gauss, C.-M., Soenen, D.R., Boger, D.L. Fostriecin: chemistry and biology. Curr. Med. Chem. 9:2005, 2002. Marsilje, T.H., Labroli, M.A., Hedrick, M.P., Jin, Q., Desharnais, J., Baker, S.J., Gooljarsingh, L.J., Ramcharan, J., Tavassoli, A., Zhang, Y., Wilson, I.A., Beardsley, G.P., Benkovic, S.J., Boger. D.L. 10-Formyl-5,10-dideaza-acyclic-5,6,7,8tetrahydrofolic acid (10-formyl-DDACTHF): a potent cytotoxic agent acting by selective inhibition of human GAR Tfase and the de novo purine biosynthetic pathway. Bioorg. Med. Chem. 10:2739, 2002. McAtee, J.J., Castle, S.L., Jin, Q., Boger, D.L. Synthesis and evaluation of vancomycin and vancomycin aglycon analogues that bear modifications of the residue 3 asparagine. Bioorg. Med. Chem. Lett. 12:1319, 2002. Wilkie, G.D., Elliott, G.I., Blagg, B.S.J., Wolkenberg, S.E., Soenen, D.R., Miller, M.M., Pollack, S., Boger, D.L. Intramolecular Diels-Alder and tandem intramolecular Diels-Alder/1,3-dipolar cycloaddition reactions of 1,3,4-oxadiazoles. J. Am. Chem. Soc. 124:11292, 2002. Wolkenberg, S.E., Boger, D.L. Mechanisms of in situ activation of DNA targeting antitumor agents. Chem. Rev. 102:2477, 2002. Wolkenberg, S.E., Boger, D.L. Total synthesis of anhydrolycorinone utilizing sequential intramolecular Diels-Alder reactions of a 1,3,4-oxadiazole. J. Org. Chem. 67:7361, 2002. Woods, C.R., Faucher, N., Eschgfaller, B., Bair, K.W., Boger, D.L. Synthesis and DNA-binding properties of saturated distamycin analogues. Bioorg. Med. Chem. Lett. 12:2647, 2002. Woods, C.R., Ishii, T., Boger, D.L. Synthesis and DNA binding properties of iminodiacetic acid-linked polyamides: characterization of cooperative extended 2:1 side-by-side parallel binding. J. Am. Chem. Soc. 124:10676, 2002. 20 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Woods, C.R., Ishii, T., Wu, B., Bair, K.W., Boger, D.L. Hairpin versus extended DNA binding of a substituted β-alanine linked polyamide. J. Am. Chem. Soc. 124:2148, 2002. Zhang, Y., Desharnais, J., Greasley, S.E., Beardsley, G.P., Boger, D.L., Wilson, I.A. Crystal structures of human GAR Tfase at low and high pH and with substrate β-GAR. Biochemistry 41:14206, 2002. Structural Biology of Membrane-Bound Transporters and Receptors A. Ma, T. Nguyen, P. Szewczyk, Y. Yin, C. Reyes, S. Wada, important drug targets. Nearly all G protein–coupled receptors are predicted to have 7 transmembrane helices with domains that bind ligand and activate G proteins. A structure of these receptors will reveal the mechanisms of cell signaling on the molecular level. PUBLICATIONS Chang, G. Structure of MsbA from Vibrio cholerae: a multidrug resistance ABC transporter homolog in a closed conformation. J. Mol. Biol. 330:419, 2003. Chang, G., Roth, C.B. Signal transduction and integral membrane proteins. In: Handbook of Cell Signaling. Bradshaw, R.A., Dennis, E.A. (Eds.). Academic Press, San Diego, 2003, p. 115. Roth, C.B., Chang, G. X-ray structure of an intact ABC-transporter, MsbA. In: ABC Proteins: From Bacteria to Man. Holland, I.B., et al. (Eds.). Academic Press, San Diego, 2003, p. 135. O. Pornillos, M. Grant, G. Chang -ray crystallography of receptors and multidrugresistance (MDR) efflux pumps embedded in the cell membrane is an important frontier in structural biology and medicine. We are interested in the structural basis for the transport of hydrophobic substrates across the cell membrane by MDR transporters and the signal transduction by receptors. We use several biophysical techniques, including molecular cloning, detergent/lipid protein chromatography, membrane protein crystallization, and protein x-ray crystallography. Methods for determining structure include the overexpression, large-scale purification, and 3-dimensional crystallization of these integral membrane proteins. Recently, we determined the structures of the open and closed conformations of an MDR transporter from the ATP-binding cassette family called MsbA. MsbA is a bacterial homolog of human MDR1, which causes MDR in the treatment of cancers. In collaboration with Ronald Milligan, The Scripps Research Institute, we are using electron cryomicroscopy to study other possible conformational changes. Through the help of the Skaggs Institute, we are expanding our structure determination efforts to incorporate mammalian ATP-binding cassette transporters that confer the MDR phenotype and other ATP-binding cassette transporters that are involved in human diseases, including cystic fibrosis and macular dystrophy. In addition, we are elucidating the structures of H +-drug antiporters from both the major facilitator superfamily and the small multidrug transporter family. We are also interested in the structures of membrane-bound receptors that are important models for signal transduction across the lipid bilayer. One of the most widely studied families of receptors is the family of G protein–coupled receptors, which are pharmaceutically X Chemical Physiology B.F. Cravatt, G.C. Adam, K.T. Barglow, M.H. Bracey, K. Chiang, M. Evans, G. Hawkin, M. Humphrey, N. Jessani, A. Joseph, D. Leung, K. Masuda, M. McKinney, A. Saghatelian, A. Speers e are interested in understanding complex physiology and behavior at the level of chemistry and molecules. At the center of cross talk between different physiologic processes are endogenous compounds that act as a molecular mode for intersystem communication. However, many of these molecular messages remain unknown, and even in the instances in which the participating molecules have been defined, the mechanisms by which these compounds function are for the most part still a mystery. Currently, we are focusing on a family of chemical messengers termed the fatty acid amides, which affect many physiologic functions, including sleep and pain. In particular, one member of this family, oleamide, accumulates selectively in the cerebrospinal fluid of tired animals. This finding suggests that oleamide may be a molecular indicator of the organism’s need for sleep. Another fatty acid amide, anandamide, is a postulated endogenous ligand for the cannabinoid receptor in the brain. The in vivo levels of chemical messengers such as the fatty acid amides must be tightly regulated to maintain proper control over the influence of the messengers on brain and body physiology. We are characterizing a mechanism by which the level of fatty acid amides can be regulated in vivo. The enzyme fatty acid amide hydrolase (FAAH) degrades the fatty acid amides to inactive metabolites. Thus, FAAH effectively terminates the sig- W THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 naling messages conveyed by fatty acid amides, possibly ensuring that these molecules do not generate physiologic responses in excess of their intended purpose. We are using transgenic and synthetic chemistry techniques to study the role of FAAH in the regulation of fatty acid amide levels in vivo. We created transgenic mice that lack the hydrolase and found that these animals have highly elevated brain levels of fatty acid amide that correlate with reduced pain behavior, suggesting that FAAH may be a new therapeutic target for the treatment of pain and related neural disorders. In collaboration with R.C. Stevens, The Scripps Research Institute, we recently solved the first 3-dimensional structure of FAAH. The structure will serve as a template for the design of potent and selective inhibitors of the enzyme. In collaboration with D.L. Boger, the Skaggs Institute, we identified potent FAAH inhibitors, and using a functional proteomic screen developed by our group, we showed that these inhibitors are highly selective for this enzyme. We are also interested in proteins responsible for the biosynthesis of fatty acid amides. A second major focus in the laboratory is the design and use of chemical probes for the global analysis of protein function. The evolving field of proteomics, defined as the simultaneous analysis of the complete protein content of given cell or tissue, encompasses considerable conceptual and technical challenges. We hope to enhance the quality of information obtained from proteomics experiments by using chemical probes that read out the collective catalytic activities of entire classes of enzymes. Using activity-based probes that target the serine hydrolases, we identified several enzymes with altered activities in human cancer. Additionally, we are developing chemical probes that target many other enzyme families. To date, we have identified activity-based proteomics probes for more than 10 mechanistically distinct enzyme classes. PUBLICATIONS Adam, G.C., Vanderwal, C.D., Sorensen, E.J., Cravatt, B.F. (–)-FR182877 is a potent and selective inhibitor of carboxylesterase-1. Angew. Chem. Int. Ed. 42:5480. 2003. Egertova, M., Cravatt, B.F., Elphick, M.R. Comparative analysis of fatty acid amide hydrolase and CB1 cannabinoid receptor expression in the mouse brain: evidence of a widespread role for fatty acid amide hydrolase in regulation of endocannabinoid signaling. Neuroscience 119:481, 2003. Kustedjo, K., Deechongkit, S., Kelly, J.W., Cravatt, B.F. Recombinant expression, purification, and comparative characterization of torsinA and its torsion dystoniaassociated variant ∆E-torsinA. Biochemistry 42:15333, 2003. Leung, D., Hardouin, C., Boger, D.L., Cravatt, B.F. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat. Biotechnol. 21:687, 2003. McKinney, M.K., Cravatt, B.F. Evidence for distinct roles in catalysis for residues of the serine-serine-lysine catalytic triad of fatty acid amide hydrolase. J. Biol. Chem. 278:37393, 2003. Speers, A.E., Adam, G.C., Cravatt, B.F. Activity-based protein profiling in vivo using a copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 125:4686, 2003. Synthetic Protein Chemistry P. Dawson, R. Balambika, J. Blankenship, M. Churchill, J. Offer, C. Neidre, F. Topert ur focus is the development and use of methods to incorporate unnatural chemical groups into proteins. We developed a chemical approach for producing the large polypeptide chains that make up protein molecules, enabling us to change the structure of a protein in ways impossible by natural means. This chemical ligation approach greatly facilitates the synthesis of proteins of moderate size and has opened the world of proteins to the synthetic tools of organic chemistry. Chemical ligation can be extended to biologically expressed proteins, enabling the semisynthesis of proteins of unlimited size that contain fluorophores or cross-linking agents at defined positions. Our goal is to introduce noncoded amino acids and other chemical groups into proteins to better understand the molecular basis of protein function. O SYNTHESIS OF PROTEINS Proteins synthesized by the chemical ligation approach are assembled from several smaller polypeptides. The modular nature of this approach provides the opportunity to assemble large numbers of modified proteins through combinatorial assembly of the individual peptide components (Fig. 1). We are developing a sys- Aebersold, R., Cravatt, B.F. Proteomics: advances, applications, and the challenges that remain. Trends Biotechnol. 20(12 Suppl.):S1, 2002. Clement, A.B., Hawkins, E.G., Lichtman, A.H., Cravatt, B.F. Increased seizure susceptibility and proconvulsant activity of anandamide in mice lacking fatty acid amide hydrolase. J. Neurosci. 23:3916, 2003. Cravatt, B.F., Lichtman, A.H. The enzymatic inactivation of the fatty acid amide class of signaling lipids. Chem. Phys. Lipids 121:135, 2002. Cravatt, B.F., Lichtman, A.H. Fatty acid amide hydrolase: an emerging therapeutic target in the endocannabinoid system. Curr Opin. Chem. Biol. 7:469. 2003. 21 F i g . 1 . A, Combinatorial assembly of polypeptide fragments. B, Array of folded proteins on a solid support. 22 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 tem in which proteins can be assembled in parallel on a sheet of cellulose by using our chemical ligation reactions. After these proteins are removed from the cellulose, they can be assayed in solution or covalently attached to glass slides to facilitate rapid screening. T O TA L S Y N T H E S I S O F A V I R A L C O AT Many viruses encapsulate themselves in a protein coating composed of multiple copies of a small number of protein subunits. The process through which individual protein-protein interactions grow into a spherical protein assembly is essential to the life cycle of these viruses and may be an important target of antiviral therapies. To study these interactions in more detail, we are synthesizing the coat protein from the MS2 bacteriophage, which self-assembles with 180 copies of itself to encapsulate the genetic material of the bacterial virus. We plan to introduce site-specific fluorophores and cross-linking agents into this synthetic viral coat in order to understand viral coat assembly. PUBLICATIONS Blankenship, J.W., Balambika, R., Dawson, P.E. Probing backbone hydrogen bonds in the hydrophobic core of GCN4. Biochemistry 41:15676, 2002. Blankenship, J.W., Dawson, P.E. Thermodynamics of a designed protein catenane. J. Mol. Biol. 327:537, 2003. Brik, A., Dawson, P.E., Keinan, E. The product of the natural reaction catalyzed by 4-oxalocrotonate tautomerase becomes an affinity label of its mutant. Bioorg. Med. Chem. 10:3891, 2002. Dawson, G., Dawson, S.A., Marinzi, C., Dawson, P.E. Anti-tumor promoting effects of palmitoyl:protein thioesterase inhibitors against a human neurotumor cell line. Cancer Lett. 187:163, 2002. Zwick, M.B., Parren, P.W.H.I., Saphire, E.O., Church, C., Wang, M., Scott, J.K., Dawson, P.E., Wilson, I.A., Burton, D.R. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 77:5863, 2003. Fundamental Processes in Neural Development G.M. Edelman, S. Aschrafi, A. Atkins, S. Chappell, G.W. Rogers, F. Smart, T. Stevens, M. Tsatmali, W. Zhou e focus our efforts on primary cellular processes of development, with emphasis on the development of the vertebrate nervous system. Our recent work, which originated from the study of the structure, function, and regulation of cell adhesion molecules, led to approaches for examining the fundamental mechanisms that control transcription and translation. We are continuing our research on the W structure and function of adhesion molecules in neural development and at synapses. The training grant from the Skaggs Institute supports the work of several postdoctoral fellows. Their efforts in various projects with senior investigators are summarized here. Development and morphogenesis require multiple rounds of differential gene expression. A variety of factors regulates this expression, including elements within the genes themselves and protein factors that bind to the elements. In examining such elements in genes for cell adhesion molecules, Fred Jones and his colleagues discovered a homeobox protein, Barx2, that affects a wide variety of differentiation processes. Tracy Stevens is working with Robyn Meech to explore the activity of Barx2 and to identify its target genes. These studies have opened new front lines related to breast cancer, muscle development, and chondrogenesis. In examining the messenger RNAs expressed in response to neural cell adhesion, Vince Mauro noted that many mRNAs had sequences that matched or were complementary to ribosomal RNA. By analyzing a subset of these sequences, he and his colleagues, Stephen Chappell, Wei Zhou, and George Rogers, Jr., defined the characteristics of novel sequences within mRNAs that can connect the translation machinery to the message in the absence of the traditional cap sequence. These internal ribosome entry sites (IRESs) were known for viral RNAs but had not been extensively defined in cellular mRNAs. Dr. Mauro and his colleagues went on to show that cellular and viral IRESs differ; the cellular sites are made up of small modules, and the action of cellular IRESs does not necessarily depend on specific secondary structures in the mRNA. Regulation of translation appears to play an important role in the nervous system, and IRESs may play a special role in the functioning of the synapse. Peter Vanderklish and Fiona Smart have been studying the factors involved in synaptic events thought to provide the basis of memory and learning in the area of the brain called the hippocampus. Translation of mRNA in dendrites plays a special role in this process. Dr. Vanderklish and his colleagues found that specific messages at the synapse are differentially translated, depending on whether the messages are regulated by cap- or IRESdependent processes. In addition, Ann Atkins and Armaz Aschrafi are working with Bruce Cunningham to define and characterize granules containing ribosomes, proteins, and mRNAs that are transported from the cell body to the synaptic area in order to allow translation to occur THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 in response to synaptic activity. They are also characterizing the mechanisms involved in neural cell adhesion molecule binding and signaling. The development of the nervous system begins with the differentiation of stem cells to neural progenitors, which in turn develop at the proper time and place into neurons and supporting glia. Moreover, neural progenitor cells remain in the brain in adults, where they presumably repopulate specific brain regions. Kathryn Crossin and her colleagues found that the neural cell adhesion molecule can act as a neurotrophin to differentiate the neural stem cells preferentially into neurons. Moreover, using special multielectrode plates, Dr. Crossin and her colleagues defined conditions under which a stem cell population in culture can be converted into neurons capable of exchanging action potentials in a functioning neural network. Marina Tsatmali is following up these observations and is investigating the influence of oxidative state on neuronal differentiation and activity. All of these activities are aimed at the study of the molecular and cellular events that define and regulate the development of the nervous system. Our efforts remain focused on fundamental processes rather than on specific diseases. This strategy is based on the belief that understanding even a single primary process can provide the necessary framework for defining the mechanisms underlying not just one but a variety of diseases. 23 Chemical Etiology of the Structure of Nucleic Acids A. Eschenmoser, R. Krishnamurthy, Z. Wang, G.R. Vavilala, C. Mang, V. Rajwanshi, J. Nandy, T. George D uring the past year we worked on the following projects. NITROGENOUS ANALOGS OF (L)-α-THREOFURANOSYL(3′→2′) OLIGONUCLEOTIDES We continued our comprehensive experimental scrutiny of informational oligomeric systems containing threose-derived backbones. To that end, we synthesized the 2′,3′-diamino analogs of (L)-α-threofuranosyl-(3′→2′) oligonucleotide (TNA) building blocks containing adenine and thymine as nucleobases. We are investigating the chemistry of the analogs in order to make oligoamides; the information will be used in studies of the base-pairing properties of the oligoamides (Fig. 1). A LT E R N AT I V E N U C L E O B A S E S We studied the chemistry of 7,9-dicarba-5,8diaza-2,6-diaminopurine, a constitutional isomer of 2,6-diaminopurine. This isomer smoothly undergoes C-nucleosidations with cyclic iminium ions derived from L-threose derivatives to form nucleosides at the C-9 carbon of the nucleobase that are isosteric to normal N-nucleosides. PUBLICATIONS Chappell, S.A., Mauro, V.P. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J. Biol. Chem. 278:33793, 2003. Jones, F.S., McKean, D.M., Meech, R., Edelman, D.B., Oakey, R.J., Jones, P.L. Regulation of vascular smooth muscle cell growth and adhesion by paired-related homeobox genes. Chest 121(3 Suppl.):89S, 2002. Makarenkova, H.P., Meech, R., Edelman, D.B., Jones, F.S. The homeobox transcription factor Barx2 is expressed during limb development, modulates chondrogenesis, and is regulated by GDF5. Development, in press. Meech, R., Makarenkova, H., Edelman, D.B., Jones, F.S. The homeodomain protein Barx2 promotes myogenic differentiation and is regulated by myogenic regulatory factors. J. Biol. Chem. 278:8269, 2003. Smart, F., Edelman, G.M., Vanderklish, P.W. BDNF induces translocation of initiation factor 4E to mRNA granules in neurons by a mechanism involving synaptic microfilaments and integrins. Proc. Natl. Acad. Sci. U. S. A. 100:14403, 2003. Zhou, W., Edelman, G.M., Mauro, V.P. Isolation and identification of short nucleotide sequences that affect translation initiation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 100:4457, 2003. F i g . 1 . The oligoamide trimer ATT derived from 2′,3′-dideoxy- 2′,3′-diamino-TNA. A indicates adenine; T, thymine. 24 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 5,8-Diaza-7,9-dicarba-2,6-diaminopurine is just one member of a family of alternative purine nucleobases that potentially can undergo C-nucleosidation with iminium ions and aldosugars. We are expanding our investigation to other members of this family, including an extension of the C-nucleosidation chemistry to pyrimidines and still another family of purinoids. Jungmann, O., Beier, M., Luther, A., Huynh, H.K., Ebert, M.O., Jaun, B., Krishnamurthy, R., Eschenmoser, A. Pentopyranosyl oligonucleotide systems: communication No. 13. The α-L-arabinopyranosyl-(4′→2′)-oligonucleotide system: synthesis and pairing properties. Helv. Chim. Acta 86:1259, 2003. Schöning, K-.U., Scholz, P., Wu, X., Guntha, S., Delgado, G., Krishnamurthy, R., Eschenmoser, A. The α-(L)-threofuranosyl-(3′→2′)-oligonucleotide system (‘TNA’): synthesis and pairing properties. Helv. Chim. Acta. 85:4111, 2002. Wang, Z., Huynh, H.K., Han, B., Krishnamurthy, R., Eschenmoser, A. 2,6-Diamino5,8-diaza-7,9-dicarba-purine. Org. Lett. 5:2067, 2003. STRUCTURAL PROPERTIES OF TNAS In collaboration with M. Egli, Vanderbilt University, Nashville, Tennessee, we determined the crystal structure of the B-form DNA duplex [d(CGCGAA)T*d(TCGCG)]2 containing a single (L)-α-threofuranosyl-(3′→2′)-thymine (T*) per strand. The structure reveals that single TNA thymine nucleotides can be easily accommodated in a B-form of DNA with little change overall relative to the reference DNA structure (Fig. 2). Nuclear magnetic resonance analysis of the TNA duplex containing the self-complementary sequence t-(CGAATTCG) was carried out by B. Jaun and H. Ebert at the Swiss Federal Institute of Technology Zürich in Switzerland. The analysis shows that TNA forms a right-handed double helix resembling that of RNA. F i g . 2 . The x-ray structure analysis of the modified Dickerson Drew dodecamer duplex [d(CGCGAA)T*d(TGCGCG)]2 shows the C4′-exo conformation of the L-(α)- threofuranose nucleoside (T*7) in the modified duplex. PUBLICATIONS Ebert, M.O., Luther, A., Huynh, H.K., Krishnamurthy, R., Eschenmoser, A., Jaun, B. NMR solution structure of the duplex formed by self-pairing of α-(L)-arabinopyranosyl-(4′→2′)-(CGAATTCG). Helv. Chim. Acta. 85:4055, 2002. Han, B., Wang, Z., Jaun, B., Krishnamurthy, R., Eschenmoser, A. C-Nucleosidations with 2,6-diamino-5,8-diaza-7,9-dicarba-purine. Org. Lett. 5:2071,2003. Wilds, C.J., Wawrzak, Z., Krishnamurthy, R., Eschenmoser, A., Egli, M. Crystal structure of a B-form DNA duplex containing (L)-α-threofuranosyl (3′→2′) nucleosides: a four-carbon sugar is easily accommodated into the backbone of DNA. J. Am. Chem. Soc. 124:13716, 2002. Intracellular RNA Assembly and Catalysis M.J. Fedor, K.F. Baban, C.P. Da Costa, Y.I. Kuzmin, E.M. Mahen, S.B. Voytek, R.S. Yadava NAs are essential components of the molecular machinery that underlies growth and development. Virtually all important RNA-mediated reactions, such as those involved in gene regulation, splicing, and protein synthesis, involve assembly and dissociation of RNA complexes. Formation of RNA complexes often occurs through recognition between complementary RNA nucleobases and annealing of base-paired helices. Reaction pathways for many RNA enzymes also involve assembly and dissociation of ribozyme complexes that contain intermolecular base-paired helices. Unlike the complicated RNA-mediated reactions that participate in gene expression, however, ribozyme cleavage of RNA substrates gives a simple, quantitative signal that a functional ribozyme-substrate complex has formed. Enzymological studies of ribozymes have been used to dissect RNA assembly processes in vitro and have defined how reaction conditions and structural features of ribozyme and substrate RNAs influence the kinetics and equilibria of assembly and dissociation in ribozyme reaction pathways. However, establishing how the properties of RNA complex formation that can be measured in simple solutions relate to the behavior of RNA molecules in the complicated environment inside living cells has been difficult. By exploiting the simple RNA cleavage reactions mediated by the hairpin ribozyme, we devised a way to learn how evidence from studies of RNA enzymes in vitro translates to the complex environment of a living R THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 cell. Our early studies established which features of ribozyme structure are important for self-cleavage inside cells. Now we are focusing on recognition and cleavage of separate RNA targets in reactions that require assembly of functional ribozyme-substrate complexes. To examine intracellular ribozyme activity, we express hairpin ribozymes in yeast as chimeric U3 small nucleolar RNAs (snoRNAs) (Fig. 1). The snoRNAs are useful carriers of ribozyme and substrate sequences because they are stable RNAs that accumulate at high concentrations in yeast. Chimeric snoRNAs containing hairpin ribozymes are coexpressed in yeast with chimeric substrate snoRNAs that contain the cleavage site. Chimeric ribozyme snoRNAs are expressed under the control of a galactose-inducible promoter so that intracellular ribozyme concentrations can be modulated by changing the galactose concentration in yeast growth media. Formation of a functional complex between ribozyme and substrate snoRNAs leads to substrate RNA cleavage. 25 through ribozyme-mediated cleavage. Consequently, substrate RNAs that associate with ribozymes and undergo cleavage disappear faster from yeast than do substrate RNAs that are expressed in yeast without ribozymes. The difference in decay rates between substrates expressed alone and substrates coexpressed with ribozymes corresponds quantitatively to the intracellular cleavage rate. By using the intrinsic snoRNA degradation rate as a “clock” in this way, we can evaluate the effects of RNA structure and concentration on intracellular cleavage kinetics. We found that intermolecular cleavage increases with increasing RNA concentrations, as expected if formation of the ribozyme-substrate complex is rate determining. A second-order rate constant for the formation of RNA complexes in vivo can be calculated from the dependence of cleavage kinetics on intracellular concentrations of ribozyme. The second-order rate constant in yeast is significantly lower than rate constants measured for cleavage reactions under physiologic salt conditions in vitro, but it has a similar dependence on the length of the intermolecular basepaired helices that form during complex formation. These studies provide the first quantitative description of the kinetics of the formation of RNA complexes in vivo. Understanding this simple example of an interaction between ribozyme and target RNAs will provide insights into more complicated RNA interactions that perform essential roles in biology but are not as easily studied in the laboratory. PUBLICATIONS Fedor, M.J. Determination of kinetic parameters for hammerhead and hairpin ribozymes. Methods Mol. Biol., in press. Fedor, M.J. RNA biochemistry from A to Z. Cell 109:20, 2002. F i g . 1 . Chimeric ribozyme-substrate snoRNA complex. Chimeric snoRNAs that contain either ribozyme or substrate sequences are coexpressed in yeast to evaluate the reactions involved in the formation of RNA complexes that must precede intermolecular cleavage. Assembly of a functional ribozyme complex requires base pairing between complementary ribozyme and substrate sequences to form H1 and H2 helices. In the absence of chimeric ribozymes, chimeric substrates disappear from yeast over time through the endogenous snoRNA degradation pathway. Normal degradation of snoRNA occurs at a characteristic rate. When substrates are coexpressed with ribozymes, substrates disappear through the normal degradation pathway, as do all snoRNAs, but the substrates also can be destroyed Fedor, M.J., Westhof, E. Ribozymes: the first 20 years. Mol. Cell 10:703, 2002. Viruses as Molecular Building Blocks M.G. Finn, D.D. Díaz, W.G. Lewis, J.-C. Meng, S. Meunier, D. Prasuhn, S. Punna, K.S. Raja, Q. Wang tarting with plant and insect viruses that are innocuous to mammals but well understood structurally, our goals are to create entities useful for the detection and treatment of disease, for the construction of functional materials, and for the catal- S 26 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 ysis of important chemical reactions. We bring the techniques of both chemistry and biology to this endeavor, an appropriate strategy because the size of virus particles makes them a natural bridge between these worlds. The combination is powerful. In the past year we made the following major advances. F U N D A M E N TA L C H E M I S T R Y We developed 2 new methods for making chemical connections to viral coat proteins (and, by extension, many other biomolecules). The more exciting of these is the result of a collaboration with K.B. Sharpless and V.V. Fokin, the Skaggs Institute, in which we used a copper-catalyzed connection between azides and alkynes. C A R B O H Y D R AT E - D E C O R AT E D PA R T I C L E S We further explored the chemistry and biology of carbohydrate-decorated virus particles. We quantified the enhancements obtained in interactions with cellular receptors by displaying the binding agents in multiple copies over the viral surface. This “polyvalency” is at the heart of prospective uses of virus particles for drug delivery and tissue imaging. Our measurements indicate that the effective polyvalencies of virus particles are as good as those of the best organic polymers made for this purpose, and the viruses have other advantages relative to these known systems. In collaboration with S. Schmid and coworkers, The Scripps Research Institute, we showed endocytosis of virus particles into liver cells mediated by carbohydrate-specific receptors. PA R T I C L E S D E C O R AT E D W I T H A R G I N I N E – G LY C I N E – A S PA R T I C A C I D The tripeptide arginine–glycine–aspartic acid (RGD) is recognized by a large family of proteins (integrins) that are crucial to a wide variety of cellular recognition and signaling processes. Many RGD-receptor interactions are thought to involve the simultaneous contact of more than one RGD sequence with the target cell, and this polyvalency has been the subject of much investigation and debate. We expressed RGD sequences corresponding to those used by the common cold virus (adenovirus) on the surface of cowpea mosaic virus (CPMV) and found that these particles bind selectively to the cells that adenovirus itself targets. This work, performed in collaboration with G. Nemerow, The Scripps Research Institute, will be expanded in the coming year to include the chemical synthesis and attachment of active peptides. ANTIBODY INTERACTIONS AND THE IMMUNE RESPONSE We measured the response of the mouse immune system to CPMV and showed that the particle can be made less immunogenic by covering it with polymeric chains attached by chemical ligation. Novel polymer synthesis and attachment strategies are currently in development; our goal is to selective modulate or enhance the antigenicity of tailored virus particles. CPMV has been found in the blood, lymphatic system, spleen, and mucosal tissues of mice after oral administration, setting the stage for the use of CPMV as an oral delivery agent for compounds used to report on and treat disease. This work is done in collaboration with M. Manchester and coworkers, The Scripps Research Institute. N E W V I R U S P L AT F O R M S We performed the first surveys of the chemical stabilities and reactivities of 2 insect viruses: flock house virus and nudaurelia capensis ω virus. Both are potentially more useful than CPMV for biological and catalytic purposes because of the following properties. They are composed of 180 and 240, rather than 60, identical protein subunits, respectively, allowing a higher density of connection points to the structure. Mutants of these viruses can be generated in a matter of days, rather than weeks, as is the case with CPMV. Flock house virus has far fewer reactive groups on its exterior than does CPMV, and nudaurelia capensis ω virus undergoes a unique and dramatic structural change in response to pH. PUBLICATIONS Lewis, W.G., Shen, Z., Finn, M.G., Siuzdak, G. Desorption-ionization on silicon (DIOS) mass spectrometry: background and applications. Int. J. Mass Spectrom. 226:107, 2003. Raja, K.S., Wang, Q., Finn, M.G. Icosahedral virus particles as polyvalent carbohydrate display platforms. Chembiochem 4:1348, 2003. Raja, K.S., Wang, Q., Gonzalez, M.J., Manchester, M., Johnson, J.E., Finn, M.G. Hybrid virus-polymer materials, 1: synthesis and properties of PEG-decorated cowpea mosaic virus. Biomacromolecules 4:472, 2003. Ripka, A.S., Díaz, D.D., Sharpless, K.B., Finn, M.G. First practical synthesis of formamidine ureas and derivatives. Org. Lett. 5:1531, 2003. Smith, J.C., Lee, K.-B., Wang, Q., Finn, M.G., Johnson, J.E., Mrksich, M., Mirkin, C.A. Nanopatterning the chemospecific immobilization of cowpea mosaic virus capsid. Nano Lett. 3:883, 2003. Wang, Q., Chan, T., Hilgraf, R., Fokin, V.V., Sharpless, K.B., Finn, M.G. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 125:3192, 2003. Wang, Q., Raja, K.S., Janda, K.D., Lin, T., Finn, M.G. Blue fluorescent antibodies as reporters of steric accessibility in virus conjugates. Bioconjug. Chem. 14:38, 2003. Yao, S., Meng, J.-C., Siuzdak, G., Finn, M.G. New catalysts for the asymmetric hydrosilylation of ketones discovered by mass spectrometry screening. J. Org. Chem. 68:2540, 2003 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 27 Insights Into Protein Chemistry and Biology From Protein Structure E.D. Getzoff, M. Aoyagi, A.S. Arvai, D.P. Barondeau, R.M. Brudler, J.M. Castagnetto, D. Cerutti, T. Cross, M. DiDonato, E.D. Garcin, U.K. Genick, S.W. Hennessy, C. Hitomi, K. Hitomi, C.J. Kassmann, I. Li, S.J. Lloyd, M.E. Pique, R.J. Rosenfeld, M.E. Stroupe, M.M. Thayer, M.J. Thompson, J.L. Tubbs, T.I. Wood C H E M I C A L B I O L O G Y A N D R E G U L AT I O N O F N I T R I C OXIDE SYNTHASES e are studying nitric oxide synthases (NOSs), key therapeutic targets important for blood pressure regulation (endothelial NOS), neurotransmission (neuronal NOS), and the immune response (inducible NOS). These 3 similar, but differentially regulated, isozymes synthesize the diatomic molecule nitric oxide, which is paradoxically both a molecular signal (at low concentrations) and a cytotoxin (at high concentrations). Isozyme-specific NOS inhibitors are highly desirable for medicinal purposes and to advance our understanding of basic human physiology. Each of the 3 isozymes has 2 major modules: (1) a catalytic oxygenase with binding sites for heme, tetrahydrobiopterin, and substrate and (2) an electron-supplying reductase with binding sites for NADPH, FAD, and FMN. The 2 modules are connected by a central linker that binds the calcium-regulated protein calmodulin. Electron transfer from the reductase to the oxygenase and consequent production of nitric oxide are triggered by the binding of calmodulin. Our recent crystallographic structure of calcium-loaded calmodulin bound to a 20-residue peptide from the endothelial NOS linker reveals how regulatory phosphorylation can counteract this calmodulin activation (Fig. 1). The goals of our ongoing mutational, biochemical, and structural studies of NOS are to (1) determine how the many domains of the dimeric holoenzyme assemble, bind their cofactors, and perform complex catalytic and redox chemistry; (2) identify and apply the features distinguishing the 3 isozymes to the design of isozyme-specific inhibitors; and (3) understand the diverse regulatory mechanisms that differentially control the 3 isozymes. W ENZYME-COFACTOR INTERACTIONS New realms of activities are made available to proteins by bound cofactors, which include metal ions, vitamins, and other small molecules. Macrocyclic metal- F i g . 1 . The solvent-exposed threonine phosphorylation site (T495) on the endothelial NOS linker (dark gray ball-and-stick model) is surrounded by glutamic acid residues (E7, E11, E127, and E498) provided by both lobes of calmodulin (pale gray), explaining why phosphorylation here counteracts calmodulin activation of nitric oxide synthesis. chelating cofactors, such as vitamin B 12 (cobalamin), chlorophyll, heme, and siroheme, share a common framework constructed during the initial steps of the branched tetrapyrrole biosynthetic pathway. We determined crystallographic structures of (1) sulfite reductase, the enzyme that uses siroheme to prepare reduced sulfur for incorporation into biomolecules, and (2) siroheme synthase, the enzyme that generates the siroheme cofactor from uroporphyrinogen. With our high-resolution structures of the sulfite reductase hemoprotein, we measured the geometric distortions in the coupled siroheme and 4Fe4S cluster cofactors that tune their redox properties for enzyme function. Our crystallographic structures of siroheme synthase, in conjunction with biochemical results, yielded insights for the 4 separate reactions catalyzed by this enzyme: 2 distinct S-adenosyl-L-methionine–dependent transmethylations, NAD +-dependent dehydrogenation, and ferrochelation. We identified an unexpectedly phosphorylated serine residue (Fig. 2) poised to inhibit the dehydrogenation reaction and thus modulate metabolic flux between the siroheme and cobalamin biosynthetic pathways. Photoactive chromophores enable proteins to translate light energy into defined conformational changes 28 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 analyses suggest that this cryptochrome functions in light-dependent transcriptional regulation. For green and red fluorescent proteins, our high-resolution crystallographic structures provide insights into the mechanism by which 3 component amino acid residues are spontaneously converted after translation into fluorescent chromophores of different colors. We determined structures of trapped intermediates in fluorophore synthesis, identified features key to fluorophore formation and spectral tuning, and proposed a novel conjugation-trapping mechanism for fluorophore synthesis. MACROMOLECULAR ASSEMBLIES IN HUMAN H E A LT H A N D D I S E A S E F i g . 2 . Electron density contours (cages) reveal phosphorylated serine (Ser128, bottom) deep within the dehydrogenase/ferrochelatase active site of siroheme synthase. Located about 11 Å away from the NAD+ cofactor (top), this unexpected phosphoserine appears poised to inhibit the dehydrogenation reaction and thus modulate metabolic flux between the siroheme and cobalamin biosynthetic pathways. or fluorescence to send biological signals. We are studying the mechanisms of light-induced protein activities in the blue-light receptor photoactive yellow protein; the cryptochrome flavoproteins, which are components of circadian clocks in animals and humans; and the family of green and red fluorescent proteins used as biological markers. By combining ultrahigh-resolution crystallography of photoactive yellow protein with different types of computational chemistry, we defined mechanisms for photochemical tuning, elucidated intermediates in the light cycle, resolved controversies arising from spectroscopic studies, and identified active-site dynamics that favor photoisomerization of the chromophore. For cryptochrome, we sequenced a new gene, identified a new cryptochrome protein family, and determined the first crystal structure. The structure contains the redox-active FAD cofactor bound in an unusual U-shaped conformation with a surrounding positive electrostatic surface consistent with a function in DNA binding. Microarray We are studying the consequences for human health of appropriate macromolecular recognition and assembly. In collaboration with J. Tainer, the Skaggs Institute, we focus on enzymes (NOS and superoxide dismutase) that control reactive oxygen species and on the pilus virulence factors responsible for the attachment of pathogenic bacteria to human hosts. For human superoxide dismutase, we are investigating the mechanisms by which many different, naturally occurring, single-site mutations cause the fatal neurodegenerative disease amyotrophic lateral sclerosis. Through structural and biochemical studies of recombinantly produced superoxide dismutase proteins that incorporate these human genetic defects, we showed that the mutant proteins are architecturally destabilized and can form amyloidlike aggregates, resembling the aggregates found in the motor neurons of patients with amyotrophic lateral sclerosis. For pili and their component pilin proteins from bacteria that cause gonorrhea, meningitis, pneumonia, and cholera, we are characterizing how pili assemble, function, and help these bacteria evade the immune response in humans. PUBLICATIONS Aoyagi, M., Arvai, A.S., Tainer, J.A., Getzoff, E.D. Structural basis for endothelial nitric oxide synthase binding to calmodulin [published correction in EMBO J. 22:1234, 2003]. EMBO J. 22:766, 2003. Barondeau, D.P., Putnam, C.D., Kassmann, C.J., Tainer, J.A., Getzoff, E.D. Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures. Proc. Natl. Acad. Sci. U. S. A. 100:12111, 2003. Brudler, R., Hitomi, K., Daiyasu, H., Toh, H., Kucho, K., Ishiura, M., Kanehisa, M., Roberts, V.A., Todo, T., Tainer, J.A., Getzoff, E.D. Identification of a new cryptochrome class: structure, function, and evolution. Mol. Cell 11:59, 2003. Craig, L., Taylor, R.K., Pique, M.E., Adair, B.D., Arvai, A.S., Singh, M., Lloyd, S.J., Shin, D.S., Getzoff, E.D., Yeager, M., Forest, K.T., Tainer, J.A. Type IV pilin structure and assembly: x-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11:1139, 2003. DiDonato, M., Craig, L., Huff, M.E., Thayer, M.M., Cardoso, R.M., Kassmann, C.J., Lo, T.P., Bruns, C.K., Powers, E.T., Kelly, J.W., Getzoff, E.D., Tainer, J.A. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J. Mol. Biol. 332:601, 2003. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Getzoff, E.D., Gutwin, K.N., Genick, U.K. Anticipatory active-site motions and chromophore distortion prime photoreceptor PYP for light activation. Nat. Struct. Biol. 10:663, 2003. Meyer, T.E., Devanathan, S., Woo, T., Getzoff, E.D., Tollin, G., Cusanovich, M.A. Site-specific mutations provide new insights into the origin of pH effects and alternative spectral forms in the photoactive yellow protein from Halorhodospira halophila. Biochemistry 42:3319, 2003. Stroupe, M.E., Leech, H.K., Daniels, D.S., Warren, M.J., Getzoff, E.D. CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis. Nat. Struct. Biol. 10:1064, 2003. Sung, N.S., Gordon, J.I., Rose, G.D., Getzoff, E.D., Kron, S.J., Mumford, D., Onuchic, J.N., Scherer, N.F., Sumners, D.L., Kopell, N.J. Science education: educating future scientists. Science 301:1485, 2003. Thompson, M.J., Bashford, D., Noodleman, L., Getzoff, E.D. Photoisomerization and proton transfer in photoactive yellow protein. J. Am. Chem. Soc. 125:8186, 2003. Wei, C.C., Wang, Z.Q., Arvai, A.S., Hemann, C., Hille, R., Getzoff, E.D., Stuehr, D.J. Structure of tetrahydrobiopterin tunes its electron transfer to the heme-dioxy intermediate in nitric oxide synthase. Biochemistry 42:1969, 2003. Supramolecular Approaches to Drug Design, Cell Diagnostics, and Complex System Engineering 29 dynamics and coupling architecture is currently unanswered. This status is largely due to the lack of a powerful self-organized model system that is subject to de novo design, predictable topological organization, experimental characterization at the level of the kinetics of its individual pathways, and direct manipulation such that the overall effects in the system of the changes (e.g., input variations, network reorganization) can be rigorously studied and understood. We are attempting to bridge the gap between the theoretical graph-based theories of network organization and the experimental realm. We developed a system that is simple in its basic molecular components to allow rational design and networking predictions but large in sequence space to provide the basis for the construction of complex networks. The system is based on template-directed coiled-coil peptide fragment condensation reactions that are substrate selective in their ligase and replicase activities (Fig. 1). For our studies, we designed 81 peptides and calculated the free energies of all A 2B-type ensembles to determine plausible network connectivities via autocatalysis and cross-catalysis pathways. The figure shows clustered M.R. Ghadiri, M. Al-Sayah, G. Ashkenasy, A. Chavochi, J. Fletcher, V. Haridas, W.S. Horne, R. Jagasia, L. Leman, S. Rahimipour, M. Yadav e seek to develop novel chemical processes for use in the design of complex functional systems. Our multidisciplinary research program ranges from the design, prediction, and study of complex peptide networks to synthesis and selection of antimicrobial, anticancer, and optical cell diagnostic agents. These ongoing efforts are expected to contribute to a better understanding of the complex protein networks of living systems, assist in developing simple and effective cell diagnostic tools for early detection of disease states, and provide a diverse supramolecular therapeutic approach for treating cancer and combating multidrug-resistant microbial infections. W DESIGN OF COMPLEX SELF-ORGANIZED CHEMICAL NETWORKS In several recent theoretical studies, researchers probed the distinctive statistical properties of complex systems in social, technological, and biological networks. The results revealed some of the unifying principles that underlie the topology of complex self-organized networks. However, the central question of how interacting dynamic systems (whether animate or inanimate) can produce collective behavior from their individual F i g . 1 . Top, Schematic representation of the template-directed peptide fragment ligation. The electrophilic peptide fragment E i bearing a C-terminal thiolester moiety and the nucelophilic peptide N bearing an N-terminal cysteine residue can preorganize on the complementary peptide template Tj to form a coiled-coil ternary (not shown) or quaternary (depicted) complex. The productive juxtaposition and enhanced effective concentration of the reactive functionalities facilitate the Kent ligation process, proceeding through the transthiolester intermediate followed by intramolecular aminolysis, to give product T i. The reaction is autocatalytic when T i = T j and crosscatalytic when T i ≠ T j. Bottom, The experimentally derived network architecture. The arrows (edges) designate template-assisted ligation pathways pointing from the template to the product. Numbers along the edges are the theoretical –∆∆G free energy values (kcal mol–1). 30 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 nodes with an overall hierarchical architecture. Nine nodes were selected for experimental evaluation of their capacity to establish the predicted network connectivity. The resulting self-organized chemical network has 25 directed edges, including 3 autocatalytic processes, in good agreement with predictions. Moreover, we showed that by varying the system inputs (substrates and templates), we can alter its network topology, even to the extremes of turning pathways on or off. In our opinion, the system described provides the first tool for designing and analyzing complex systems in a highly rational and informative manner. This approach can offer a means to study and to better understand the emergent, collective behaviors of networks. New Vistas in Immunopharmacotherapy SELF-ASSEMBLING PEPTIDE NANOTUBES AS A ANTIBODY LIBRARIES NEW CLASS OF CELL DIAGNOSTIC, ANTIMICROBIAL, hage display antibody technology is a powerful method for the design and discovery of antibodies that bind targets with high specificity and affinity. Because the use of combinatorial antibody libraries displayed on the surface of phage particles is an entirely in vitro procedure, it obviates animal-, labor-, and time-intensive immunization and hybridoma protocols required to isolate monoclonal antibodies. We extended the usefulness of phage display methods, most notably by developing fully human single-chain Fv (scFv) and Fab antibody library formats displayed on phage. These libraries allow the direct selection of antigenspecific, fully human antibodies that can be moved more expeditiously into clinical trials. The phage coat proteins pIII and pVIII have been used extensively for the display of antibody libraries; pIII in particular has been used to select numerous highly specific human antibodies, which continue to be the focus of a great deal of research in biotechnology and medicine. We expanded the sphere of phage display antibody technology to develop antibody libraries that are combinatorial not just in composition but also in conformational space. We accomplished this step by using pVII and pIX, which are constitutively separate but physically near each other, for the simultaneous display of scFv variable heavy-chain region (V H) and variable light-chain region (VL) chains. This past year, we extended the usefulness of in vitro antibody library design by developing a novel phage display format in which only the pIX coat protein is used. In our model studies, this display system allowed the selection of highly specific antibodies that recognized a number of complex antigens (e.g., cholera toxin and ricin) important in both medicine and defense against AND ANTICANCER AGENTS Supramolecular systems constitute an emerging frontier in drug design and diagnostics. The capacity for self-organization and cellular self-targeting offered by self-assembling peptide nanotubes can provide a range of functional attributes that are not generally feasible by the traditional molecular approaches. Peptide nanotubes are thought to operate on targeted cellular membranes by membrane absorption of small cyclic peptides followed by self-assembly into hole-punching supramolecular tubular complexes. The class of selfassembling cyclic peptides can provide an unprecedented level of membrane discrimination via the capability to sense and respond to the membrane environment. We exploited these characteristics to design self-assembling peptide nanotubes with anticancer and potent antimicrobial activities against multidrug-resistant infections. In our studies, we use various combinatorial cyclic peptide library approaches to select and optimize desired biological activities against a variety of cellular targets. Using fluorescent-labeled cyclic peptide libraries, we exploited the capacity of self-assembling peptide nanotubes to discriminate cell membranes to design optical systems for detecting pathogens. The new avenue of research in the design of photonic cell recognition agents is expected to yield a series of diagnostic tools that complement our ongoing efforts directed at developing antimicrobial and anticancer therapeutic agents. K.D. Janda, J.-M. Ahn, J. Ashley, G. Boldt, R. Carrera, Y. Chen, B. Clapham, A. Coyle, T. Dickerson, Y. Ding, L. Eubanks, T. Fujimori, R. Galve, C. Gambs, C. Gao, T. Hoffman, L. Hom, G. Kaufmann, Y.-S. Kim, E. Laxman, B.-S. Lee, S.-H. Lee, S.-J. Lee, M. Lillo, S. Mahajan, H. Matsushita, M. Matsushita, J. McDunn, G. McElhaney, J. Mee, M. Meijler, J. Moss, J.-I. Park, N. Reed, A. Shafton, C. Sun, R. Troseth, A.D. Wentworth, P. Wentworth, Jr., P. Wirsching, Y. Xu, N. Yamamoto, K. Yoshida, B. Zhou IN VITRO DESIGN OF HUMAN P THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 bioterrorism. We are investigating the scope and advantages of this new antibody display technology as an adjunct and alternative to existing methods. In parallel with our work on the basic science of antibody phage display technology, we continue to explore new biomedical applications for in vitro antibody libraries. For example, we used an in vitro human scFv antibody library to select for antibodies that not only specifically bound to but were internalized into cancer cells. This finding is clinically relevant, because many of the side effects of cancer chemotherapy stem from the inherent nonspecificity of the cytotoxic drugs used to kill cancer cells. By attaching such drugs to cancer cell–specific, internalizing antibodies, the deleterious side effects of many of these chemotherapeutic agents could be attenuated, improving the prognoses for cancer patients. Although attaching a single drug molecule to a cancer cell–specific, internalizing antibody clearly is of therapeutic interest, we prepared dendritic carriers for attaching multiple drug copies to a single scFv antibody (Fig. 1). Through the use of these hybrid macromolecules, which harness the exquisite specificity and drug delivery potential of these antibodies to the potent toxicity of synthetic chemotherapeutic dendrimers, the outlook for cancer chemotherapy in the coming years could be all the brighter. F i g . 1 . Preparation of a site specifically modified scFv antibody– drug immunoconjugate by using a multifunctional dendritic drug carrier. VA C C I N AT I O N F O R S E P T I C S H O C K Septic shock is 1 of the 10 leading causes of infant and adult mortality in the United States. According to the Centers for Disease Control and Prevention, this condition was directly linked to more than 30,000 deaths in 1999 alone. The prime pathogenic determinant of septic shock is lipopolysaccharide, a class of oligosaccharides liberated by lysis of bacteria that have multiple O-linked fatty acyl substituents. The Escherichia coli isoform of lipid A is the most well-studied form of lipopolysaccharide and is responsible for the toxic effects associated with most documented cases of septic shock. 31 One promising therapeutic tactic is use of a passive vaccine, that is, monoclonal antibodies that could, in principle, be used to bind lipid A and mediate its clearance from the body during instances of infection. Although this strategy showed promise at an early stage of research, it was not effective in clinical trials, underscoring the need for alternative therapeutic strategies. We developed an immunotherapeutic approach for treatment of septic shock that involves the use of an active vaccine, a designed synthetic variant of lipid A (compound 1 in Fig. 2), that could induce an innate immune response to future exposure to lipopolysaccharide, thereby abrogating the toxic effects. We designed F i g . 2 . Bisphosphonate TSAs 2a and 2b were synthesized as immunogens for active immunization to treat lipid A–mediated endotoxicosis. The native E coli lipid A structure is depicted in 1. a structural homolog of E coli lipid A with 2 key features. First, a bisphosphonate transition-state analog (TSA) motif, a mimic for hydroxide-catalyzed ester hydrolysis, was incorporated in place of the 2 acyloxyacyl motifs present in the native lipid A structure. This modified structure was used to elicit antibodies that not only bound lipid A but also catalytically deactivated it through catalysis of base-catalyzed fatty ester hydrolysis. Second, 2 different immunostimulatory linkers were used to attach the final TSA structure to a carrier protein for immunization. These linkers were included to circumvent the notoriously low immunogenicity of carbohydrate-based antigens, which would otherwise have severely hampered the selection of a lipid A–specific antibody binder or catalyst. Immunization of 3 different mouse strains with compounds 2a and 2b (Fig. 2), as conjugates to the carrier protein keyhole limpet hemocyanin (KLH), led to quantitatively different immune responses, suggesting that the choice of linker directly affected the immunogenicity of the TSA structure. These differences notwithstanding, the obtained antibodies were qualitatively similar, as evinced by significant cross-reactivity of the antibodies elicited to immunogens 2a and 2b. To investigate the protective effect of the TSA conjugates, we gave the immunized mice sublethal bolus 32 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 doses of lipid A. We measured the efficacy of the active vaccine by using the known upregulatory effect of lipid A on the production of TNF-α. Normalizing to TNF-α production in control mice not immunized with KLH conjugates of 2a or 2b, we observed between a 4- to 20-fold reduction in TNF-α production (depending on the mouse strain) after lipid A challenge in mice protected by previous immunization with KLH conjugates of 2a or 2b. It is unclear as yet whether this immunoprotection against the toxic effects of E coli lipid A is due to the activity of antibody catalysts or binders. Nonetheless, these results indicate the therapeutic potential of rationally designed phosphonate TSAs for harnessing the native immune system for immunotherapy of septic shock in an active vaccine program. We are isolating and characterizing the responsible antibodies for mechanistic characterization as quick-acting therapeutic entities themselves in a complementary passive vaccine approach. IMMUNOPHARMACOTHERAPY FOR NICOTINE ABUSE Cigarette smoking is still the leading cause of avoidable death due to cancer worldwide; as such, it has the unique distinction of being an elective malady of pandemic proportions. Increasingly, evidence suggests that chronic use of cigarettes and other tobacco products is due in large part to the addictive psychoactive and reinforcing effects of nicotine on the brain. Although socalled nicotine replacement therapies have grown in popularity in recent years, these are not true therapies per se; rather they are merely alternative forms of nicotine delivery. Because these strategies do not attack the underlying problem, the neurostimulatory effects of nicotine and concomitant user dependence on it, they have done little to decrease tobacco dependence on the whole. In recent years, we and other researchers have explored immunopharmacotherapy as an alternative route to tackle the problem of chronic nicotine dependence. Several different approaches have been used to elicit antibodies that bind and thereby block nicotine from exerting its neurostimulatory effects on the brain. Although the structure of nicotine does not suggest that it poses any significant difficulties in the generation of antibody binders, all synthetic nicotine haptens prepared thus far have unexplainably fallen short of their promise. This situation led us to explore the possibility of some fundamental, yet hidden, chemical foundation for this problem. Recently, new data indicated that in aqueous solution, nicotine exists in multiple conformations arising from rotation about the pyrrolidine-pyridine linkage axis. This finding is of key relevance for our research because it suggests a possible rationale for the unvaryingly low affinity of nicotine-binding antibodies, irrespective of protein conjugation chemistry. Whereas we and others viewed nicotine as a static, torsionally constrained 2-dimensional molecule, the immune system constantly samples all accessible solution-phase conformations. In exchange for high specificity and affinity, immunization with nicotine conjugates prepared to date—by us as well as by others—inevitably leads to a heterogenous antibody response. The isolated antibodies are characterized by low conformational specificity and an affinity that is averaged over a broad sample space of nicotine conformers. We surmised that the solution-phase conformations of nicotine might be partly responsible for our otherwise unexplainably low success in generating nicotinebinding antibodies with our first-generation nicotine hapten (compound 3 in Fig. 3). Therefore, we prepared conformationally constrained nicotine analogs 4 and 5 (Fig. 3) for immunization to elicit improved antibody binders. The haptens were attached to KLH by using the same linkage chemistry previously used for 3 to allow meaningful side-by-side comparison. We were gratified to find that the serum antibody titers resulting from immunization with haptens 4 and 5 were 7to 8-fold higher than those obtained with the conformationally free hapten 3. F i g . 3 . First-generation nicotine hapten 3 and constrained trans analogs 4 and 5 used to elicit improved nicotine-binding antibodies. We are isolating and characterizing the affinity of monoclonal antibodies generated in response to these conformationally constrained nicotine haptens. We envision that these antibodies could be useful in improving the efficacy of immunopharmacotherapy strategies as treatments for nicotine addiction. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 PUBLICATIONS Ahn, J.-M., Wentworth, P., Jr., Janda, K.D. Soluble polymer-supported convergent parallel library synthesis. Chem. Commun. (Camb.) 4:480, 2003. Cannizzaro, C.E., Ashley, J.A., Janda, K.D., Houk, K.N. Experimental determination of the absolute enantioselectivity of an antibody-catalyzed Diels-Alder reaction and theoretical explorations of the origins of stereoselectivity. J. Am. Chem. Soc. 125:2489, 2003. Clapham, B., Lee, S.-H., Koch, G., Zimmermann, J., Janda, K.D. The preparation of polymer bound β-ketoesters and their conversion into an array of oxazoles. Tetrahedron Lett. 43:5407, 2002. 33 Wentworth, P., Jr., McDunn, J.E., Wentworth, A.D., Takeuchi, C., Nieva, J., Jones, T., Bautista, C., Ruedi, J.M., Gutierrez, A., Janda, K.D., Babior, B.M., Eschenmoser, A., Lerner, R.A. Evidence for antibody-catalyzed ozone formation in bacterial killing and inflammation. Science 298:2195, 2002. Wentworth, P., Jr., Wentworth, A.D., Zhu, X., Wilson, I.A., Janda, K.D., Eschenmoser, A., Lerner, R.A. Evidence for the production of trioxygen species during antibody-catalyzed chemical modification of antigens. Proc. Natl. Acad. Sci. U. S. A. 100:1490, 2003. Zhu, X.Y., Heine, A., Monnat, F., Houk, K.N., Janda, K.D., Wilson, I.A. Structural basis for antibody catalysis of a cationic cyclization reaction. J. Mol. Biol. 329:69, 2003. Delgado, M., Janda, K.D. Polymeric supports for solid phase organic synthesis. Curr. Org. Chem. 6:1031, 2002. Dickerson, T.J., Janda, K.D. A previously undescribed chemical link between smoking and metabolic disease. Proc. Natl. Acad. Sci. U. S. A. 99:15084, 2002. Dickerson, T.J., Janda, K.D. Glycation of the amyloid β-protein by a nicotine metabolite: a fortuitous chemical dynamic between smoking and Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 100:8182, 2003. Dickerson, T.J., Reed, N.N., Janda, K.D. Soluble polymers as scaffolds for recoverable catalysts and reagents. Chem. Rev. 102:3325, 2002. Gambs, C., Dickerson, T.J., Mahajan, S., Pasternack, L.B., Janda, K.D. High-resolution diffusion-ordered spectroscopy to probe the microenvironment of JandaJel and Merrifield resins. J. Org. Chem. 68:3673, 2003. Gao, C., Mao, S., Ditzel, H.J., Farnaes, L., Wirsching, P., Lerner, R.A., Janda, K.D. A cell-penetrating peptide from a novel pVII-pIX phage-displayed random peptide library. Bioorg. Med. Chem. 10:4057, 2002. Gao, C., Mao, S., Kaufmann, G., Wirsching, P., Lerner, R.A., Janda, K.D. A method for the generation of combinatorial antibody libraries using pIX phage display. Proc. Natl. Acad. Sci. U. S. A. 99:12612, 2002. Gao, C., Mao, S., Ronca, F., Zhuang, S., Quaranta, V., Wirsching, P., Janda, K.D. De novo identification of tumor-specific internalizing human antibody-receptor pairs by phage-display methods. J. Immunol. Methods 274:185, 2003. Jones, L.H., Altobell, L.J. III, MacDonald, M.T., Boyle, N.A., Wentworth, P., Jr., Lerner, R.A., Janda, K.D. Active immunization with a glycolipid transition state analogue protects against endotoxic shock. Angew. Chem. Int. Ed. 41:4241, 2002. Lee, K.J., Mao, S., Sun, C., Gao, C., Blixt, O., Arrues, S., Hom, L.G., Kaufmann, G.F., Hoffman, T.Z., Coyle, A.R., Paulson, J., Felding-Habermann, B., Janda, K.D. Phage-display selection of a human single-chain Fv antibody highly specific for melanoma and breast cancer cells using a chemoenzymatically synthesized GM3carbohydrate antigen. J. Am. Chem. Soc. 124:12439, 2002. Lee, S.-H., Clapham, B., Koch, G., Zimmermann, J., Janda, K.D. Rhodium carbenoid N-H insertion reactions of primary ureas: solution and solid-phase synthesis of imidazolones. Org. Lett. 5:511, 2003. Liu, C., Sun, C., Huang, H., Janda, K., Edgington, T. Overexpression of legumain in tumors is significant for invasion/metastasis and a candidate enzymatic target for prodrug therapy. Cancer Res. 63:2957, 2003. Meijler, M.M., Matsushita, M., Altobell, L.J. III, Wirsching, P., Janda, K.D. A new strategy for improved nicotine vaccines using conformationally constrained haptens. J. Am. Chem. Soc. 125:7164, 2003. Redwan, el-R.M., Larsen, N.A., Zhou, B., Wirsching, P., Janda, K.D., Wilson, I.A. Expression and characterization of a humanized cocaine-binding antibody. Biotechnol. Bioeng. 82:612, 2003. Shimomura, O., Clapham, B., Spanka, C., Mahajan, S., Janda, K.D. Application of microgels as polymer supports for organic synthesis: preparation of a small phthalide library, a scavenger, and a borohydride reagent. J. Comb. Chem. 4:436, 2002. Sun, C., Wirsching, P., Janda, K.D. Enabling scFvs as multi-drug carriers: a dendritic approach. Bioorg. Med. Chem. 11:1761, 2003. Wang, Q., Raja, K.S., Janda, K.D., Lin, T., Finn, M.G. Blue fluorescent antibodies as reporters of steric accessibility in virus conjugates. Bioconjug. Chem. 14:38, 2003. Catalytic Nucleic Acids for Treating the Molecular Basis of Disease G.F. Joyce, J.T. Hillman, R.M. Kumar, N. Paul, W.M. Shih n recent years, nucleic acid molecules have shown promise for the treatment of a variety of diseases. These molecules can be used to downregulate the expression of disease-related genes by targeting either the genetic material itself or the corresponding RNA or protein product. Antisense agents are one type of therapeutic nucleic acid. These agents usually consist of short, single-stranded DNA molecules that bind to particular cellular RNAs, leading to inactivation of the RNAs. Small interfering RNAs are another type of potential therapeutic nucleic acid that bind to specific cellular RNAs, resulting in cleavage of those RNAs through the “RNA interference” pathway. We are developing catalytic nucleic acids, composed of either RNA or DNA, that bind to target RNAs and inactivate them by carrying out a cleavage reaction that destroys the target. We obtain these catalysts through a process of test-tube evolution that mimics natural evolution but occurs much more rapidly and according to selection constraints that we impose. I R N A - C L E AV I N G D N A E N Z Y M E S We have developed DNA enzymes that can be made to cleave almost any targeted RNA under cellular conditions. Our most widely used DNA enzyme is the “10-23” motif, which contains approximately 30 deoxynucleotides and operates with high catalytic efficiency and sequence specificity. We and scientists in many other laboratories have used this enzyme to cleave disease-related mRNAs, both in cultured cells and in whole animals. In collaboration with R. Smith, a Skaggs Clinical Scholar, we focused on the DNA-catalyzed cleavage of 34 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 superoxide dismutase (SOD1) mRNA as a potential treatment for amyotrophic lateral sclerosis. Recent studies indicated that amyotrophic lateral sclerosis is caused by mutation or inappropriate expression of SOD1, suggesting that decreased expression of SOD1 might have therapeutic benefit. We tested the ability of the 10-23 DNA enzyme to inhibit expression of mutant human SOD1 mRNA in transgenic rats, seeking to delay the rapid onset of disease in these animals. Two different DNA enzymes were constructed and formulated for delivery to the cerebral ventricles by continuous infusion. The compounds were delivered at a dose of 0.1 mg/kg per day for 30 days. These animals were compared with control animals that received the same volume of isotonic sodium chloride solution. Both the treated and the untreated animals had severe muscle wasting by the end of the treatment period, indicating that the DNA enzymes had no therapeutic benefit. Unlike the situation in humans with amyotrophic lateral sclerosis, the transgenic rats express high concentrations of mutant human SOD1 mRNA and succumb to the disease by 4 months of age. This model of the disease may be an overly aggressive one and one that provides little opportunity for validation of treatments that depend on downregulation of the overexpressed SOD1 mRNA. Currently, using both normal and transgenic rats, we are examining whether the DNA enzymes can significantly reduce mRNA levels. length. The other 5 struts are based on double crossovers, which have the structure of 2 adjacent double helices that are joined at 2 crossover points. Each of these motifs is about twice as rigid as a simple DNA double helix, lending stiffness to the overall octahedron. The structure of the DNA octahedron was confirmed by using electron cryomicroscopy. Approximately 1000 raw images were obtained and averaged to produce the 3-dimensional reconstruction shown in Figure 1. Even though the overall structure has a regular appearance at this resolution (~30 Å), each strut has a different sequence and therefore is independently addressable by other molecules that bind DNA in a sequence-specific manner. The interior of the octahedron has a diameter of about 14 nm, and the 8 triangular “windows” have an opening of about 8 nm. Thus, it should be possible to trap macromolecules that are between 8 and 14 nm in size within the cavity of the octahedron. Furthermore, because it can be constructed from a long, single-stranded DNA molecule, the DNA octahedron can be cloned into bacteria or other cells, where it can be amplified and produced in large quantity. DNA-BASED NANOTECHOLOGY Reader, J.S., Joyce, G.F. A ribozyme composed of only two different nucleotides. Nature 420:841, 2002. We recently began a project to construct rigid DNAbased nanostructures that could be used to encapsulate macromolecules or to direct the assembly of materials on a submicrometer scale. We designed and synthesized a 1.7-kb, single-stranded DNA molecule that folds into a regular octahedron structure upon heating and cooling. The octahedron consists of 12 struts, each 14 nm long, that are joined together at 6 four-way junctions (Fig. 1). Seven of the struts are based on paranemic crossovers, which have the structure of 2 adjacent double helices that exchange strands 6 times along their PUBLICATIONS Kuhns, S.T., Joyce, G.F. Perfectly complementary nucleic acid enzymes. J. Mol. Evol. 56:711, 2003. Kumar, R.M., Joyce, G.F. A modular, bifunctional RNA that integrates itself into a target RNA. Proc. Natl. Acad. Sci. U. S. A. 100:9738, 2003. Shih, W.M., Quispe, J.D., Joyce, G.F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature, in press. Biomolecular Computing, Catalytic Antibodies, Organic Synthesis, and Synthetic Enzymes E. Keinan, H.C. Lo, H. Han, S. Sasmal, M. Arifuddin, A. Brik, S. Saphier, G. Sklute, N. Metanis, M. Soreni, D. Vebenov, L. Kosoy, M.K. Sinha, A. Alt, I. Ben-Shir, R. Piran, P.E. Dawson BIOMOLECULAR COMPUTING F i g . 1 . Structure of a DNA octahedron based on 3-dimensional reconstruction of images obtained by electron cryomicroscopy. The single-stranded DNA folds into a regular octahedron upon heat denaturation and gradual cooling. lectronic computers can only process data encoded electronically, yet data for many valuable computations occur naturally as biomolecules. For example, modern medicine often involves procedures that can be viewed as computational processes, with molecules obtained from a patient as input E THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 and drug molecules to be administered as output. In collaboration with E. Shapiro and Z. Livneh, Weizmann Institute of Science, and Y. Shoham, Technion, Haifa, Israel, we developed a biomolecular computing machine that is a task-oriented, nanoscale, autonomous programmable computer made of molecules (Fig. 1). It can process biomolecules as data and produce biomolecules as output. We constructed in vitro biomolecular finite automatons that compute autonomously when their “hardware,” “software,” and “input” are mixed in solution. The hardware consists of a restriction nuclease and a ligase. Programming the device amounts to choosing the transition molecules (encoded by double-stranded DNA) to be mixed. F i g . 1 . A biomolecular computing machine made of molecules. The hardware consists of a restriction nuclease and a ligase; the input, transition molecules (software), and detection molecules are all made of double-stranded DNA. Each automaton processes an input double-stranded DNA molecule via a cascade of hybridization, ligation, and restriction cycles, producing an output doublestranded DNA molecule that encodes the final state of the computation. The computation runs at a combined rate of 109 transitions per second with accuracy greater than 99.8% per transition. C ATA LY T I C A N T I B O D I E S In our studies on catalytic antibodies, we focus on antibody-catalyzed photochemical reactions, oxidation reactions, and organometallic reactions, as illustrated in the following examples. Although the solution photochemical reaction of ketone 1 (Fig. 2) yields only the cleavage products 2 and 3, in the presence of 20F10, an antibody to 5a 35 F i g . 2 . The photochemical Norrish type II reaction of ketone 1 produces in solution the cleavage products 2 and 3. Antibody 20F10, which was elicited against a mixture of 5a and 5b, catalyzes enantioselective formation of cis-cyclobutanol (4). and 5b, a Norrish type II reaction occurred. leading to the selective formation of cis-cyclobutanol (compound 4 in Fig. 2). Furthermore, the fact that compound 4, which consists of 2 asymmetric centers, is obtained as a single diastereomer, makes this photoproduct a valuable building block for the synthesis of natural products. Other photochemical reactions catalyzed by 20F10 afford products of cyclopropanol derivatives from 1,3diaryl-1-propanones and formation of phenanthrenes from substituted stilbenes. In addition, an aldolase antibody, 24H6, obtained by immunization with large 1,3 diketone haptens, has an active-site lysine residue with a perturbed pK a of 7.0. This antibody catalyzes both the aldol addition and the retrograde aldol fragmentation with a broad range of substrates that differ structurally from the haptens. This antibody also catalyzes the oxidation of α-hydroxyketones to α-diketones and deuterium exchange at the α position of many ketones and aldehydes. During our ongoing efforts to develop catalytic antibodies for organometallic reactions, we discovered a new chemistry of platinum. A novel, air- and moisturestable dihydrido(methyl)platinum(IV) complex, TpPtH2Me (compound 7 in Fig. 3), was formed as the sole product by the reaction of TpPtMeCO (compound 6 in Fig. 3) with water. We found that although TpPtH 2Me has a high energy barrier for the liberation of methane, it F i g . 3 . Isotopic scrambling of the methylplatinum complex 7 proceeds via a σ-methane intermediate. 36 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 readily forms a σ-methane platinum(II) complex. This complex is the first example of a hydridoalkyl metal complex that undergoes isotopic scrambling at elevated temperatures without concomitant liberation of either alkane or dihydrogen. Later, we discovered that the methyl protons in compound 6 underwent proton exchange with the hydrogen atoms in water molecules. The proton-exchange reaction was proposed to proceed via a pathway similar to the catalytic triad found in natural enzymes. ORGANIC SYNTHESIS The single-step, tandem oxidative polycyclization reaction with rhenium(VII) reagents is a powerful method by which polyene alcohols can be converted into polytetrahydrofuran products in a single step. A set of rules has been deduced to predict the product’s configuration in this reaction. Annonaceous acetogenins, particularly those with adjacent bis-tetrahydrofuran rings, have remarkable cytotoxic, antitumor, antimalarial, immunosuppressive, pesticidal, and antifeedant activities. We developed synthetic approaches that can be used to generate chemical libraries of stereoisomeric acetogenins. These efforts resulted in the total synthesis of several naturally occurring acetogenins, including asimicin, bullatacin, trilobacin, rolliniastatin, solamin, reticulatacin, rollidecins C and D, goniocin, cyclogoniodenin, and mucocin, and many nonnatural stereoisomers. The 34-hydroxyasimicin and its derivatives were synthesized for photoaffinity labeling studies of bovine mitochondrial NADH-ubiquinone oxidoreductase (complex I). These studies may shed light on the structure and function of this intricate enzyme and on the origin of the high antitumor activity of the annonaceous acetogenins. This work is being done in collaboration with S.C. Sinha, the Skaggs Institute. SYNTHETIC ENZYMES Efforts to generate new enzymatic activities from existing protein scaffolds may not only provide biotechnologically useful catalysts but also lead to a better understanding of the natural process of evolution. We profoundly changed the catalytic activity and mechanism of 4-oxalocrotonate tautomerase via a rationally designed single amino acid substitution that corresponds to a mutation in a single base pair. Although the wildtype enzyme catalyzes only the tautomerization of oxalocrotonate to 2-oxo-3E-hexenedioate, the P1A mutant catalyzes 2 reactions: the original tautomerization reaction via a general acid-base mechanism and the decarboxylation of oxaloacetate via a nucleophilic mechanism. The observation that a single catalytic group in an enzyme can catalyze 2 reactions by 2 different mechanisms supports the theory that new enzymatic activity can evolve in a continuous manner. Highly evolved enzymes are optimized not only to catalyze a desired reaction but also to avoid undesired processes. We showed that P1A, which catalyzes the isomerization of 4-oxalocrotonate tautomerase to 2-oxo3E-hexenedioate, also undergoes specific 1,4-addition to the tautomerization product to form a stable covalent adduct. This research on synthetic enzymes is being done in collaboration with P.E. Dawson, the Skaggs Institute. PUBLICATIONS Han, H., Sinha, M.K., D’Souza, L., Keinan, E., Sinha S.C. Total synthesis of 34hydroxy asimicin and its photoactive derivative for affinity labeling of the mitochondrial complex I. Chemistry, in press. Keinan, E., Sinha, S.C. Oxidative polycyclizations with rhenium(VII) oxides. Pure Appl. Chem. 74:93, 2002. Lo, H.C., Iron, M.A., Martin, J.M.L., Keinan, E. Organometallic catalytic triad: proton walk in aqueous platinum complex via a sticky methane. J. Am. Chem. Soc., in press. Saphier, S., Sinha, S.C., Keinan, E. Antibody-catalyzed enantioselective Norrish type II cyclization. Angew. Chem. Int. Ed. 42:1378, 2003. Sklute, G., Oiserowitz, R., Shulman, H., Keinan, E. Antibody-catalyzed benzoin oxidation as a mechanistic probe for nucleophilic catalysis by an active-site lysine. Chemistry, in press. Metabolite-Initiated Protein Misfolding J.W. Kelly, S. Deechongkit, M.A. Dendle, T. Foss, D. Fowler, K. Frankenfeld, N. Green, A. Hurshman, M.B. Huff, S. Johnson, H.-J. Lim, E. Powers, A. Sawkar, Y. Sekijima, S. Werner, L. Wiseman, S.-L. You, Q. Zhang he central theme of our research under the auspices of The Skaggs Institute for Chemical Biology during the past year was the initiation of new projects related to human health that if successful would generate sufficient preliminary results to make the projects competitive for funding from the National Institutes of Health. Two of the projects proposed 2 years ago resulted in highly relevant discoveries. One of the discoveries was patented and should be translatable to the clinic. In 1972, C.B. Anfinsen won the Nobel Prize for showing that the fold of a protein is specified by the protein’s primary amino acid sequence. This tenet T THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 makes it difficult to rationalize how the same amino acid sequence can adopt a native structure in most persons and amyloidogenic structures in patients with Alzheimer’s disease in what appears to be a common extracellular environment. One possibility is that a unique posttranslational modification makes the β-amyloid peptide amyloidogenic, but evidence for this possibility is sparse. In work supported by the Skaggs Institute, and in collaboration with R.A. Lerner, The Scripps Research Institute, we found that the newly discovered abnormal metabolites associated with chronic inflammation modify β-amyloid and other amyloidogenic polypeptides in a covalent fashion, initiating the amyloidogenesis of the peptides under physiologic concentrations. A subset of oxidative metabolites containing an aldehyde group is unique in that the metabolites enable the attachment of a large hydrophobic structure to polypeptides by formation of a Schiff base (a covalent bond), very rapidly initiating aggregation, a process that typically requires the formation of an energetically unfavorable nucleus. This research provides evidence that the amyloid diseases associated with β-amyloid and transthyretin, which do not involve any known mutations—the socalled sporadic amyloid diseases—can be initiated by the covalent attachment of a small number of aberrant metabolites. Metabolite-initiated misfolding may also explain the onset of familial amyloidosis of the Finnish type caused by gelsolin amyloidogenesis. We are optimistic that this finding could be the long-sought explanation for most human amyloid diseases in which the cause of the disease is not linked to a mutation. In a collaboration with D. Burton and A. Williamson, The Scripps Research Institute, we are studying the mechanism of how an antibody they produced cures systemic prion disease, that is, how it prevents the conversion of the prion protein from its normal functional conformation into the disease-associated scrapie conformation. Although much remains to be accomplished, these results are encouraging and provide hope that small molecules may be discovered that function similarly, allowing the brain component of prion diseases to be ameliorated. PUBLICATIONS Calarese, D.A., Scanlan, C.N., Zwick, M.B., Deechongkit, S., Mimura, Y., Kunert, R., Zhu, P., Wormald, M.R., Stanfield, R.L., Roux, K.H., Kelly, J.W., Rudd, P.M., Dwek, R.A., Katinger, H., Burton, D.R., Wilson, I.A. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065, 2003. Cohen, F., Kelly, J.W. Therapeutic approaches to protein-misfolding diseases. Nature 426:905, 2003. 37 Colfer, S., Kelly, J.W., Powers E.T. Factors governing the self-assembly of a β-hairpin peptide at the air-water interface. Langmuir 19:1312, 2003. DiDonato, M., Craig, L., Huff; M.E., Thayer, M.M., Cardosa, R.M.F., Kassmann, C.J., Lo, T.P., Bruns, C.K., Powers, E.T., Kelly, J.W., Getzoff, E.D., Tainer, J.A. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization [published correction in J. Mol. Biol. 334:175, 2003]. J. Mol. Biol. 332:601, 2003. Ferrao-Gonzales, A.D., Palmieri, L., Valory, M., Silva, J.L., Lashuel, H., Kelly, J.W., Foguel, D. Hydration and packing are crucial to amyloidogenesis as revealed by pressure studies on transthyretin variants that either protect or worsen amyloid disease. J. Mol. Biol. 328:963, 2003. Foguel, D., Suarez, M.C., Ferrao-Gonzales, A.D., Porto, T.C.R., Palmieri, L., Einsiedler, C.M., Andrade, L.R., Lashuel, H.A., Lansbury, P.T., Kelly, J.W., Silva, J.L. Dissociation of amyloid fibrils of α-synuclein and transthyretin by pressure reveals their reversible nature and the formation of water-excluded cavities. Proc. Natl. Acad. Sci. U. S. A. 100:9831, 2003. Green, N.S., Palaninathan, S.K., Sacchettini, J.C., Kelly, J.W. Synthesis and characterization of potent bivalent amyloidosis inhibitors that bind prior to transthyretin tetramerization. J. Am. Chem. Soc. 125:13404, 2003. Hammarström, P., Sekijima, Y., White, J.T., Wiseman, R.L., Lim, A., Costello, C.E., Atland, K., Garzuly, F., Budka, H., Kelly, J.W. D18G transthyretin is monomeric, aggregation prone, and not detectable in plasma and cerebrospinal fluid: a prescription for central nervous system amyloidosis? Biochemistry 42:6656, 2003. Hammarström, P., Wiseman, R.L., Powers, E.T., Kelly, J.W. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science 299:713, 2003. Huff, M.E., Page, L., Balch, W.E., Kelly, J.W. Gelsolin domain 2 Ca2+ affinity determines susceptibility to furin proteolysis and familial amyloidosis of Finnish type. J. Mol. Biol. 334:119, 2003. Kelly, J.W. Towards an understanding of amyloidogenesis. Nat. Struct. Biol. 9:323, 2002. Kelly, J.W., Balch, W.E. Amyloid as a natural product. J. Cell Biol. 161:461, 2003. Lu, J.R., Perumal, S., Powers, E.T., Kelly, J.W., Webster, J.R.P., Penfold, J. Adsorption of β-hairpin peptides on the surface of water: a neutron refection study. J. Am. Chem. Soc. 125:3751, 2003. Nguyen, H., Jager, M., Moretto, A., Gruebele, M., Kelly, J.W. Tuning the freeenergy landscape of a WW domain by temperature, mutation, and truncation. Proc. Natl. Acad. Sci. U. S. A. 100:3948, 2003. Razavi, H., Palaninathan, S.K., Powers, E.T., Wiseman, R.L., Purkey, H.E., Mohamedmohaideen, N.N., Deechongkit, S., Chiang, K.P., Dendle, M.T.A., Sacchettini, J.C., Kelly, J.W. Benzoxazoles as transthyretin amyloid fibril inhibitors: synthesis, evaluation and mechanism of action. Angew. Chem. Int. Ed. 42:2758, 2003. Sekijimi, Y., Hammarström, P., Matsumura, M., Shimizu, Y., Iwata, M., Tokuda, T., Ikeda, S., Kelly, J.W. Energetic characteristics of the new transthyretin variant A25T may explain its atypical central nervous system pathology. Lab. Invest. 83:409, 2003. Zhang, Q., Kelly, J.W. Cys 10 mixed disulfides make transthyretin more amyloidogenic under mildly acidic conditions. Biochemistry 42:8756, 2003. Zhang, Q., Powers, E.T.. Wentworth, P., Jr., Lerner, R.A., Kelly, J.W. Metabolite initiated protein misfolding in Alzheimer’s disease. Proc. Natl. Acad. Sci., in press. 38 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Expression of Therapeutic Proteins in Eukaryotic Algae S.P. Mayfield, E. Brown, M. Beligni, A. Manuell, K. Yamaguchi, S.E. Franklin herapeutic proteins, including hormones, enzymes, and monoclonal antibodies, account for approximately 10% of the total number of pharmaceutical agents on the market and are the fastest growing area of drug development. With the advent of genomics and proteomics technologies, the rate of discovery of new protein therapeutic agents is predicted to increase in the years ahead. Although this prediction bodes well for our ability to treat a myriad of diseases, many of the relevant diseases are chronic and require several grams of therapeutic protein per patient annually. This requirement presents a challenge to the pharmaceutical industry because existing technologies for producing these therapeutic proteins are extremely expensive. Thus, we appear to be at a crossroads where the ability of medical science to treat human disease may outpace the ability of industry to produce therapeutic agents at a reasonable cost and in sufficient amounts. In addition, healthcare providers, both public and private, are increasingly reticent to pay for therapies that although efficacious are cost prohibitive. Hence, an ever-increasing demand exists for systems that can be used to produce therapeutic proteins economically and at scales not previously achieved. Our current research is focused on developing just such a system for the expression of monoclonal antibodies in the eukaryotic alga Chlamydomonas reinhardtii. Currently, monoclonal antibodies are produced primarily by culture of mammalian cells, and capital costs for the production facilities can run into the hundreds of millions of dollars. Because of these high capital costs and the complexity of mammalian production systems, the production capacity for monoclonal antibodies is expected to fall substantially short of demand during the next 5 years. In addition to capital costs, media and other costs for mammalian cell culture are also quite expensive, making monoclonal antibodies some of the most expensive drugs on the market. Clearly, systems with reduced costs of capitalization that allow rapid scale-up and economical production can greatly affect the way in which protein therapeutic agents are manufactured. We constructed strains of C reinhardtii that express a number of variants of a human antibody directed against herpes simplex virus. We showed that these T antibodies assemble in the algae to form fully functional molecules that bind herpes simplex proteins. Antibody binding to the coat protein of herpes simplex virus blocks viral propagation, making these algally expressed antibodies potential therapeutic agents. In a related project, we are assessing the ability of an antibody Fab fragment that has catalytic activity to assemble into a functional molecule in vivo. The ability to produce catalytic antibodies in a cost-effective manner and on a large scale could revolutionize the pharmaceutical and chemical industries. We are also exploring the ability of this system to produce therapeutic proteins such as IL-10. Evidence suggests that IL-10 may be an effective oral therapy for irritable bowel syndrome. Here again, current production strategies make oral delivery of IL-10 cost prohibitive, whereas production and delivery in algae might be quite cost effective. Finally, we developed 2 reporter gene systems for eukaryotic algae, 1 based on green fluorescent protein and 1 on bacterial luciferase, that will allow us to better define the transcriptional and translational elements required to obtain even higher levels of expression of therapeutic proteins. PUBLICATIONS Franklin, S., Ngo, B., Efuet, E., Mayfield, S.P. Development of a GFP reporter gene for Chlamydomonas reinhardtii chloroplast. Plant J. 30:733, 2002. Mayfield, S.P., Franklin, S.E., Lerner, R.A. Expression and assembly of a fully active antibody in algae. Proc. Natl. Acad. Sci. U. S. A. 100:438, 2003. Mayfield, S.P., Schultz, J. Development of a luciferase reporter gene, luxCt, for Chlamydomonas reinhardtii chloroplast. Plant J., in press. Chemical Synthesis and Chemical Biology K.C. Nicolaou, R. Baati, T. Bando, M. Bella, F. Bernal, W. Brenzovich, S. Bulat, D. Chen, J. Chen, E. Couladouros, P. Dagneau, P. Diamandis, A. Estrada, M. Follmann, T. Francis, M. Frederick, L. Gomez Paloma, M. Govindasamy, D. Gray, P. Guntupalli, S. Harrison, J. Hao, X. Huang, R. Hughes, M. Jennings, F. Kaiser, D. Kim, T. Koftis, S. Lee, Y. Li, T. Ling, D. Longbottom, F. Marr, C. Mathison, T. Montagnon, M. Nevalainen, A. Nalbandian, R. Noronha, B. Paraselli, B. Pratt, W. Qian, G. Rassias, M. Reddy, M. Rodriguez, A. Roecker, B. Safina, P. Sasmal, D. Seigel, S. Snyder, X. Sun, G. Vasilikogiannakis, S. Vyskocil, H. Xu, Y. Yamada, M. Zak O ne of the most vigorous research frontiers in chemistry is total synthesis. Considered by many the engine that drives organic synthesis THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 forward, this scientific endeavor combines elements of art, science, and technology and provides a strong foundation for drug discovery. The field is fueled by the pharmaceutical and biotechnology industries because of potential applications in medicine and by the discovery of naturally occurring novel molecules with biological properties relevant to disease. The examples of aspirin, penicillin, and paclitaxel come to mind, but these are only a few of the myriad of naturally occurring lead compounds that provided direct or indirect medical breakthroughs. With the advances of molecular and cell biology, the science of chemical synthesis promises to become an even sharper tool in the hands of the medicinal chemists who are poised to design and synthesize the next generation of medicines. Our research efforts under the funding auspices of The Skaggs Institute for Chemical Biology are directed at the total synthesis of biologically active natural products and the application of the gained knowledge to the design, chemical synthesis, and biological evaluation of new molecules. Thus, our aim is to enable and facilitate studies in biology and drug discovery and development, and our programs are designed to optimize the educational and training opportunities for young men and women. In the past year, we made marked progress in the areas of molecular design, chemical synthesis, and chemical biology. Specifically, we designed and synthesized a group of new and potent epothilone analogs, one of which is now in clinical trials as an anticancer drug. Our efforts in the anticancer field also included a new total synthesis of the marine-derived antitumor agent diazonamide A and of 1-O-methyllalteriflorone, a derivative of the plant-derived antitumor agent lateriflorone. Our studies on the potent antitumor lomaiviticins and our efforts to synthesize the halipeptins, a new class of marine-derived antitumor agents, are continuing (Fig. 1). In the area of antibiotics, we extended our progress toward the highly complex antibiotic thiostrepton with the synthesis of a number of truncated analogs. In the field of cardiovascular disease, in collaboration with R.M. Evans, the Salk Institute, La Jolla, California, we designed, synthesized, and tested several high-affinity ligands to FXR, a steroid receptor related to bile acid metabolism. Our work on the environmentally dangerous marine neurotoxin azaspiracid-1 and the powerful insecticide azadirachtin resulted in the first synthesis of the proposed structure of the former (proving the proposed 39 F i g . 1 . Selected target molecules. structure was incorrect) and considerable progress toward reaching synthesis of the latter. We also made further advances in new synthetic technologies that are expected to enable drug discovery and development. PUBLICATIONS Bocci, G., Nicolaou, K.C., Kerbel, R.S. Protracted low-dose effects on human endothelial cell proliferation and survival in vitro reveal a selective antiangiogenic window for various chemotherapeutic drugs. Cancer Res. 62:6938, 2002. Downes, M., Verdecia, M., Roecker, A.J., Hughes, R., Hogenesch, J.B., KastWoelbern, H.R., Bowman, M.E., Ferrer, J.L., Anisfeld, A.M., Edwards, P.A., Rosenfeld, J.M., Alvarez, J.G.A., Noel, J.P., Nicolaou, K.C., Evans, R.M. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol. Cell 11:1079, 2003. Giannakakou, P., Nakano, M., Nicolaou, K.C., O’Brate, A., Yu, J., Blagosklonny, M.V., Greber, U.R., Fojo, T. Enhanced microtubule-dependent trafficking and p53 nuclear accumulation by suppression of microtubule dynamics. Proc. Natl. Acad. Sci. U. S. A. 99:10855, 2002. Nicholas, G.M., Eckman, L.L., Ray, S., Hughes, R.O., Pfefferkorn, J.A., Barluenga, S., Nicolaou, K.C., Bewley, C.A. Bromotyrosine-derived natural and synthetic products as inhibitors of mycothiol-S-conjugate amidase. Bioorg. Med. Chem. Lett. 12:2487, 2002. Nicolaou, K.C. Perspectives in total synthesis: a personal account. Tetrahedron 59:6683, 2003. 40 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Nicolaou, K.C., Chen, D.Y.-K., Li, Y., Qian, W., Ling, T., Vyskocil, S., Koftis, T.V., Govindasamy, M., Uesaka, N. Total synthesis of the proposed azaspiracid-1 structure, 2: coupling of C1-C20, C21-C27 and C28-C40 fragments and completion of the synthesis. Angew. Chem. Int. Ed. 42:3649, 2003. Molecular Recognition and Encapsulation Nicolaou, K.C., Evans, R.M., Roecker, A.J., Hughes, R., Downes, M., Pfefferkorn, J.A. Discovery and optimization of non-steroidal FXR agonists from natural product-like libraries. Org. Biomol. Chem. 1:908, 2003. J. Rebek, Jr., T. Amaya, P. Ballester,* S. Biros, W.-D. Cho, Nicolaou, K.C., Jennings, M.P., Dagneau, P. An expedient entry into the fused polycyclic skeleton of vannusal A. Chem. Commun. (Camb.) 2480, 2002, Issue 21. L. Kröck, Y. Kim, H. Onagi, L. Palmer, B. Purse, Nicolaou, K.C., Kim, D.W., Baati, R., O’Brate, A., Giannakakou, P. Total synthesis and biological evaluation of (–)-apicularen A and analogues thereof. Chemistry 9:6177, 2003. E. Ullrich, M. Yamanaka Nicolaou, K.C., Koumbis, A.E. Chemistry, biology and medicine in the 21st century. In: Science, Frontier Technology, and Orthodoxy: Proceedings of the Biomedical Ethics Symposium. Holy Church of Greece, Athens, 2002, p. 177. ALKANES WRUNG OUT TO DRY Nicolaou, K.C., Li, Y., Uesaka, N., Koftis, T.V., Vyskocil, S., Ling, T., Govindasamy, M., Qian, W., Bernal, F., Chen, D.Y.-K. Total synthesis of the proposed azaspiracid-1 structure, 1: construction of the enantiomerically pure C1-C20, C21C27 and C28-C40 fragments. Angew. Chem. Int. Ed. 42:3643, 2003. Nicolaou, K.C., Mathison, C.J.N., Montagnon, T. New reactions of IBX: oxidation of nitrogen- and sulfur-containing substrates to afford useful synthetic intermediates. Angew. Chem. Int. Ed. 42:4077, 2003. Nicolaou, K.C., Mitchell, H.J., Snyder, S.A. Synthesis of complex carbohydrates: everninomicin 13,384-1. In: Carbohydrate-Based Drug Discovery. Wong, C.-H. (Ed.). Wiley-VCH, New York, 2003, p. 948. Nicolaou, K.C., Montagnon, T., Snyder, S.A. Tandem reactions, cascade sequences, and biomimetic strategies in total synthesis. Chem. Commun. (Camb.) 551, 2003, Issue 5. Nicolaou, K.C., Montangon, T., Vassilikogiannakis, G. Natural products and chemical synthesis at the forefront of the fight against cancer [in Greek]. Bio 22, 2003, Issue 5. Nicolaou, K.C., Nevalainen, M., Zak, M., Bulat, S., Bella, M., Safina, B.S. Synthetic studies on thiostrepton: construction of thiostrepton analogues with the thiazoline-containing macrocycle. Angew. Chem. Int. Ed. 42:3418, 2003. Nicolaou, K.C., Rao, P.B., Hao, J., Reddy, M.V., Rassias, G., Huang, X., Chen, D.Y.-K., Snyder S.A. The second total synthesis of diazonamide A. Angew. Chem. Int. Ed. 42:1753, 2003. T. Dale, J. Friese, A. Gissot, S. Gu, F. Hof, D.W. Johnson, D. Rachavi-Robinson, A. Scarso, A. Shivanyuk, L. Trembleau, * Universitat de les Illes Balears, Palma de Mallorca, Spain lkanes are the simplest structures made up of repeating units, and their apolar surfaces are the ultimate hydrophobic molecules. Akanes typically adopt extended, straight-chain conformations in solution that minimize strain but maximize surface area. Reduction of the hydrophobic surface in water can be achieved by folding to more compact forms, but unfavorable interactions result. We found that the long alkyl chains of common surfactants such as sodium dodecyl sulfate adopt a coiled conformation when bound within a synthetic receptor in aqueous solution. The coiled alkane (Fig. 1) complements the size, shape, and chemical surface of the receptor better than the extended conformation does, even though strain is introduced by twisting the backbone. The proper filling of space and burial of hydrophobic surface are thought to drive the molecular recognition between receptor and the alkane: the alkane experiences a dry environment inside the receptor. The A Nicolaou, K.C., Ritzén, A., Namoto, K., Buey, R.M., Fernando Díaz, J., Andreu, J.M., Wartmann, M., Altmann, K.-H., O’Brate, A., Giannakakou, P. Chemical synthesis and biological evaluation of novel epothilone B and trans-12,13-cyclopropyl epothilone B analogues. Tetrahedron 58:6413, 2002. Nicolaou, K.C., Roecker, A.J., Hughes, R., van Summeren, R., Pfefferkorn, J.A., Winssinger, N. Novel strategies for the solid phase synthesis of substituted indolines and indoles. Bioorg. Med. Chem. 11:465, 2003. Nicolaou, K.C., Roecker, A.J., Monenschein, H., Guntupalli, P., Follmann, M. Studies towards the synthesis of azadirachtin: enantioselective entry into the azadirachtin framework through cascade reactions. Angew. Chem. Int. Ed. 42:3637, 2003. Nicolaou, K.C., Sasmal, P.K., Rassias, G., Reddy, M.V., Altmann, K.-H., Wartmann, M., O’Brate, A., Giannakakou, P. Design, synthesis and biological properties of highly potent epothilone B analogues. Angew. Chem. Int. Ed. 42:3515, 2003. Nicolaou, K.C., Sasmal, P.K., Xu, H., Namoto, K., Ritzén, A. Total synthesis of 1O-methyllateriflorone. Angew. Chem. Int. Ed. 42:4225, 2003. Nicolaou, K.C., Snyder, S.A. Cascade reactions in total synthesis: recent advances. Actual. Chim. 81:April-May, 2003. F i g . 1 . Model of an alkane (dodecane) coiled inside a vase-shaped synthetic receptor. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 helical conformation adopted by the alkane shows that even the simplest molecules have enough information built into their structures to fold properly in response to their surroundings. T H E N AT U R E O F E N C L O S E D S PA C E S We have long been interested in the synthesis and characterization of chemical containers, large molecular hosts that can completely surround smaller guests. We use these molecules to explore “inner space,” the inside of a molecule. This environment appears be very different from that of bulk solvent. One host forms by spontaneous assembly of 2 identical but self-complementary modules and is held together by hydrogen bonds (Fig. 2, left) The interior cavity of this assembly provides enough space to surround 1, 2, or even 3 guest molecules. Using nuclear magnetic resonance techniques, we examined the behavior of the molecules inside and discovered the response of the guest to the cramped quarters: chemical reactions between guests are accelerated, weak interactions between guests are isolated and magnified, and the arrangements of guests in the space create new forms of stereochemistry. 41 Large proteins, such as enzymes or receptors, often recognize smaller proteins through a well-defined binding site, and the interaction between the large and small proteins is often compared with that of a lock and its key: only keys containing the correct notches and grooves will fit and function. Regulation of these recognition events via small, synthetic molecules is desirable because protein-protein interactions elicit specific responses within the cell. The ability to block certain pathways and enhance others lies at the center of medicinal chemistry (Fig. 3). F i g . 3 . Left, Model structure of a small peptide adopting an α- helix conformation. The functional groups used to recognize other proteins are shown as spheres. Right, An example of a small molecule that displays groups in a similar orientation to that found in peptides and can interfere with protein-protein interactions. F i g . 2 . Left, a cylindrical, dimeric capsule encapsulates 2 molecules of toluene. Right, a large hexameric assembly surrounds 8 molecules of benzene. Recent advances with even larger hosts such as hexameric assemblies enabled us to study ever larger inner spaces (Fig. 2, right). In this system, we can observe simultaneous encapsulation of many small guests (up to 8 or more). Many of these larger molecules have unique optical and electrochemical properties that can be used to probe the molecular environment of the capsule interior. The encapsulation of small- and medium-sized guests revealed never before seen trends in the packing of molecules into confined spaces. PROTEOMIMETICS Recognition events between molecules in biology are often mediated by many weak attractive forces. The aim of our research is to manipulate specific protein-protein interactions with carefully crafted mimics. These molecules contain structural features similar to those of natural protein surfaces but lack the undesirable aspects of proteins, that is, vulnerability to enzymes and poor adsorption. The synthetic structures can display chemical groups identical to those found on protein surfaces. We are building a collection of synthetic molecules that act as scaffolds and that can be used repeatedly to imitate protein surfaces. The molecules are prepared in modular form through standard organic synthetic methods enhanced by combinatorial chemistry techniques. DYNAMICS We are also blending molecular recognition with catalysis to recruit weak, noncovalent forces that cause changes in covalent bonds. The concave, inner surface of the host must be outfitted with functional groups that can be in contact with the guests held inside. We synthesized a vase-shaped host molecule with a methyl ester group directed toward an interior 42 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 cavity. With certain amine guests, transfer of the carbon atom of the methyl ester takes place from host to guest (Fig. 4). The size of the cavity allows selectivity for different amine guests, and for ideally suited amines, the reaction rate is at least 105 times faster than the comparable reaction in solution. Potential Medical Consequences of Ambiguity in the Genetic Code P. Schimmel, J. Bacher, A. Bates, K. Beebe, V. de Crécy-Lagard, K. Ewalt, J. Liu, M. Lovato, D. Metzgar, C. Myers, L. Nangle, B. Nordin, F. Otero, J. Reader, L. Ribas de Pouplana, B. Slike, M. Swairjo, K. Tamura, W. Waas, X. Yang mbiguity in the genetic code may have a significant impact on human health and disease. The code is an algorithm that relates each amino acid to specific nucleotide triplets that are found in genes and in mRNAs. The code is established in reactions catalyzed by a group of enzymes known as aminoacyl tRNA synthetases. In these reactions, each of the 20 natural amino acids is matched with its cognate tRNA. The amino acid is linked to its cognate tRNA through an ester linkage. Each specific tRNA harbors the triplet of the genetic code for a specific amino acid. Thus, by matching each amino acid with its cognate tRNA, the aminoacyl tRNA synthetases relate each amino acid to the appropriate triplet. Each of the 20 amino acids has a distinct, separate aminoacyl tRNA synthetase. If any of these enzymes makes an error, that is, attaches the wrong amino acid to a given tRNA, then the algorithm of the code is broken. As a consequence of the error, the wrong amino acid is incorporated at a specific codon during translation of an mRNA. The result is the synthesis of proteins that have small imperfections, for example, a change of just a single amino acid of the many (hundreds to even thousands) that the protein is made up of. These imperfections can accumulate and be detrimental to cell viability. The effect on viability has been shown in bacterial cells, but limited information is available on this effect in mammalian cell systems. Ambiguity in the genetic code occurs when, as stated, a mismatch occurs between a given amino acid and a tRNA. Normally, mismatches are prevented by a specialized editing activity that is embedded in many of the aminoacyl tRNA synthetases. This activity corrects errors of mismatching amino acids with their tRNAs by hydrolyzing the ester linkage that joins the 2 components, amino acid and tRNA. The activity is powerful and can rapidly clear mistakes that lead to ambiguity. However, simple point mutations in the active site for editing are sufficient to disrupt editing and cause ambiguity in the genetic code. In bacterial cells, the A F i g . 4 . Model of a guest quinuclidine molecule inside a host cavitand. The ester group at the top of the host is poised to transfer its methyl group to the amine of the guest. PUBLICATIONS Amrhein, P., Wash, P.L., Shivanyuk, A., Rebek, J., Jr. Metal ligation regulates conformational equilibria and binding properties of cavitands. Org. Lett. 4:319, 2002. Bartfai, T., Behrens, M.M., Gaidarova, S., Pemberton, J., Shivanyuk, A., Rebek, J., Jr. A low molecular weight mimic of the Toll/IL-1 receptor/resistance domain inhibits IL-1 receptor-mediated responses. Proc. Natl. Acad. Sci. U. S. A. 100:7971, 2003. Johnson, D.W., Hof, F., Palmer, L.C., Martin, T., Obst, U., Rebek, J., Jr. Glycoluril ribbons tethered by complementary hydrogen bonds. Chem. Commun. (Camb.) 14:1638, 2003. Purse, B.W., Ballester, P., Rebek, J., Jr. Reactivity and molecular recognition: amine methylation by an introverted ester. J. Am. Chem. Soc. 125:14682, 2003. Shivanyuk, A., Friese, J.C., Döring, S., Rebek, J., Jr. Solvent-stabilized molecular capsules. J. Org. Chem. 68:6489, 2003. Shivanyuk, A., Rebek, J., Jr. Assembly of resorcinarene capsules in wet solvents. J. Am. Chem. Soc. 125:3432, 2003. Trembleau, L., Rebek, J., Jr. Helical conformation of alkanes in a hydrophobic cavitand. Science 301:1219, 2003. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 ambiguity can be so great that cell death occurs. Lethality happens in part because a single mutation in an aminoacyl tRNA synthetase can lead to imperfections in all of the cellular proteins. As these imperfections build up, among others, the enzymes, signaling peptides, structural proteins, and replication and transcription factors needed for cell growth are made less and less effective (Fig. 1). 43 PUBLICATIONS Beebe, K., Merriman, E., Schimmel, P. Structure-specific tRNA determinants for editing a mischarged amino acid. J. Biol. Chem. 278:45056, 2003. Beebe, K., Ribas de Pouplana, L., Schimmel, P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 22:668, 2003. Bishop, A.C., Beebe, K., Schimmel, P. Interstice mutations that block site-to-site translocation of a misactivated amino acid bound to a class I tRNA synthetase. Proc. Natl. Acad. Sci. U. S. A. 100:490, 2003. Cen, S., Javanbakht, H., Kim, S., Shiba, K., Craven, R., Rein, A., Ewalt, K., Schimmel, P., Musier-Forsyth, K., Kleiman, L. Retrovirus-specific packaging of aminoacyl tRNA synthetases with cognate primer tRNAs. J. Virol. 76:13111, 2002. Ewalt, K.L., Schimmel, P. Activation of angiogenic signaling pathways by two human tRNA synthetases. Biochemistry 41:13344, 2002. Frugier, M., Giegé, R., Schimmel, P. RNA recognition by designed peptide creates “artificial” tRNA synthetase. Proc. Natl. Acad. Sci. U. S. A. 100:7471, 2003. Hendrickson, T.L., Schimmel, P. Transfer RNA-dependent amino acid discrimination by aminoacyl-tRNA synthetases. In: Translation Mechanisms. LaPointe, J., Brakier-Gringas, L. (Eds.). Kluwer Academic, New York, and Landes Bioscience, Georgetown, TX, 2003, p. 34. A volume in the series Molecular Biology Intelligence Unit. Also available at Eurekah.com. F i g . 1 . Mischarged tRNAs generated by an editing-deficient aminoacyl tRNA synthetase lead to incorrect incorporation of amino acids. The proteins generated would carry small imperfections, resulting in a statistical pool of proteins. Possible fates for these statistical proteins include altered specificity (or activity) and inability to fold correctly. Such imperfections would have diverse ramifications relevant to disease. ER indicates endoplasmic reticulum. To date, no human disease has been associated with an editing-defective aminoacyl tRNA synthetase. However, a concerted effort to make this sort of connection has not occurred. Thus, our objective is to establish editing-defective aminoacyl tRNA synthetases in an animal model such as the mouse. In preliminary experiments, we altered the editing site of a human aminoacyl tRNA synthetase by mutation. When the gene for the altered mutant enzyme was introduced into mammalian cells, it affected the phenotype of these cells, even though the native, nonmutant enzyme was also present. (Thus, the phenotypic changes are due to a “trans-dominant” effect of the mutant protein that overrides the native, wild-type enzyme.) In future experiments, a similar construction will be placed in mice. Analysis of phenotypes associated with specific diseases can then be undertaken. These phenotypes would be the result of imperfections introduced into specific proteins that, for example, affect pathways related to insulin signaling (diabetes), oncogenesis, and autoimmunity. Once a connection can be made between an editing defect and a specific disease, therapeutic approaches can be explored. Kushiro, T., Schimmel, P. Trbp111 selectively binds a noncovalently assembled tRNA-like structure. Proc. Natl. Acad. Sci. U. S. A. 99:16631, 2002. Liu, J., Yang, X.-L., Ewalt, K.L., Schimmel, P. Mutational switching of a yeast tRNA synthetase into a mammalian-like synthetase cytokine. Biochemistry 41:14232, 2002. Nordin, B.E., Schimmel, P. Isoleucyl tRNA synthetase. In: Aminoacyl tRNA Synthetases. Francklyn, C. (Ed.). Kluwer Academic, New York, and Eurekah.com, in press. Nordin, B.E., Schimmel, P. Transiently misacylated tRNA is a primer for editing of misactivated adenylates by class I aminoacyl-tRNA synthetases. Biochemistry 42:12989, 2003. Ribas de Pouplana, Schimmel, P. Alanyl tRNA synthetases. In: Aminoacyl tRNA Synthetases. Francklyn, C. (Ed.). Kluwer Academic, New York, and Eurekah.com, in press. Schimmel, P., Tamura, K. tRNA structure goes from L to λ. Cell 113:276, 2003. Tamura, K., Schimmel, P. Peptide synthesis with a template-like RNA guide and aminoacyl phosphate adaptors. Proc. Natl. Acad. Sci. U. S. A. 100:8666, 2003. Tamura, K., Schimmel, P. Ribozyme programming extends the protein code. Nat. Biotechnol. 20:669, 2002. Wang, C.-C., Chang, K.J., Tang, H.-L., Hsieh, C.-J., Schimmel, P. Mitochondrial form of a tRNA synthetase can be made bifunctional by manipulating its leader peptide. Biochemistry 42:1646, 2003. Yang, X.-L., Liu, J., Skene, R.J., McRee, D.E., Schimmel, P. Crystal structure of an EMAP-II-like cytokine released from a human tRNA synthetase. Helv. Chim. Acta 86:1246, 2003. Yang, X.-L., Skene, R.J., McRee, D.E., Schimmel, P. Crystal structure of an aminoacyl tRNA synthetase cytokine. Proc. Natl. Acad. Sci. U. S. A. 99:15369, 2002. 44 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Expanding the Genetic Code P.G. Schultz, J.C. Anderson, M. Bose, C. Cho, J. Gildersleeve, J. Hong, Q. Huang, J. Liu, K.H. Min, M. Mukherji, S. Santoro, F. Tian, L. Wang, P. Yang, J. Yin, C. Yu, Z. Zhang, L.X. Zheng he genetic codes of every known organism encode 20 amino acid building blocks with triplet codons generated from 4 nucleotides. These 20 amino acids contain a limited number of functional groups, including acids, amides, alcohols, basic amines, and thiols. Clearly, additional functional groups are needed in proteins as indicated by the large number of posttranslational modifications and cofactors and by the rare occurrence of the unnatural amino acids selenocysteine and pyrrololysine. Why then are there only 20 genetically encoded amino acids? Is this number the ideal one, or would additional amino acids allow the generation of proteins or even entire organisms with enhanced functions? To address this fundamental question, we developed a method that allows us to genetically encode, in bacteria, novel amino acids beyond the common 20. This method involves the generation of an orthogonal tRNA that is not a substrate for any natural aminoacyl synthetases and that inserts its cognate amino acid in response to the amber nonsense codon. An orthogonal synthetase is then generated that recognizes this unique tRNA and no other. The substrate specificity of this synthetase is then evolved to recognize a desired “21st” amino acid and no endogenous amino acid. We showed that this method can be used to efficiently incorporate a large number of amino acids into proteins in Escherichia coli with fidelity rivaling that of the common amino acids. In the past year, we added glycosylated amino acids, redox-active amino acids, and keto-containing amino acids to the growing list of novel genetically encoded amino acids. In addition, we showed that acetylenecontaining amino acids can be used to selectively modify proteins with fluorophores and polyethylene glycol methods that may facilitate cellular studies of protein function and the development of therapeutic proteins. We also generalized our method to yeast. Using an E coli–derived orthogonal tRNA synthetase pair and a 2-step positive and negative selection based on suppression of a nonsense codon in the gene gal4, we added 5 novel amino acids independently to the genetic code of Saccharomyces cerevisiae. We are using these amino acids to probe signal transduction in yeast. The T yeast system also should allow us to apply this method to higher eukaryotes. Indeed, we showed that 6-hydroxytryptophan can be selectively incorporated into proteins in mammalian cells in response to the opal codon, and we are attempting to genetically encode novel amino acids in the multicellular organism Caenorhabditis elegans. We also developed a consensus-based strategy that allows us to generate additional orthogonal tRNA synthetase pairs for incorporating multiple distinct amino acids into a single protein, including lysine, tyrosine, and glutamate pairs. Importantly, we showed that the lysine pair can be used to genetically encode an unnatural amino acid (homoglutamine) in response to the 4-base codon AGGA. In collaboration with F. Romesberg and colleagues, The Scripps Research Institute, we are also involved in efforts to expand the genetic lexicon to include a third base pair in addition to the Watson-Crick adenine-thymine and guanine-cytosine base pairs. Such bases would allow us to encode additional genetic information and to evolve nucleic acids with novel properties. We have focused on the use of hydrophobic and metalmediated base-pairing interactions. We synthesized more than 50 such bases and identified a number of homo and hetero base pairs with thermodynamic stability and selectivity rivaling or exceeding those of the Watson-Crick base pairs. In the past year, we focused largely on the development of simple methylated and fluorinated 5- and 6-membered ring “bases.” Pairs derived from these nucleosides are quite selective and stable, and a number are good substrates for DNA polymerases. Currently, we are attempting to crystallize duplex DNAs containing these unnatural base pairs and to evolve in vitro polymerases that can incorporate these bases into DNA with high fidelity. PUBLICATIONS Alfonta, L., Zhang, Z., Uryu, S., Loo, J.A., Schultz, P.G. Site-specific incorporation of a redox-active amino acid into proteins. J. Am. Chem. Soc. 125:14662, 2003. Chin, J.W., Cropp, T.A., Anderson, J.C., Mukherji, M., Zhang, Z., Schultz, P.G. An expanded eukaryotic genetic code. Science 301:964, 2003. Liu, H., Wang, L., Brock, A., Wong, C.-H., Schultz, P.G. A method for the generation of glycoprotein mimetics. J. Am. Chem. Soc. 125:1702, 2003. Mehl, R.A., Anderson, J.C., Santoro, S.W., Wang, L., Martin, A.B., King, D.S., Horn, D.M., Schultz, P.G. Generation of a bacterium with a 21 amino acid genetic code. J. Am. Chem. Soc. 125:935, 2003. Wang, L., Zhang, Z., Brock, A., Schultz, P.G. Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 100:56, 2003. Zhang, Z., Smith, B.A., Wang, L., Brock, A., Cho, C., Schultz, P.G. A new strategy for site-specific modification of proteins in vivo. Biochemistry 42:6735, 2003. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Click Chemistry and the Search for Biological Activity K.B. Sharpless, Y. Baba, S. Boral, J. Cappiello, T. Chan, A. Converso, M. David, A. Feldman, V. Fokin, R. Hilgraf, P. Holzer, C. Kappe, T. Kitayama, A. Krasinski, I. Lewis, V. Litosh, R. Manetsch, A. McPherson, J. Muldoon, S. Narayan, V. Rostovtsev, S. Silverman, N. Stuhr-Hansen, E. Van der Eycken, P. Wu, Y. Xie he primary goals of our research are the discovery and development of selective, powerful, and reliable transformations that allow rapid preparation of compounds with desired biological function from readily available starting materials. During the past year, we developed several such reactions and applied them to targets of chemical diversity and biological activity. Additionally, we focused on performing these and related transformations in water, the lingua franca of life on Earth. In addition to this pursuit of developing novel synthetic transformations, we refined a “Trojan horse” approach to identification of drug candidates in which a biologically active substance is selectively synthesized by the target enzyme itself. The highlights of our research are described here. Water is rarely used or even considered as a solvent for organic reactions. The 2 foremost reasons are the lack of solubility of most organic compounds in this medium and concerns that the high “acid-base” reactivity will interfere with the desired reaction. However, it is hard to ignore water as a solvent. Beyond being the most abundant natural solvent, it has extraordinary physical properties, and although the traditional concerns seem well founded, many examples indicate that reactions commonly performed with organic solvents work equally well, if not better, in water. During the past several years, we encountered numerous examples of water as the “perfect” solvent. These include (1) osmium-catalyzed dihydroxylation and aminohydroxylation, (2) nucleophilic opening of epoxides and aziridines, (3) cycloaddition reactions of all kinds, (4) most oxime ether, hydrazone, and aromatic heterocycle condensation processes, and (5) the formation of an amide from a primary amine and an acid chloride when aqueous Schotten-Baumann conditions are used. In many of these instances, the starting material, product, or both are relatively insoluble in water; in others, the reagents are water sensitive, yet the reac- T 45 tions proceed with high yield and fewer side products than when organic solvents are used. Thus encouraged, we envision a special style of organic synthesis, one based on an entire family of reactions for which water is the best solvent. In our continuing quest for new, supremely reliable reactivity, we uncovered several convenient approaches to 1H-tetrazole functionality, which is commonly found in a variety of drugs and biologically active compounds. The most convenient route to 5-substituted-1H-tetrazoles is the addition of azide ion to nitriles. Although many methods can be used for this transformation, appealing synthetic routes are scarce. All of the reported syntheses have one or more of the following drawbacks: a requirement for expensive and toxic metals, severe water sensitivity, or the presence of hydrazoic acid, which is highly toxic and explosive as well as volatile. In addition, all of the known methods require the use of organic solvents, which are expensive and environmentally unfriendly. Therefore, we were pleased to find a safer and exceptionally efficient process for transforming nitriles into tetrazoles in water; the only other reagents are sodium azide and a zinc salt (Fig. 1). F i g . 1 . Zinc-catalyzed synthesis of 1H-tetrazoles from nitriles and sodium azide. Thus, formation of tetrazoles proceeds with excellent yields and scope in refluxing water. Thanks to the low pK a of 1H-tetrazoles and their highly crystalline nature, a simple acidification is usually sufficient to provide the pure products. Thus, we prepared a wide variety of tetrazoles by using an exceedingly simple procedure, often involving little more than mixing the starting materials in water, heating, and filtering off pure products. To further expand the usefulness of this transformation, we applied this method to the synthesis of tetrazole analogs of amino acids. Such compounds are important in peptide chemistry as mimics for terminal carboxylic acid residues and for cis-amide bonds. In many cases, these analogs are as active as the native 46 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 compounds but are more metabolically stable, making them attractive medicinal targets. Simple heating of the α-amino nitrile in a mixture of water and isopropanol at reflux (about 80°C) in the presence of sodium azide and zinc bromide effects clean conversion to the α-amino tetrazole. These solvents are both environmentally benign and easily isolated; acidification and extraction are usually sufficient to provide the product as a pure solid (Fig. 2). F i g . 2 . Synthesis of tetrazole analogs of amino acids. As a result of collaborations with colleagues at The Scripps Research Institute, many of our synthetic products are currently being tested for a wide range of activities. These include screening for antibacterial agents, in our laboratory; inhibition of HIV protease, in collaboration with J. Elder, The Scripps Research Institute, and C.-H. Wong, the Skaggs Institute; amyloid binding, with J. Kelly, the Skaggs Institute; RNA binding, with J. Williamson, the Skaggs Institute; and inhibition of anthrax lethal factor, with P. Vogt, The Scripps Research Institute. PUBLICATIONS Himo, F., Demko, Z.P., Noodleman, L., Sharpless, K.B. Why is tetrazole formation by addition of azide to organic nitriles catalyzed by zinc (II) salts? J. Am. Chem. Soc. 125:9983, 2003. Lee, L.V., Mitchell, M.L., Huang, S.-J., Fokin, V.V., Sharpless, K.B., Wong, C.-H. A potent and highly sensitive inhibitor of human α-1,3-fucosyltransferase via click chemistry. J. Am. Chem. Soc. 125:9588, 2003. Ripka, A.S., Diaz, D.D., Sharpless, K.B., Finn, M.G. First practical synthesis of formamidine ureas and derivatives. Org. Lett. 5:1531, 2003. Wang, Q., Chan, T.R., Hilgraf, R., Fokin, V.V., Sharpless, K.B., Finn, M.G. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 125:3192, 2003. Antibody Catalysis, Organic Synthesis, and Prodrug and Targeting Therapies S.C. Sinha, R.A. Lerner, L.-S. Li, S. Das, S. Dutta ur research interests involve synthetic methods, total synthesis of biologically important natural products, antibody catalysis, and development of prodrug and targeting therapies. In the past year, we mainly focused on the development of new synthetic strategies, including the stereoselective synthesis of the substituted oxacyclic ketones by the intramolecular aldol-type reaction. We also reported a new method for the intermolecular aldol-type reaction between a ketone and an acetal or a ketal. Using the aldol-type reaction, in combination with aldolase monoclonal antibodies, we are developing chemoabzymatic syntheses of a number of macrocyclic compounds, including epothilones and sorangiolides. We are collaborating with E. Keinan, the Skaggs Institute, in the synthesis of annonaceous acetogenins and in reactions mediated by rhemium(VII) oxides. In the joint effort, we synthesized an analog of asimicin, an annonaceous acetogenin, for the photoaffinity labeling of the corresponding receptor. In the prodrug therapy project, we synthesized and evaluated a number of prodrugs from doxorubicin and paclitaxel. In the targeting approach, we used SCS-873, a small-molecule dual antagonist of the integrins αvβ3 and α v β 5 to transform the function of the catalytic aldolase monoclonal antibody 38C2 into a targeting antibody. The conjugate composed of SCS-873 and 38C2 targets cancer cells that overexpress the integrins α v β 3 and α v β 5 and inhibits the growth of the cells. The prodrug and targeting therapies are done in collaboration with C.F. Barbas and C. Rader, The Scripps Research Institute. O SYNTHETIC METHODS AND THE SYNTHESIS OF N AT U R A L P R O D U C T S Our intramolecular and intermolecular aldol-type reaction is an alternative 1-step Mukaiyama reaction. In the classic Mukaiyama reaction, a carbonyl compound is first transformed into the corresponding silyl enol ether, which then reacts with an acetal to form β-alkoxy carbonyl compounds. In our 1-step process, a boron enolate of a ketone, generated in situ by reacting a ketone with dibutylboron triflate and diisopropyl- THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 ethyl amine, reacts with an acetal or a ketal to provide the corresponding β-alkoxy carbonyl compounds. The intramolecular reaction is highly stereoselective although limited in scope. Thus, only substrates containing methyl ketone react effectively to provide the 4-tetrahydropyrone derivatives with very high cis-selectivity. The intermolecular reaction is, on the other hand, versatile but less stereoselective (Fig. 1). F i g . 1 . Dibutylboron triflate/i-Pr 2NEt–mediated aldol-type reaction of ketones with acetals. In the past year, we also focused on the synthesis of macrolide natural products, such as sorangiolides A and B (Fig. 2). Although, these macrolides have a weak antibiotic activity against gram-positive bacteria, the synthesis will provide access to their analogs. 47 CBI analogs, and epothilones. We are developing new linkers for the production of the prodrugs. We are also synthesizing new haptens to elicit aldolase antibodies. Both monoclonal antibodies and small-molecule antagonists will be used to direct the aldolase monoclonal antibodies to cancer cells. We synthesized a number of integrin-targeting compounds, including SCS-563 and SCS-572. These compounds are based on the SKB compounds. SCS-563 and SCS-572 are equipped with a linker, which allowed us to prepare the corresponding biotin and diketone derivatives. All of these compounds bound to cells expressing the integrins αvβ3 and αvβ5. The diketone compounds SCS-864, SCS-873, and SCS-913 and many other diketone-containing integrin-targeting compounds that we synthesized, react covalently with the reactive lysine residues in the binding site of an aldolase monoclonal antibody. The resultant conjugates have several merits, including prolongation of the half-life of the antagonist and in vivo assembly of the conjugate. We used the conjugate composed of SCS-873 and the antibody 38C2 to study the effect of the conjugate on the growth of the Kaposi sarcoma and melanoma, which express the integrin αvβ3. In mice with human cancer xenografts, compared with SCS-873 alone, the SCS-873–38C2 conjugate significantly inhibited tumor growth. PUBLICATIONS Das, S., Li, L.S., Sinha, S.C. Stereoselective aldol-type cyclization reaction mediated by dibutylboron triflate/diisopropylethylamine. Org. Lett. 6:123, 2004. Han H., Sinha, M.K., D’Souza, L., Keinan, E., Sinha, S.C. Total synthesis of 34hydroxyasimicin and its photoactive derivative for affinity labeling of the mitochondrial complex I. Chemistry, in press. Li, L.S., Das, S., Sinha, S.C. Efficient one-step aldol-type reaction of ketones with acetals and ketals mediated by dibutylboron triflate/diisopropylethyl amine. Org. Lett. 6:127, 2004. Rader, C., Sinha, S.C., Popkov, M., Lerner, R.A., Barbas, C.F. III. Chemically programmed monoclonal antibodies for cancer therapy: adaptor immunotherapy based on a covalent antibody catalyst. Proc. Natl. Acad. Sci. U. S. A. 100:5396, 2003. Rader, C., Turner, J.M., Heine, A., Shabat, D., Sinha, S.C., Wilson, I.A., Lerner, R.A,, Barbas, C.F. A humanized aldolase antibody for selective chemotherapy and adaptor immunotherapy. J. Mol. Biol. 332:889, 2003. F i g . 2 . Structures of sorangiolides A and B. Saphier, S., Sinha, S.C., Keinan, E. Antibody-catalyzed enantioselective Norrish type II cyclization. Angew. Chem. Int. Ed. 42:1378, 2003. P R O D R U G A N D TA R G E T I N G T H E R A P I E S Prodrug therapy provides a unique approach that can be used to minimize the toxic effects of a drug. Ideally, a nontoxic prodrug selectively releases the cytotoxic form of the drug at an appropriate site. We are developing new prodrugs of cytotoxic molecules, including paclitaxel, doxorubicin analogs, enediynes, 48 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Macromolecular Machines as Master Keys in Genome Integrity, the Cell Cycle, Control of Reactive Oxygen Species, and Pathogenesis J.A. Tainer, A.S. Arvai, D.P. Barondeau, M. Bjoras, R. Brudler, R.M. Cardoso, J.M. Castagnetto, B.R. Chapados, C. Chahwan, L. Craig, D.S. Daniels, M. DiDonato, G. Divita, L. Fan, S. Han, C. Hitomi, K. Hitomi, J.L. Huffman, K.-P. Hopfner, D. Hosfield, C.D. Mol, G. Moncalian, S.S. Parikh, C.D. Putnam, D.S. Shin, M.E. Stroupe, O. Sundheim, T.I. Wood, A. Yamagata he goal of our research funded by the Skaggs Institute is to bridge the gaps from molecular structure to cell biology to therapeutics. We proceed by providing insights on the structure and function of macromolecular machines, by designing novel drugs and proteins, and by creating a molecular-based understanding of and intervention into human diseases. Importantly, the Skaggs funding appropriately leverages our Computational Center for Macromolecular Structure at The Scripps Research Institute and our synchrotron beamline, Structurally Integrated Biology for Life Sciences, at the University of California/Lawrence Berkeley National Laboratory, Berkeley, California, which is designed to characterize macromolecular machines and conformational changes. This year we published results of research on machines relevant to cellular protection via control of reactive oxygen species, pathogenesis, and cancerrelated aspects of genome maintenance. We made substantial progress in understanding the molecular machinery that acts in all 3 major processes related to cancer susceptibility: DNA repair, DNA replication, and DNA recombination. Our work on control of reactive oxygen species by the protein superoxide dismutase addresses the central paradox of how mutations that are spread over the protein sequence cause a single disease phenotype. We found that mutations in superoxide dismutase that cause the fatal neurodegenerative disease familial amyotrophic lateral sclerosis are positioned at sites expected to affect formation of the dimer and the integrity of the subunits. This instability promotes the formation of amyloid-like filamentous aggregates that resemble those present in the neurons of patients who have the disease (Fig. 1). As a result of our studies on pathogenesis, we published x-ray crystal structures of type IV pilins from Vib- T F i g . 1 . Mutations that destabilize the structural architecture of Cu,Zn superoxide dismutase promote formation of amyloid-like fibrils. A, Stereo pair shows structural changes at the H43 mutation site and differences in the molecular surface (brick texture). B, Electron micrograph of negatively stained mutant superoxide dismutase shows formation of amyloid-like fibrils after 22 days’ incubation at 37°C, pH 3.5, in the presence of EDTA. No fibrils were observed under these conditions for the wild-type enzyme. Scale bar = 40 nm. rio cholerae and Pseudomonas aeruginosa and a model for their assembly based on our additional electron microscopy and image analysis. Because type IV pilins are virulence factors, understanding their structure and function is critical to controlling cholera, pneumonia, gonorrhea, meningitis, and severe diarrhea. We showed that the conserved N termini of type IV pilins provide a hydrophobic surface for self-assembly, allowing each pilin subunit to interact with multiple neighboring subunits along an extensive surface area to form a thin flexible filament with remarkable tensile strength. Variable regions in the C terminus form specific subunit interaction sites and expose a variable surface that imparts highly specialized pilus functions of colonization, self-aggregation, twitching motility, DNA uptake, and immune escape. In our studies on the pathogenesis of schistosomiasis, we characterized the mechanism by which glutathione S-transferase from the parasite Schistosoma japonicum catalyzes the conjugation of glutathione with toxic secondary products of membrane lipid peroxidation. In DNA repair, we analyzed the complex evolutionary, functional, and structural relationships between light-dependent DNA repair photolyase enzymes and cryptochromes. We calculated a comprehensive phylogenetic tree for the photolyase/cryptochrome protein family, determined the first crystal structure of a cryptochrome (Synechocystis cryptochrome), and performed microarray analyses on the whole Synechocystis genome. Our results suggest a new cryptochrome protein class (cryptochrome DASH) in plants and bacteria and indicate its involvement in transcriptional control. The cryptochrome structure reveals commonalities with DNA THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 repair photolyases in DNA binding and redox-dependent function, despite distinct active-site and interaction surface features. Cytidine deamination to uracil is crucial for antibody hypervariation as part of the normal immune response, but outside lymphocytes uracil is mutagenic in replicating DNA. Ubiquitous enzymes called dUTPases remove deoxyuridine triphosphate (dUTP) from the nucleotide pool to prevent its misincorporation, and another ubiquitous pathway exists to remove uracil from DNA upon spontaneous deamination of cytidine. We determined crystal structures of the bifunctional deoxycytidine triphosphate (dCTP) deaminase and dUTPase from Methanococcus jannaschii, which converts dCTP or dUTP to deoxyuridine monophosphate (dUMP) and has the added metabolic role of initiating the conversion of dCTP to deoxythymidine triphosphate (dTTP) to adjust intracellular levels. These structures revealed the specificity for both dCTP and dUTP, the likely mechanism for deaminase activity, and how the protein might be subject to feedback inhibition by dTTP. DNA recombination is responsible for repair of the most serious form of genome damage: breaks in double-stranded DNA. We successfully used comparative structural genomics, mutational analyses, and cell biology to understand the molecular basis of the role of the breast cancer susceptibility protein BRCA2 in the control of Rad51-mediated DNA recombination. The Rad51 protein forms a polymeric filament around broken DNA ends as part of its mechanism to catalyze DNA strand exchange, the precursor reaction of DNA recombination. By solving the full-length polymeric structure of the Pyrococcus furiosus Rad51 homolog (Fig. 2), we identified the key Rad51 polymerization motif (Rad51-PM) that forms the basis for intermolecular stabilization of active polymeric Rad51 nucleoprotein filaments and oligomeric Rad51 inactive storage rings. Moreover, this motif is found in 8 repetitive sequences within BRCA2, which we found mimic and disassemble inactive Rad51 rings and also chaperone Rad51 subunits to sites of double-stranded breaks to initiate formation of Rad51 polymers on DNA. Further support of our hypothesis that Rad51-mediated DNA recombination is controlled by interface exchange via Rad51-PM is supported by the identification of the motif in Rad52, a mediator protein also involved in the formation of Rad51-DNA polymers (Fig. 2). PUBLICATIONS Abdalla, A.M., Bruns, C.M., Tainer, J.A., Mannervik, B., Stenberg, G. Design of a monomeric human glutathione transferase GSTP1, a structurally stable but catalytically inactive protein. Protein Eng. 15:827, 2002. 49 F i g . 2 . Breast cancer and the repair of DNA damage. The Rad51- PM consists of an intermolecular β-sheet formed by β0 and β3 of adjacent Rad51 subunits and serves as a flexible hinge to stabilize Rad51 heptameric rings as revealed by the x-ray crystal structure (A) of full-length Rad51 and the crystal structure of DNA-bound helical filaments docked into an electron microscopy 3-dimensional reconstruction (B). C, Rad51-PM also contains conserved phenylalanine and alanine residues on one subunit and hydrophobic pockets on the adjacent subunit. This motif is mimicked by BRCA2 for Rad51 disassembly and DNA loading. D, By identifying a second critical Rad51-BRCA2 binding element composed of an α-helical clamp, we were able to design an archaeal Rad51 mutant that could be targeted to damaged DNA by human BRCA2 within human cells upon γ-irradiation. Fluorescent foci in human cell nuclei formed by green fluorescent protein fused to the designed archaeal Rad51 mutant are shown. Adak, S., Bilwes, A.M., Panda, K., Hosfield, D., Aulak, K.S., McDonald, J.F., Tainer, J.A., Getzoff, E.D., Crane, B.R., Stuehr, D.J. Cloning, expression, and characterization of a nitric oxide synthase protein from Deinococcus radiodurans. Proc. Natl. Acad. Sci. U. S. A. 99:107, 2002. Aoyagi, M., Arvai, A.S., Tainer, J.A., Getzoff, E.D. Structural basis for endothelial nitric oxide synthase binding to calmodulin [published correction appears in EMBO J. 22:1234, 2003]. EMBO J. 22:766, 2003. Brudler, R., Hitomi, K., Daiyasu, H., Toh, H., Kucho, K., Ishiura, M., Kanehisa, M., Roberts, V.A., Todo, T., Tainer, J.A., Getzoff, E.D. Identification of a new cryptochrome class: structure, function, and evolution. Mol. Cell 11:59, 2003. Cardoso, R.M., Daniels, D.S., Bruns, C.M., Tainer, J.A. Characterization of the electrophile binding site and substrate binding mode of the 26-kDa glutathione Stransferase from Schistosoma japonicum. Proteins 51:137, 2003. Cardoso, R.M., Thayer, M.M., DiDonato, M., Lo, T.P., Bruns, C.K., Getzoff, E.D., Tainer, J.A. Insights into Lou Gehrig’s disease from the structure and instability of the A4V mutant of human Cu,Zn superoxide dismutase. J. Mol. Biol. 324:247, 2002. Chapados, B.R., Hosfield, D.J., Han, S., Qiu, J., Yelent, B., Shen, B., Tainer, J.A. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell 116:39, 2004. 50 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Craig, L., Taylor, R.K., Pique, M.E., Adair, B.D., Arvai, A.S., Singh, M., Lloyd, S.J., Shin, D.S., Getzoff, E.D., Yeager, M., Forest, K.T., Tainer, J.A. Type IV pilin structure and assembly: x-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11:1139, 2003. DiDonato, M., Craig, L., Huff, M.E., Thayer, M.M., Cardoso, R.M.F., Kassmann, C.J., Lo, T.P., Bruns, C.K., Powers, E.T., Kelly, J.W., Getzoff, E.D., Tainer, J.A. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization [published correction appears in J. Mol. Biol. 334:175, 2003]. J. Mol. Biol. 332:601, 2003. of HIV that is involved in transport of viral mRNAs from the cell nucleus, where mRNA is made, into the cytoplasm, where the mRNA is used to make the viral proteins (Fig. 1). The RRE is the binding site for the HIV Rev protein, and the Rev-RRE interaction is critical for viral replication. If we can find small molecules that block this interaction, they would impair HIV replication. Hearn, A.S., Stroupe, M.E., Cabelli, D.E., Ramilo, C.A., Luba, J.P., Tainer, J.A., Nick, H.S., Silverman, D.N. Catalytic and structural effects of amino acid substitution at histidine 30 in human manganese superoxide dismutase: insertion of valine C gamma into the substrate access channel. Biochemistry 42:2781, 2003. Hopfner, K.P., Tainer, J.A. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 13:249, 2003. Huffman, J.L., Li, H., White, R.H., Tainer, J.A. Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii. J. Mol. Biol. 331:885, 2003. McMurray, C.T., Tainer, J.A. Cancer, cadmium and genome integrity. Nat. Genet. 34:239, 2003. Shin, D.S., Pellegrini, L., Daniels, D.S., Yelent, B., Craig, L., Bates, D., Yu, D.S., Shivji, M.K., Hitomi, C., Arvai, A.S., Volkmann, N., Tsuruta, H., Blundell, T.L., Venkitaraman, A.R., Tainer, J.A. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 22:4566, 2003. Wang, M., Howell, J.M., Libbey, J.E., Tainer, J.A., Fujinami, R.S. Manganese superoxide dismutase induction during measles virus infection. J. Med. Virol. 70:470, 2003. Targeting HIV RNA J.R. Williamson, A. Bunner, J. Chao, S. Edgcomb, P.M. Funke, M. Hennig, E. Johnson, K.A. Lehmann, P.K. Radha, M. Recht, S.P. Ryder, L.G. Scott, E. Sperling, M.W. Trevathan lthough a number of widely used therapeutic strategies exist to combat HIV infection and AIDS, HIV still has a strong propensity to mutate, becoming resistant to drug therapies. The 2 main classes of anti-HIV drugs are protease inhibitors and reverse transcriptase inhibitors. These drugs target 2 of the critical viral enzymes required for HIV replication. Most known drugs target proteins, as is the case for the currently prescribed anti-HIV drugs. HIV is a retrovirus, and its genetic material is composed of RNA, in contrast to the DNA of humans. In addition to serving as the genetic material, HIV RNA contains several important regions critical for programmed reproduction if the virus inside the host cell. If these key RNA regulatory elements could be blocked by drugs, we would have a novel therapeutic approach to combat HIV infection. We are working toward developing small molecules as drug candidates for blocking a particular RNA element in HIV. The Rev response element (RRE) is a part A F i g . 1 . Function of the HIV Rev protein and the RRE in HIV repli- cation. Binding of Rev to the RRE in the nucleus facilitates transport of viral mRNA into the cytoplasm. Early in infection, a set of regulatory proteins is synthesized, whereas late in infection, structural proteins are synthesized. Binding of Rev to RRE in the nucleus is thus a critical switch in the viral life cycle and an important therapeutic opportunity. We are developing libraries of chemical compounds that are more appropriate for binding RNA targets than are most known drugs, which bind to protein targets. Most proteins have hydrophobic pockets, whereas RNA molecules are very polar and bear charged residues. The types of molecules that will preferentially bind RNAs are expected to be somewhat different from the druglike molecules developed on the basis of medicinal chemistry. The libraries of compounds for our first screening effort were made from a collection of commercially available compounds, selected for both their druglike character and the diversity of their chemical structures. Each compound in the library consists of a different molecular framework that corresponds to frameworks typically found in known drug molecules. A representative set of molecules from the library is shown in Figure 2A. We are using these compound libraries and nuclear magnetic resonance spectroscopy to identify compounds that bind to the RRE. We record a nuclear magnetic resonance spectrum of a potential RRE-binding compound in the presence and absence of the RRE target (Fig. 2B), and from the changes in the spectrum, we can determine if the molecule is binding to the RNA. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 51 Gonsalvez, G.B., Lehmann, K.A., Ho, D.K., Stanitsa, E.S., Williamson, J.R., Long, R.M. RNA-protein interactions promote asymmetric sorting of the ASH1 mRNA ribonucleoprotein complex. RNA 9:1383, 2003. Hoggan, D.B., Chao, J.A., Prasad, G.S., Stout, C.D., Williamson, J.R. Combinatorial crystallization of an RNA-protein complex. Acta Crystallogr. D Biol. Crystallogr. 59(Pt. 3):466, 2003. Crystallographic Studies of Immune Recognition and Therapeutic Targets I.A. Wilson, D.A. Calarese, R.M.F. Cardoso, C.-G. Cheong, A.L. Corper, M.D.M. Crispin, M.-A. Elsliger, S. Ferguson, A. Gakhal, P.A. Horton, S. Ito, N.A. Larsen, J.G. Luz, E. Ollmann Saphire, J.B. Reiser, R. Sanders, D.A. Shore, R.L. Stanfield, R.S. Stefanko, J. Stevens, P. Verdino, D.W. Wolan, D. Wu, L. Xu, D.M. Zajonc, Y. Zhang, X. Zhu main focus of our research is determining the crystal structure of proteins involved in human diseases; our goal is to design structure-based therapies. The proteins of interest include antibodies that neutralize HIV type 1 (HIV-1), antibodies that catalyze the degradation of cocaine and heroin, anticancer target enzymes in the purine biosynthesis pathway, and molecules involved in the innate and adaptive immune responses to microbial pathogens. A F i g . 2 . A, Small molecules for screening against RNA targets. These molecules were selected by using a chemical informatics procedure to be small soluble druglike molecules with chemical frameworks different from the frameworks typically found in drug molecules. B, Nuclear magnetic resonance spectroscopy screening for a compound that specifically binds to the RRE. The experiment performed is termed water-LOGSY. The bottom tracing is a reference spectrum of the compound alone. The inverted signals in the top tracing indicate that the compound tested is binding to the RRE. In the middle tracing, binding to simple double-stranded RNA (the control) is much less, indicating that the compound binds specifically to the RRE. We obtained several initial hits from our first round of screening against the RRE. These compounds bind too weakly to be inhibitors of viral replication, so we will need to identify analogs of our initial hits that have increased potency. In addition, we should be able to obtain structural information on how these first-generation inhibitors bind to the RRE RNA and use this information to guide synthesis of the next generation of compounds. PUBLICATIONS Chao, J.A., Prasad, G.S., White, S.A., Stout, C.D., Williamson, J.R. Inherent protein structural flexibility at the RNA-binding interface of L30e. J. Mol. Biol. 326:999, 2003. Cilley, C.D., Williamson, J.R. Structural mimicry in the phage ϕ21 N peptide-boxB RNA complex. RNA 9:663, 2003. H I V VA C C I N E D E S I G N The HIV-1 envelope glycoproteins gp120 and gp41 are the targets of neutralizing antibodies and are of interest as potential components of an HIV-1 vaccine. However, only a handful of antibodies against these proteins have been identified that can potently neutralize a wide array of primary HIV-1 isolates. We carried out x-ray structural studies for several of these broadly neutralizing human antibodies to understand their mechanism of action. We are examining b12, 2G12, and 447-52D, which are antibodies to gp120, and 4E10, an antibody to gp41. The human antibody 2G12 neutralizes a broad range of HIV-1 isolates by binding an unusually dense cluster of carbohydrate moieties on the gp120 envelope glycoprotein. Crystal structures of the Fab fragment of 2G12 in complex with the dimannose and the oligomannose revealed that 2 Fabs assemble into an interlocked V H domain-swapped dimer. The dimerized Fabs provide an extended surface for multivalent interaction with a conserved cluster of oligomannose sugars on the 52 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 surface of gp120. This extraordinary domain-swapped configuration was not observed previously in more than 200 antibody structures and therefore can provide a novel template for engineering antibodies with higher affinities against molecular clusters. Crystal structures were also determined for human antibodies 447-52D (Fig. 1) and 4E10 in complex with peptides corresponding to the antibodies’ linear epitopes. These structures not only help explain how these potent antibodies recognize their antigens but also provide templates for the design of structurally constrained immunogens. The HIV-1 studies are done in collabora- is unknown. In collaboration with N.C. Bruce, University of York, York, England, we determined the crystal structures of heroin esterase alone and in complex with the inhibitor dimethylarsinic acid. The findings provide a structural framework for improving the specificity and sensitivity of a recently reported heroin biosensor. In collaboration with R. Lerner and K. Janda, the Skaggs Institute, and P. Wirsching, The Scripps Research Institute, we are investigating a novel approach to treat cocaine abuse by administering catalytic antibodies that metabolize cocaine. The crystal structures of these antibodies (3A6, 7A1, and 4D3) bound to the substrate cocaine, to transition-state analogs, or to product complexes will be crucial in elucidating the catalytic mechanism and for redesigning haptens and engineering proteins to increase the efficiency and specificity of the antibodies. THE ADAPTIVE IMMUNE RESPONSE AGAINST M I C R O B I A L PAT H O G E N S F i g . 1 . 447-52D is a human antibody to HIV-1 that recognizes the V3 loop region of gp120. The antibody light and heavy chains are shown as light and dark gray ribbons; the V3 peptide, as a ball-and-stick representation. The V3 peptide binds primarily via the H3 complementarity-determining region loop, forming a 3stranded β-sheet, with most of the V3-Fab interactions through main chain–main chain hydrogen bonds. This mode of binding is restricted in sequence solely at the crown region of the V3 loop, one reason why 447-52D may be unusually broadly neutralizing. tion with D. Burton, The Scripps Research Institute; H. Katinger, University of Agriculture, Vienna, Austria; and S. Zolla-Pazner, New York University Medical Center, New York, New York. COCAINE AND HEROIN ANTIBODIES AND ENZYMES Heroin esterase hydrolyzes the acetyl groups from heroin and phenyl acetate to yield morphine and phenol, respectively, although the natural substrate for the enzyme Binding of the murine T-cell receptor (TCR) 2C to the non-self H-2K bm3 MHC molecule presenting the self-peptide dEV8 elicits an alloresponse, which in tissue graft recipients can lead to graft rejection and organ failure. Comparison of the structure of this complex (2C–H-2K bm3 –dEV8) with that of the corresponding TCR–self-antigen complex revealed subtle rearrangements in the binding interface that mediate the alloreactivity. These rearrangements alter the surface complementarity and number of atomic contacts between the TCR and the MHC-peptide antigen. In collaboration with D. Kranz, University of Illinois, Urbana, Illinois, we are investigating the T-cell selection process by studying the interaction of engineered, high-affinity TCRs with MHC molecules. We crystallized one of these TCRs, M40, in complex with the H-2K b–SIYR superagonist antigen. We are also investigating the structural basis of recognition of glycosylated and nonglycosylated peptides by MHC class I molecules for cancer immunotherapy. A successful anticancer vaccine is an important goal, and currently MUC1 (a tumor-associated antigen) is a favored target antigen. Low-affinity MUC1 peptides, which do not contain canonical anchor residues, induce strong T-cell responses and protect mice against a tumor challenge. In collaboration with V. Apostolopoulos, Austin Research Institute, Melbourne, Australia, we previously found that a MUC1 peptide, SAPDTRPA, without the standard anchor residues, still bound to the MHC class I molecule H-2K b . This research is being extended to include the crystal structures of low-affin- THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 ity peptides, short peptides (5-mer), and glycopeptides, all of which induce T-cell responses in mice. An inflammatory joint disease with many similarities to human rheumatoid arthritis develops spontaneously in KRN TCR transgenic mice. Presentation of the class II MHC molecule I-A g7 in complex with a self-peptide derived from glucose-6-phosphate isomerase to the KRN TCR of these mice is a critical step in the initiation of the disease. The crystal structures of I-A g7 in complex with the isomerase and of the TCR KRN will enhance our understanding of how autoimmune disease is mediated at the molecular level. T H E I N N AT E I M M U N E R E S P O N S E A G A I N S T M I C R O B I A L PAT H O G E N S Members of the CD1 family of nonpolymorphic, nonclassical MHC molecules bind lipids and glycolipids for presentation to specialized T cells. We recently determined the structure of the human CD1a isoform in complex with a sulfatide self-antigen (Fig. 2). Compared with healthy subjects, patients with multiple sclerosis have greater numbers of T cells that react with these selfantigens, a condition that contributes to the pathogen- F i g . 2 . View into the binding groove of CD1a. The glycosphingolipid sulfatide is shown in the binding pocket of CD1a between the α1-α2 helices. The tail end of the fatty acid and the sulfate group of the galactose sugar moiety are exposed for T-cell recognition. esis of this autoimmune disease. We are testing an array of TCRs, against both self-antigens and foreign antigens, for expression and crystallization in complex with CD1-glycolipids. Analysis of the presentation of self-antigens and foreign antigens by various CD1 isotypes will greatly enhance our understanding of the initiation of the cellular immune response and the triggering of autoimmune diseases. Lipid and glycolipid ligands are provided by our collaborators, D.B. Moody and M.B. Brenner, Harvard Medical School, Boston, 53 Massachusetts, and C.H. Wong, the Skaggs Institute. Collaborators in our studies on CD1 and TCRs include L. Teyton, The Scripps Research Institute; M. Kronenberg, La Jolla Institute for Allergy and Immunology, San Diego, California; and R. Dwek and P. Rudd, University of Oxford, Oxford, England. A N T I F O L AT E S F O R C H E M O T H E R A P Y Glycinamide ribonucleotide transformylase is a key folate-dependent enzyme in the de novo purine biosynthesis pathway and is a target for the design of antitumor drugs. We determined structures for both apo and complexed forms of the human transformlyase at various pH values. The 1.7-Å structure of the enzyme in complex with an epoxide-based folate analog provided by D. Boger, The Scripps Research Institute, provided important new insights into the mechanism of inhibition of human gycinamide ribonucleotide transformylase and for the design of inhibitors of the enzyme. Aminoimidazole carboxamide ribonucleotide (AICAR) transformylase inosine monophosphate cyclohydrolase (ATIC) is a homodimeric folate-dependent enzyme that encompasses both AICAR transformylase and inosine monophosphate cyclohydrolase activities responsible for the catalysis of the penultimate and final steps of the purine de novo synthesis pathway. ATIC is also a potential target for the development of anticancer drugs. Structure-based development of novel inhibitors aimed at reducing nonspecific interactions with other folatedependent enzymes within the cell are being designed in collaboration with D. Boger, The Scripps Research Institute; G. Beardsley, Yale University, New Haven, Connecticut; and S. Benkovic, Pennsylvania State University, University Park, Pennsylvania. Complexes of both human and avian ATIC with substrates and inhibitors (Fig. 3) are being studied to elucidate the mechanistic capabilities of the complexes and advance the development of inhibitors for both of the activities in ATIC. In collaboration with C. Li and A. Olson, The Scripps Research Institute, we are using virtual ligand screening via protein docking to identify potential small-molecule ligands for ATIC. Using this approach, combined with enzymatic assays, we identified several novel inhibitors that have micromolar binding affinities. A crystal structure of ATIC in complex with one of the inhibitors allowed the compound to be unambiguously identified from the electron density. The structure provided an important initial template for iterations of design and optimization of the inhibitors as novel chemotherapeutic agents. 54 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Royle, L., Roos, A., Harvey, D.J., Wormald, M.R., Van Gijlswijk-Janssen, D.J., Redwan, el-R.M., Wilson, I.A., Daha, M.R., Dwek, R.A., Rudd, P.M. Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J. Biol. Chem. 278:20140, 2003. Rudolph, M.G., Kelker, M.S., Schneider, T.R., Yeates, T.O., Oseroff, V., Heidary, D.K., Jennings, P.A., Wilson, I.A. Use of multiple anomalous dispersion to phase highly merohedrally twinned crystals of interleukin-1β. Acta Crystallogr. D Biol. Crystallogr. 59(Pt. 2):290, 2003. Rudolph, M.G., Stevens, J., Speir, J.A., Trowsdale, J., Butcher, G.W., Joly, E., Wilson, I.A. Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially with peptide transporter alleles TAP-A and TAP-B. J. Mol. Biol. 324:975, 2002. F i g . 3 . Solvent-accessible surface of the multisubstrate adduct inhibitor bound to the active site of AICAR transformylase. The structure is significant because the location of the folate moiety corroborates the formyl transfer mechanism that was previously proposed on the basis of the substrate-bound ATIC complex. This structural template can aid in future development of inhibitors to AICAR transformylase with more optimal pharmacokinetic properties. PUBLICATIONS An, Y., Shao, Y., Alory, C., Matteson, J., Sakisaka, T., Chen, W., Gibbs, R.A., Wilson, I.A., Balch, W.E. GDI-Rab GTPase recycling. Structure (Camb.) 11:347, 2003. Brinen, L.S., Canaves, J.M., Dai, X., Deacon, A.M., Elsliger, M.A., Eshaghi, S., Floyd, R., Godzik, A., Grittini, C., Grzechnik, S.K., Guda, C., Jaroszewski, L., Karlak, C., Klock, H.E., Koesema, E., Kovarik, J.S., Kreusch, A., Kuhn, P., Lesley, S.A., McMullan, D., McPhillips, T.M., Miller, M.A., Miller, M.D., Morse, A., Moy, K., Ouyang, J., Robb, A., Rodrigues, K., Selby, T.L., Spraggon, G., Stevens, R.C., van den Bedem, H., Velasquez, J., Vincent, J., Wang, X., West, B., Wolf, G., Taylor, S.S., Hodgson, K.O., Wooley, J., Wilson, I.A. Crystal structure of a zinc-containing glycerol dehydrogenase (TM0423) from Thermotoga maritima at 1.5 Å resolution. Proteins 50:371, 2003. Calarese, D.A., Scanlan, C.N., Zwick, M.B., Deechongkit, S., Mimura, Y., Kunert, R., Zhu, P., Wormald, M.R., Stanfield, R.L., Roux, K.H., Kelly, J.W., Rudd, P.M., Dwek, R.A., Katinger, H., Burton, D.R., Wilson, I.A. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065, 2003. DeSantis, G., Liu, J., Clark, D.P., Heine, A., Wilson, I.A., Wong, C.H. Structurebased mutagenesis approaches toward expanding the substrate specificity of D-2deoxyribose-5-phosphate aldolase. Bioorg. Med. Chem. 11:43, 2003. Diaz, M., Stanfield, R.L., Greenberg, A.S., Flajnik, M.F. Structural analysis, selection, and ontogeny of the shark new antigen receptor (IgNAR): identification of a new locus preferentially expressed in early development. Immunogenetics 54:501, 2002. Larsen, N.A., Heine, A., de Prada, P., Redwan, el-R., Yeates, T.O., Landry, D.W., Wilson, I.A. Structure determination of a cocaine hydrolytic antibody from a pseudomerohedrally twinned crystal. Acta Crystallogr. D Biol. Crystallogr. 58(Pt. 12):2055, 2002. Luz, J.G., Hassig, C.A., Pickle, C., Godzik, A., Meyer, B.J., Wilson, I.A. XOL-1, primary determinant of sexual fate in C elegans, is a GHMP kinase and a structural prototype for a class of developmental regulators. Genes Dev. 17:977, 2003. Pantophlet, R., Ollmann Saphire, E., Poignard, P., Parren, P.W., Wilson, I.A., Burton, D.R. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642, 2003. Pantophlet, R., Wilson, I.A., Burton, D.R. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J. Virol. 77:5889, 2003. Redwan, el-R.M., Larsen, N.A., Zhou, B., Wirsching, P., Janda, K.D., Wilson, I.A. Expression and characterization of a humanized cocaine-binding antibody. Biotechnol. Bioeng. 82:612, 2003. Schwarzenbacher, R., Canaves, J.M., Brinen, L.S., Dai, X., Deacon, A.M., Elsliger, M.A., Eshaghi, S., Floyd, R., Godzik, A., Grittini, C., Grzechnik, S.K., Guda, C., Jaroszewski, L., Karlak, C., Klock, H.E., Koesema, E., Kovarik, J.S., Kreusch, A., Kuhn, P., Lesley, S.A., McMullan, D., McPhillips, T.M., Miller, M.A., Miller, M.D., Morse, A., Moy, K., Ouyang, J., Robb, A., Rodrigues, K., Selby, T.L., Spraggon, G., Stevens, R.C., van den Bedem, H., Velasquez, J., Vincent, J., Wang, X., West, B., Wolf, G., Hodgson, K.O., Wooley, J., Wilson, I.A. Crystal structure of uronate isomerase (TM0064) from Thermotoga maritima at 2.85 Å resolution. Proteins 53:142, 2003. Stanfield, R.L., Ghiara, J.G., Ollmann Saphire, E., Profy, A.T., Wilson, I.A. Recurring conformation of the human immunodeficiency virus type 1 gp120 V3 loop. Virology 315:159, 2003. Turner, J.M., Larsen, N.A., Basran, A., Barbas, C.F. III, Bruce, N.C., Wilson, I.A., Lerner, R.A. Biochemical characterization and structural analysis of a highly proficient cocaine esterase. Biochemistry 41:12297, 2002. Wada, M., Hsu, C.C., Franke, D., Mitchell, M., Heine, A., Wilson, I., Wong, C.H. Directed evolution of N-acetylneuraminic acid aldolase to catalyze enantiomeric aldol reactions. Bioorg. Med. Chem. 11:2091, 2003. Wentworth, P., Jr., Wentworth, A.D., Zhu, X., Wilson, I.A., Janda, K.D., Eschenmoser, A., Lerner, R.A. Evidence for the production of trioxygen species during antibody-catalyzed chemical modification of antigens. Proc. Natl. Acad. Sci. U. S. A. 100:1490, 2003. Wolan, D.W., Greasley, S.E., Beardsley, G.P., Wilson, I.A. Structural insights into the avian AICAR transformylase mechanism. Biochemistry 41:15505, 2002. Zajonc, D.M., Elsliger, M.A., Teyton, L., Wilson, I.A. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 Å. Nat. Immunol. 4:808, 2003. Zhang, Y., Desharnais, J., Greasley, S.E., Beardsley, G.P., Boger, D.L., Wilson, I.A. Crystal structures of human GAR Tfase at low and high pH and with substrate β-GAR. Biochemistry 41:14206, 2002. Zhang, Y., Desharnais, J., Marsilje, T.H., Li, C., Hedrick, M.P., Gooljarsingh, L.T., Tavassoli, A., Benkovic, S.J., Olson, A.J., Boger, D.L., Wilson, I.A. Rational design, synthesis, evaluation and crystal structure of a potent inhibitor of human GAR Tfase: 10-(trifluoroacetyl)-5-10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid. Biochemistry 42:6043, 2003. Zhu, X., Heine, A., Monnat, F., Houk, K.N., Janda, K.D., Wilson I.A. Structural basis for antibody catalysis of a cationic cyclization reaction. J. Mol. Biol. 329:69, 2003. Zhu, X., Larsen, N.A., Basran, A., Bruce, N.C., Wilson, I.A. Observation of an arsenic adduct in an acetyl esterase crystal structure. J. Biol. Chem. 278:2008, 2003. Zhu, Y., Rudensky, A.Y., Corper, A.L., Teyton, L., Wilson, I.A. Crystal structure of MHC class II I-Ab in complex with a human CLIP peptide: prediction of an I-Ab peptide-binding motif. J. Mol. Biol. 326:1157, 2003. Zwick, M.B., Parren, P.W., Saphire, E.O., Church, S., Wang, M., Scott, J.K., Dawson, P.E., Wilson, I.A., Burton, D.R. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 77:5863, 2003. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Bioorganic Chemistry and Drug Discovery C.-H. Wong, F. Agnelli, C. Behrens, M. Best, A. Brik, M.C. Bryan, A. Buchnyskyy, G. Cain, A. Chang, W.C. Cheng, A. Datta, S. Duron, F. Fazio, D. Franke, J.I. Furukawa, M.C. Galan, S. Hanson, Z.-Y. Hong, J. Hsu, C.-Y. Huang, H.K. Lee, L. Lee, F.-S. Liang, D. Lin, H. Liu, J. Liu, 55 DeSantis, G., Liu, J., Clark, C.P., Heine, A., Wilson, I.A., Wong, C.-H. Structurebased mutagenesis approaches toward expanding the substrate specificity of D-2deoxy-ribose-5-phosphate aldolase. Bioorg. Med. Chem. 11:43, 2003. Fazio, F., Bryan, M.C., Blixt, O., Paulson, J.C., Wong, C.-H. Synthesis of sugar arrays in microtiter plates. J. Am. Chem. Soc. 124:14397, 2003. Lee, L.V., Mitchell, M.L., Huang, S.-J., Fokin, V.V., Sharpless, K.B., Wong, C.-H. A potent and highly selective inhibitor of human α-1,3-fucosyltransferase via click chemistry. J. Am. Chem. Soc. 125:9588, 2003. Liu, H., Ritter, T.K., Sadamoto, R., Sears, P.S., Wu, M., Wong, C.-H. Acceptor specificity and inhibition of the bacterial cell-wall glycosyltransferase Mur G. Chembiochem 4:603, 2003. M. Numa, P. Nyffeler, T. Polat, D. Thayer, S.K. Wang, G. Watt, D. Wu, Y.Y. Yang, N. Yu, G.-W. Xing, R. Xu uring the past year, our effort was directed toward carbohydrate-based drug discovery. In continuation of our interest in developing new enzymatic methods for organic synthesis, we developed new aldolases through directed evolution and structurebased mutagenesis and used the modified enzymes in the synthesis of iminocyclitol derivatives as fucosyltransferase inhibitors. We also developed a new noncovalent method to assemble saccharide microarrays in styrene microtiter plates for high-throughput screening. The saccharides prepared were attached to a C13–C16 hydrocarbon chain at the anomeric center for the noncovalent attachment to the microplate surface. The sugars were further modified enzymatically by using glycosyltransferases to expand the diversity of the array. This array was then used in a high-throughput assay for discovery of fucosyltransferase inhibitors as potential drug candidates for the treatment of inflammatory diseases. We also used the copper(I)-catalyzed triazole formation, in collaboration with K.B. Sharpless, the Skaggs Institute, to prepare lipidated oligosaccharides for attachment to styrene plates and to discover new fucosyltransferase inhibitors. The triazole-forming chemistry was carried out in the plates, and then screening in situ was used to identify potent inhibitors. This strategy was effective for discovery of new inhibitors of any enzyme and was applied to the discovery of new HIV protease inhibitors and glycosidase inhibitors by using different reaction and in situ screening methods. D PUBLICATIONS Brik, A., Muldoon, J., Lin, Y.-C., Elder, J.H., Goodsell, D., Olson, A.J., Fokin, V.V., Sharpless, K.B., Wong, C.-H. Rapid diversity-oriented synthesis in microtiter plates for in situ screening of HIV protease inhibitors. Chembiochem 4:1246, 2003. Chapman, E., Bryan, M.C., Wong, C.-H. Mechanistic studies of β-arylsulfotransferase-IV. Proc. Natl. Acad. Sci. U. S. A. 100:910, 2003. Chapman, E., Ding, S., Schultz, P.G., Wong, C.-H. A potent and highly selective sulfotransferase inhibitor. J. Am. Chem. Soc. 124:14524, 2002. Liu, H., Wang, L., Brock, A., Wong, C.-H., Schultz, P.G. A method for the generation of glycoprotein mimetics. J. Am. Chem. Soc. 125:1702, 2003. Mak, C.C., Brik, A., Lerner, D.L., Elder, J.H., Morris, G.M., Olson, A.J., Wong, C.-H. Design and synthesis of broad-based mono- and bicyclic inhibitors of HIV and FIV proteases. Bioorg. Med. Chem. 11:2025, 2003. Mong, T.-K., Huang, C.-Y., Wong, C.-H. A new reactivity-based one-pot synthesis of N-acetyllactosamine oligomers. J. Org. Chem. 68:2135, 2003. Mong, T.-K., Lee, H.K., Duron, S.G., Wong, C.-H. Reactivity-based one-pot synthesis of fucose GM1 oligosaccharide: a sialylated antigenic epitope of small-cell lung cancer. Proc. Natl. Acad. Sci. U. S. A. 100:797, 2003. Ritter, T.K., Mong, K.-K., Liu, H., Nakatani, T., Wong, C.-H. A programmable onepot oligosaccharide synthesis for diversifying the sugar domains of natural products: a case study of vancomycin. Angew. Chem. Int. Ed. 42:4657, 2003. Sawkar, A.R., Cheng, W.-C., Beutler, E., Wong, C.-H., Balch, W.E., Kelly, J.W. Chemical chaperones increase the cellular activity of N370S β-glucosidase: a therapeutic strategy for Gaucher disease. Proc. Natl. Acad. Sci. U. S. A. 99:15428, 2002. Silvestri, M.G., DeSantis, G., Wong, C.-H. Asymmetric aldol reactions using aldolases. Top. Stereochem. 23:267, 2003. Tolbert, T.J., Wong, C.-H. Glycoprotein synthesis. In: McGraw-Hill 2003 Yearbook of Science and Technology. McGraw-Hill, New York, 2003, p. 159. Wada, M., Hsu, C.-C., Franke, D., Mitchell, M., Heine, A., Wilson, I., Wong, C.-H. Directed evolution of N-acetylneuraminic acid aldolase to catalyze enantiomeric aldol reactions. Bioorg. Med. Chem. 11:2091, 2003. Wu, C.-Y., Chang, C.-F., Chen, J.S.-Y., Wong, C.-H. Lin, C.-H. Rapid diversity-oriented synthesis in microtiter plates for in situ screening: discovery of potent and selective α-fucosidase inhibitors. Angew. Chem. Int. Ed. 42:4661, 2003. Studies of Macromolecular Recognition by Multidimensional Nuclear Magnetic Resonance P.E. Wright, H.J. Dyson, M. Martinez-Yamout, J. Covalt, R. De Guzman, T. Dunzendorfer-Matt, N. Goto, B. Hudson, J. Lansing, T. Nishikawa, G. Perez-Alvarado, J. Wojciak, T. Zor, J. Hosea, M. Landes pecific interactions between molecules are of fundamental importance in all biological processes. An understanding of how biological macromolecules such as proteins and nucleic acids recognize each other is essential for understanding the S 56 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 fundamental molecular events of life. Knowledge of the 3-dimensional structures of biological macromolecules is key to understanding their interactions and functions and also forms the basis for rational design of new drugs. A particularly powerful method for mapping the 3-dimensional structures and interactions of biological macromolecules in solution is multidimensional nuclear magnetic resonance spectroscopy. We are applying this method to study a number of protein-protein and protein–nucleic acid interactions of fundamental importance in health and disease. Transcriptional regulation in eukaryotes relies on protein-protein interactions between DNA-bound factors and coactivators that, in turn, interact with the basal transcription machinery. A major effort in our laboratory is focused on elucidation of the structural principles that determine specificity of key protein-protein interactions involved in regulation of gene expression. The transcriptional coactivator CREB-binding protein (CBP) and its ortholog p300 play a central role in cell growth, differentiation, and development in higher eukaryotes. CBP mediates interactions between a number of gene regulatory proteins and viral proteins, including proteins from several tumor viruses and hepatitis B virus. Understanding the molecular mechanisms by which CBP recognizes its various target proteins is of fundamental biomedical importance. CBP has been implicated in diverse human diseases such as leukemia, cancer, and mental retardation and is a novel target for therapeutic intervention. We have initiated a major program to determine the structure of the various domains of CBP and map their functional interactions with other components of the transcriptional machinery. Some years ago, we determined the 3-dimensional structure of the kinase-inducible activation domain of the transcription factor CREB bound to its target domain, the KIX domain, in CBP. The structure provides a starting point for design of small molecules that can inhibit the CREB-KIX interactions, an important goal in development of novel therapeutics for treatment of diabetes. In the past year, we determined the structure of the complex between KIX and the transcriptional activation domain of the proto-oncogene c-Myb (Fig. 1). The Myb activation domain is intrinsically unstructured but folds into a helical conformation on binding to KIX; it uses the same hydrophobic binding groove as the CREB activation domain but makes more intimate hydrophobic contacts. The structure provides new insights into the factors that determine constitutive and inducible F i g . 1 . The c-Myb transcriptional activation domain forms a helix on binding in a hydrophobic surface groove on the KIX domain of CBP. binding to KIX. We also showed that the activation domain of the mixed lineage leukemia protein binds cooperatively with Myb and CREB to an allosteric site on KIX, suggesting a direct mechanism by which CBP can mediate transcriptional synergy, and we are refining the structure of the ternary complex formed between the KIX domain of CBP and the activation domains of Myb and CREB. We performed a structural and thermodynamic analysis of the complex formed between CBP and a p160 nuclear receptor coactivator, the activator for thyroid hormone and retinoid receptors (ACTR). CBP and the p160 coactivators play an essential role in the nuclear hormone response and are implicated in cancer and other diseases. Our structure reveals the mechanism by which p160 coactivators recruit CBP and forms the basis for design of novel therapeutic agents that could modulate the nuclear hormone response. The ACTR and CBP domains are both intrinsically unstructured, but they fold synergistically to form a cooperatively folded intermolecular complex. Mutagenesis experiments indicated that the molecular interaction is finely tuned for specificity rather than for binding affinity. We recently determined the structure of the complex formed between the carboxy-terminal activation domain of the hypoxia-inducible transcription factor HIF-1 and the Taz 1 zinc finger motif of CBP. HIF-1 activates genes that are crucial for cell survival under hypoxic conditions; this is accomplished through interactions between its α-subunit (HIF-1α) and CBP/p300. The hypoxic response, which plays an important role in tumor progression and metastasis, is tightly regulated THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 in the cell. In particular, the protein CITED2 functions as a negative feedback regulator that inhibits HIF-1α by competing for binding to CBP/p300. We recently determined the 3-dimensional structure of the complex between the activation domain of CITED2 and the Taz 1 domain of CBP (Fig. 2). The CITED2 domain is intrinsically unstructured and folds upon binding to CBP. Our structure shows that CITED2 and HIF-1α bind to partially overlapping surfaces of the Taz 1 domain and compete for binding through a highly conserved sequence 57 PUBLICATIONS De Guzman, R.N., Martinez-Yamout, M.A., Dyson, H.J., Wright, P.E. Interaction of the TAZ1 domain of CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites. J. Biol. Chem., in press. De Guzman, R.N., Martinez-Yamout, M.A., Dyson, H.J., Wright, P.E. Structure and function of the CBP/p300 TAZ domains. In: Zinc Finger Proteins: From Atomic Contact to Cellular Function. Iuchi, S., Kuldell, N. (Eds.). Landes Bioscience, Georgetown, TX, in press. Gearhart, M.D., Holmbeck, S.M.A., Evans, R.M., Dyson, H.J., Wright, P.E. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J. Mol. Biol. 327:819, 2003. Perez-Alvarado, G.C., Martinez-Yamout, M., Allen, M.M., Grosschedl, R., Dyson, H.J., Wright, P.E. Structure of the nuclear factor ALY: insights into post-transcriptional regulatory and mRNA nuclear export processes. Biochemistry 42:7348, 2003. Schnell, J.R., Dyson, H.J., Wright, P.E. Structure, dynamics and catalytic function of dihydrofolate reductase. Ann. Rev. Biophys. Biomol. Struct., in press. F i g . 2 . Structure of the activation domain of CITED2 (dark gray ribbon) bound to the Taz 1 domain of CBP. The zinc atoms are shown as gray spheres. motif. This work provides new insights into the mechanism by which intrinsically unstructured proteins can compete effectively for binding to a common target within the complex macromolecular assembly that regulates transcription. Gene expression in eukaryotes is mediated both by transcriptional regulators and by posttranscriptional events that control the lifetime of the mRNA. We recently determined the structure of the zinc-binding domain of the regulatory protein TIS11d bound to its cognate AU-rich single-stranded RNA recognition element. TIS11d is a member of the tristetraprolin family of regulatory proteins that bind the recognition element in the 3′ untranslated region of the mRNA of TNF-α and mediate its expression by promoting degradation of the message. Our structure provides novel insights into the mechanisms of site-specific recognition of singlestranded RNA. 58 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Staff Awards and Activities Boger, D.L.—Adrien Albert Medal, Royal Society of Joyce, G.F.—Member, U.S. National Academy of Sciences; Chemistry, London, England; Bachmann Lecturer, University of Michigan, Ann Arbor, Michigan; Ingersoll Lecturer, Vanderbilt University, Nashville, Tennessee; Member, Medicinal Chemistry Study Section, National Institutes of Health; Member, Electorate Nominating Committee, American Association for the Advancement of Science; Member, Drug Candidate Evaluation Committee, Institute for the Study of Aging; Editor-in-Chief, Bioorganic and Medicinal Chemistry Letters; Editorial Boards, Tetrahedron Publications, Organic Reactions, Journal of Organic Chemistry, Current Opinion in Drug Discovery and Development, Current Drugs. Head of Faculty in Chemical Biology, Faculty 1000, Biology Reports Ltd.; Associate Editor, BioSystems, Evolutionary Computation, Origins of Life and Evolution of the Biosphere. Ernest Beutler— E. Donnall Thomas Lecture and Prize, American Society of Hematology; Professional Achievement Citation, University of Chicago Alumni Association; Kent Kiekow Memorial Leukemia Lecture, Northwestern University Medical School, Chicago, Illinois; Member, National Academy of Sciences; Member, American Academy of Arts and Sciences; Scientific Advisory Boards, Baxter Healthcare, Edwards Lifesciences, iMetrikus, MPD Foundation, MyOwnMD, Optimer Pharmaceuticals, Insert Therapeutics; Contributing Editor, Blood Cells, Molecules, and Diseases; Associate Editor, Acta Haematologica, Molecular Medicine. Dawson, P.E.— Research Fellowship in Chemistry, Alfred P. Sloan Foundation; Member, Faculty in Chemical Biology, Faculty 1000, Biology Reports, Ltd.; Editorial Board, Letters in Peptide Science. Janda, K.D.— Fellow, American Association for the Advancement of Science; Sigma-Aldrich Lecturer, Milwaukee Section, American Chemical Society, Milwaukee, Wisconsin; Rayson Huang Visiting Lecturer, University of Hong Kong, Hong Kong, China; Shire BioChem Lecturer, Universite de Montreal, Montreal, Canada; Editorial Boards, Chemical Reviews, Combinatorial Chemistry Research and Applications, Bioorganic and Medicinal Chemistry Letters, Bioorganic and Medicinal Chemistry, Combinatorial Chemistry HighThroughput Screening. Lerner, R.A.—Paul Ehrlich and Ludwig Darmstaedter Prize, Paul Ehrlich Foundation, Frankfurt, Germany; Honorary Doctoral Degree, Ben-Gurion University of the Negev, Beer-Sheva, Israel; Editorial Boards, Bioorganic and Medicinal Chemistry, Bioorganic and Medicinal Chemistry Letters, Catalysis Technology, Chemistry and Biology, Drug Targeting and Delivery, Journal of Virology, Molecular Biology and Medicine, Molecular Medicine, Journal of Peptide Research, Vaccine, Angewandte Chemie. Nicolaou, K.C.—Tetrahedron Prize for Creativity in Organic Chemistry, Tetrahedron Publications; Nobel Laureate Signature Award for Graduate Education in Chemistry, American Chemical Society; Alder and Bayer Lecturer, Cologne, Germany; AstraZeneca Lecturer, University of Montreal and AstraZeneca, Montreal, Canada; Bristol Myers Lecturer, Boston College, Boston, Massachusetts; Co-Editor-in-Chief, Chemistry and Biology; Editorial Boards, Tetrahedron Publications, Synthesis, Carbohydrate Letters, Chemistry—A European Journal, Perspectives in Drug Discovery and Design, Indian Journal of Chemistr y, Section B, Combinatorial Chemistry High-Throughput Screening, Current Opinion in Bioorganic Chemistry, Current Organic Chemistry, Organic Letters, ChemBioChem. Rebek, J., Jr.— Chemical Pioneer Award, American Institute of Chemists; Woodward Scholar, Harvard University, Boston, Massachusetts; Ronald Breslow Award for Achievement in Biomimetic Chemistry, American Chemical Society; Advisory Boards, Physical Sciences Division, University of Chicago, National Cancer Institute, and Institute of Chemical Research of Catalonia, Spain; Editorial Boards, Tetrahedron Publications, Chemistry and Biology, Current Opinion in Chemical Biology, Journal of the Chemical Society— Perkin Transactions, Journal of Organic Chemistry, Bioorganic and Medicinal Chemistry Letters, Progress in Physical Organic Chemistry, Journal of Supramolecular Chemistry. THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 Schultz, P.G.—Paul Ehrlich and Ludwig Darmstaedter Award, Paul Ehrlich Foundation, Frankfurt, Germany. Sharpless, K.B.—Honorary Member, Kitasato Institute, Tokyo, Japan; Visiting Professor, Kitasato University, Tokyo, Japan; Danforth Lecturer, Grinnell College, Grinnell, Iowa; Robbins Lecturer, Pomona College, Claremont, California; Editorial Boards, Enantiomer, Advanced Synthesis and Catalysis, Chirality, Current Opinion in Drug Discovery and Development, Organic Letters; Honorary Advisory Board, Synlett. Williamson, J.R.—Associate Editor, Annual Reviews of Biophysics and Biomolecular Structure; Editorial Boards, RNA, Molecular Cell, Chemistry and Biology. Wilson, I.A.— Fellow, Royal Society of London; Fellow, American Academy of Arts and Sciences; Member, Burroughs Wellcome Career Awards Advisory Committee; Member, Scientific Advisory Board, Keystone Symposia; Associate Editor, Journal of Molecular Biology, Immunity; Editorial Boards, Science, Journal of Experimental Medicine. Wong, C.-H.—Member, National Research Council on Chemical Sciences and Technology; Scientific Advisor, Max-Planck-Institute, Dortmund, Germany; Editor-inChief, Bioorganic and Medicinal Chemistry; Editorial Boards, Tetrahedron Publications, Current Opinion in Chemical Biology, Biocatalysis, Advanced Synthesis and Catalysis. Wright, P.E.—Honorary Doctor of Science, University of Sydney, Sydney, Australia; Editor-in-Chief, Journal of Molecular Biology; Editorial Boards, Biochemistry, Current Opinion in Structural Biology, Journal of Biomolecular NMR. 59 60 THE SKAGGS INSTITUTE FOR CHEMICAL BIOLOGY 2003 SUBJECT INDEX Acetogenins 36, 46 Adaptive immunity 52 AIDS 13 Alkanes 40 Alzheimer’s disease 37 Amyloid diseases 37 Amyloidogenesis 37 Amyotrophic lateral sclerosis 28, 34, 48 Anandamide 20 Antibiotics 19 Antibodies expression in algae 38 HIV neutralizing 51 to cocaine 52 to tumors 14 Antibody libraries 30 Anticancer agents 53 Antisense agents 33 Asymmetric synthesis 13 ATIC 53 Bacterial pathogenesis 48 Biomolecular computing 34 Bioorganic chemistry 18, 55 Cancer chemotherapy 31 Cancer 13, 20 Catalysis 45 Catalytic antibodies 13, 34, 46 Cell adhesion molecules 22 Cell cycle 48 control of 16 Cell diagnostics 29 Chemical biology 38 Chemical ligation 21 ACKNOWLEDGMENTS The scientists who have contributed sections to this report wish to acknowledge the dedication and hard work of the laboratory technicians who helped bring the research to fruition, the administrative assistants who made it presentable for publication, and the support personnel who provided critical specialized services and equipment. Chemical physiology 20 Chemical synthesis 38 Click chemistry 45 Cryptochromes 28 DNA enzymes 33 DNA repair 48 DNA-binding proteins 56 Drug design 18, 29, 53 Drug discovery 50, 55 Encapsulation 40 Enzyme inhibitors 55 Enzymes synthetic 34 Erythropoietin 18 Fatty acid amide hydrolase 20 Fluorescent proteins 28 Genetic code 14, 42, 44 Genome integrity 48 Glycinamide ribonucleotide transformylase 53 Heroin esterase 52 HIV 50 vaccine to 51 Hypoxia 56 Immune recognition 51 Immunopharmacotherapy 30 Innate immunity 16, 53 Integrins 46 Lipopolysaccharides 31 Macromolecular machines 48 Major histocompatibility complex 52 Medicinal chemistry 18 Membrane proteins 20 Molecular evolution 33 Molecular recognition 40 Multidrug resistance 20 NADPH oxidase 16 Nanotechnology 34 Nanotubes 30 Natural products 19, 39, 46 Nicotine abuse 32 Nitric oxide synthases 27 Nuclear magnetic resonance spectroscopy 55 Nucleic acids 33 structure of 23 Oleamide 20 Organic synthesis 34, 46 Organocatalysis 13 Pain 20 Peptide synthesis 21 Phage display technology 30 Photoactive yellow protein 28 Platelet receptors 17 Prion diseases 37 Prodrug therapy 46 Protein-protein interactions 18 Proteins DNA binding 14 folding of 38 structure of 27 synthesis of 21 Proteomics 21 Proteomimetics 41 Reactive oxygen species 16, 48 Receptors 20 on T cells 52 Ribosomes 22 Ribozymes 24 RNA assembly and catalysis of 24 Saccharides 55 Self-organized networks 29 Septic shock 31 Serine hydrolases 21 Siroheme synthase 27 Sleep 20 Stem cells 23 Sulfite reductases 27 Superoxide dismutases 28, 48 Synapses 22 Synthetic chemistry 18 Targeting therapy 46 Therapeutic proteins expression in algae 38 Transcription 56 Transfer RNAs 42 Translation 22 Transporters 20 Ubiquitination 16 Vaccines 31, 51 Viral coats 22 Viral infection 16 Viruses as molecular building blocks 25 cowpea mosaic virus 26 flock house virus 26 hepatitis C virus 17 lymphocytic choriomeningitis virus 16 nudaurelia capensis ω virus 26 X-ray crystallography 51 Zinc fingers 14 The Skaggs Institute for Chemical Biology scientific report is published annually by The Scripps Research Institute and is available on request from Editor Barbara L. Halliburton, Ph.D. D E PA R T M E N TA L Graphic Design Pentagram Design Janette Lundgren The Skaggs Institute for Chemical Biology Cover Illustration Steve Lustig BioDesign Communications Inc. Ruby Blair Department of Molecular Biology Office of Communications TPC-20 The Scripps Research Institute 10550 North Torrey Pines Road La Jolla, CA 92037 (858) 784-2171 e-mail delasol@scripps.edu Project Manager Jann Coury Office of Communications Production Specialist Jennifer O’Sullivan Office of Communications Printing Moore Wallace-Commerical Press Photography Michael Balderas Mark Dastrup Alan McPhee, Biomedical Graphics Department C O O R D I N AT I O N Cheryl Negus Department of Cell Biology Vicky Nielsen Department of Chemistry Lynn Oleski Department of Molecular and Experimental Medicine Michelle Platero Department of Neurobiology Rigid molecular objects, approximately 22 nm in diameter, constructed from 1.7-kb strands of DNA. Their structure was determined by using 3-dimensional reconstruction of images obtained by electron cryomicroscopy. This work was performed by William M. Shih, Ph.D., in the labora tory of Gerald F. Joyce, M.D., Ph.D., with assistance from Joel D. Quispe in the Center for Integrated Molecular Biosciences. Figure courtesy of Michael Pique. THE SCRIPPS RESEARCH INSTITUTE 10550 N O RT H T O R R E Y P I N E S R O A D L A J O L L A , C A L I F O R N I A 92037 US A T E L E P H O N E : 858.784.1000 www.scripps.edu