Crystal Structures and Inhibition Kinetics Reveal a Implications for Rhomboid Proteolysis
advertisement
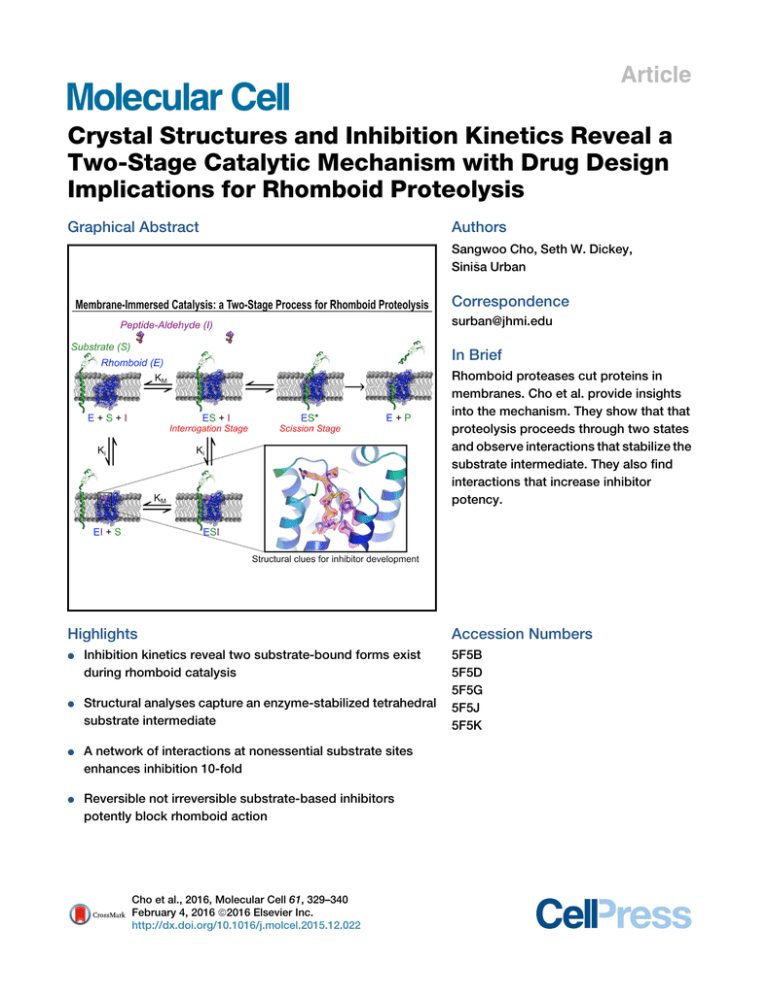
Article Crystal Structures and Inhibition Kinetics Reveal a Two-Stage Catalytic Mechanism with Drug Design Implications for Rhomboid Proteolysis Graphical Abstract Authors Sangwoo Cho, Seth W. Dickey, a Urban Sinis Correspondence surban@jhmi.edu In Brief Rhomboid proteases cut proteins in membranes. Cho et al. provide insights into the mechanism. They show that that proteolysis proceeds through two states and observe interactions that stabilize the substrate intermediate. They also find interactions that increase inhibitor potency. Highlights d Inhibition kinetics reveal two substrate-bound forms exist during rhomboid catalysis d Structural analyses capture an enzyme-stabilized tetrahedral substrate intermediate d A network of interactions at nonessential substrate sites enhances inhibition 10-fold d Reversible not irreversible substrate-based inhibitors potently block rhomboid action Cho et al., 2016, Molecular Cell 61, 329–340 February 4, 2016 ª2016 Elsevier Inc. http://dx.doi.org/10.1016/j.molcel.2015.12.022 Accession Numbers 5F5B 5F5D 5F5G 5F5J 5F5K Molecular Cell Article Crystal Structures and Inhibition Kinetics Reveal a Two-Stage Catalytic Mechanism with Drug Design Implications for Rhomboid Proteolysis a Urban1,* Sangwoo Cho,1,2 Seth W. Dickey,1,2 and Sinis 1Department of Molecular Biology & Genetics, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, 725 North Wolfe Street, Room 507 PCTB, Baltimore, MD 21205, USA 2Co-first author *Correspondence: surban@jhmi.edu http://dx.doi.org/10.1016/j.molcel.2015.12.022 SUMMARY Intramembrane proteases signal by releasing proteins from the membrane, but despite their importance, their enzymatic mechanisms remain obscure. We probed rhomboid proteases with reversible, mechanism-based inhibitors that allow precise kinetic analysis and faithfully mimic the transition state structurally. Unexpectedly, inhibition by peptide aldehydes is non-competitive, revealing that in the Michaelis complex, substrate does not contact the catalytic center. Structural analysis in a membrane revealed that all extracellular loops of rhomboid make stabilizing interactions with substrate, but mainly through backbone interactions, explaining rhomboid’s broad sequence selectivity. At the catalytic site, the tetrahedral intermediate lies covalently attached to the catalytic serine alone, with the oxyanion stabilized by unusual tripartite interactions with the side chains of H150, N154, and the backbone of S201. We also visualized unexpected substrate-enzyme interactions at the non-essential P2/P3 residues. These ‘‘extra’’ interactions foster potent rhomboid inhibition in living cells, thereby opening avenues for rational design of selective rhomboid inhibitors. INTRODUCTION Proteolysis inside the cell membrane lies at the regulatory core of many pathways that are paramount to the health of a cell (Brown et al., 2000; Chan and McQuibban, 2013; De Strooper and Annaert, 2010; Wolfe, 2009). Each of the four known families of intramembrane proteases continues to be implicated in diverse pathologies, including Alzheimer’s disease, Parkinson’s disease, cancer, malaria infection, hepatitis C virus maturation, tuberculosis virulence, and diabetes (Chan and McQuibban, 2013; De Strooper and Annaert, 2010; Manolaridis et al., 2013; Urban, 2009). In contrast to soluble proteases, which are arguably the best understood enzymes and among the most effective therapeutic targets (Drag and Salvesen, 2010), the catalytic mecha- nisms of these membrane-immersed enzymes are incompletely understood and have proved difficult to target effectively for therapeutic benefit (Golde et al., 2013). Inhibitors that chemically mimic intermediates in the reaction pathway offer a powerful means to dissect the enzymatic mechanism of a reaction (Hedstrom, 2002). Kinetic analysis of inhibition can reveal how the reaction is ordered and/or functionally organized, while structural analysis can identify the specific atomic contacts that the enzyme forges to guide substrates through the catalytic steps. However, this powerful strategy has eluded the study of intramembrane proteolysis (Nguyen et al., 2015); kinetic analysis of catalysis inside the membrane has not been possible until only recently (Dickey et al., 2013; Kamp et al., 2015). Moreover, inhibitor co-structures have been achieved thus far only with rhomboid proteases, and despite more than a dozen such high-resolution rhomboid-inhibitor structures in the Protein Data Bank (PDB) (Brooks and Lemieux, 2013), all structural information is limited to irreversible inhibitors. These agents form adducts that distort the active site and thus offer limited insights into the reaction mechanism and do not permit kinetic analysis. In fact, no reversible inhibitors of any kind have yet been developed for studying rhomboid proteolysis (Nguyen et al., 2015). For serine proteases such as rhomboid, the ‘‘committed step’’ for proteolysis under physiological conditions is formation of the covalent acyl intermediate following nucleophilic attack by the catalytic serine; once a substrate reaches this step, it is destined to complete the cleavage reaction (Hedstrom, 2002). Peptides with a C-terminal aldehyde moiety bind as substrates and, after attack on the terminal aldehyde carbon by the protease, arrest the reaction. The resulting catalytic complex faithfully mimics the key high-energy tetrahedral transition state that must be stabilized by the enzyme for catalysis to proceed (Hedstrom, 2002). Peptide aldehydes offer several notable advantages for interrogating the enzymatic mechanism of rhomboid proteases. First, because peptide aldehydes resemble substrates precisely and inhibit the protease reversibly, they allow detailed kinetic analysis. Structurally, because the only reaction is between the serine nucleophile and a true carbonyl, peptide aldehydes avoid the unnatural alkylation of the catalytic histidine base that besets isocoumarins or chloromethylketones, which ultimately distort the active site by crosslinking the catalytic serine and histidine Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. 329 Figure 1. Inhibition Kinetics of Peptide Aldehydes on Intramembrane Proteolysis by GlpG (A) Mechanisms of GlpG inhibition (inhibitor warheads are in red). Nucleophilic attack by the serine oxygen of rhomboid (blue) results in a covalent tetrahedral intermediate that differs only at one substituent between aldehydes (hydrogen, red) and natural substrates (black). Both produce an identical oxyanion (pink) that rhomboid must stabilize through electrophilic catalysis. Inhibitor adducts that deviate from natural proteolytic intermediates are shaded with red ovals (see text). (B) Summary of the 11 substrate peptides used in this study (Ac denotes acetylation, CHO is an aldehyde, NH2 is an amide, and CMK is a chloromethylketone). (C) Comparison of GlpG inhibition by various peptides. GlpG was assayed in real time while reconstituted in liposomes formed from E. coli lipids, and peptide aldehydes were added to proteoliposomes when the proteolytic reaction was initiated (without pre-incubation). vo and vi denote initial rates in the absence and presence of inhibitors, respectively. Only Ac-RKVRMA-CHO inhibition was modeled by including an inaccessibility parameter (see Figure S1A). (D) Kinetics of inhibition by Gurken peptide aldehydes was non-competitive (also see Figure S1B). Left: initial reaction rates versus substrate concentration in the presence or absence of peptide aldehydes are plotted. Right graph compares the resulting KM and Vmax parameters under different inhibitor concentrations. Note that for competitive inhibition, KM would increase in response to inhibitor concentration, while Vmax would be unaltered. For non-competitive inhibition, KM would be unaffected, but Vmax would decrease in response to inhibitor concentration. p values of parameter differences relative to no inhibitor controls are shown above each bar. residues (Figure 1A). The atom being attacked during catalysis with peptide aldehydes is a carbonyl carbon that generates a true oxyanion, unlike with phosphonates or sulfonylfluorides (Powers et al., 2002), in which the oxygen is not negatively charged and is connected to a non-carbon atom (Figure 1A). Finally, using peptide aldehydes overcomes the naturally low affinity of rhomboid for substrates by stabilizing the natural covalent attachment step that follows serine attack, while observing covalent linkage provides assurance that substrate had adopted a catalytically competent conformation. 330 Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. We recently developed an inducible, real-time assay capable of quantifying the kinetics of proteolysis directly inside the membrane (Dickey et al., 2013). Therefore, to gain a better understanding of the mechanism underlying intramembrane catalysis, we sought to adapt this approach to characterize the inhibition kinetics of substrate-mimicking peptide aldehydes. Next, because mounting evidence indicates that cell membranes affect the properties of rhomboid proteases beyond just serving as their environment (Bondar et al., 2009; Moin and Urban, 2012; Urban and Moin, 2014; Vinothkumar, 2011), we developed conditions to crystallize a catalytically active rhomboid protease in a membrane. Soaking rhomboid crystallized from bicelles with the most potent substrate peptide aldehydes produced high-resolution structures that revealed the characteristics of substrate stabilization during catalysis by a membrane-immersed rhomboid protease. Finally, we unexpectedly visualized ‘‘extra’’ substrate-enzyme interactions that we subsequently found are able to drive rhomboid inhibition to completion in living cells, thereby opening avenues for the rational and selective design of rhomboid inhibitors. RESULTS Kinetic Analysis of Peptide Aldehyde Inhibition Because the kinetics of inhibition have not been studied within the membrane, our first goal was to adapt our inducible reconstitution assay to study the inhibition kinetics of peptide aldehydes directly within the membrane. We therefore modeled a series of peptide aldehydes (Figure 1B) on three well-studied rhomboid substrates (Strisovsky et al., 2009; Urban et al., 2002), namely, TatA, Spitz, and Gurken, and examined their ability to inhibit GlpG, the rhomboid protease from Escherichia coli, reconstituted within the membrane. A key feature of our approach is the fact that both substrate and peptide aldehydes engage the active protease together at the start of the reaction (without needing to pre-incubate rhomboid with inhibitor as in prior studies). Although most peptide aldehydes had Ki values in the millimolar range, Gurken hexapeptide and tetrapeptide aldehydes proved to be more potent inhibitors of GlpG (Figure 1C). The Gurken tetrapeptide aldehyde Ac-VRMA-CHO exhibited a Ki of 113 ± 4 mM, and there was a further increase in potency with the hexapeptide aldehyde Ac-RKVRMA-CHO. As anticipated, inhibition required the aldehyde moiety, because replacing it with an amide raised Ki by >20-fold. This is consistent with rhomboid displaying weak affinity for substrates (Dickey et al., 2013) and forming a reversible covalent product with peptide aldehydes. Contrary to expectation, the mode of inhibition proved to be non-competitive (Figure 1D), with no significant change in KM (p = 0.16–0.80) but a strong and dose-dependent decrease in Vmax (p = 2.1 3 109 to 2.0 3 1013). This was also clearly reflected in the residuals when we compared fitting the kinetic data to a competitive versus a non-competitive model. Moreover, we observed non-competitive inhibition with tripeptide (Ac-RMACHO), tetrapeptide (Ac-VRMA-CHO), and hexapeptide (AcRKVRMA-CHO) aldehydes (Figure S1) and with Providencia stuartii AarA (a rhomboid protease with seven transmembrane segments that shares <15% identity with E. coli GlpG). Taken together, these observations suggest that non-competitive inhibition is the mechanism by which peptides based on substrate sequences preceding the cleavage site inhibit rhomboid proteases. Crystallization of an Active Rhomboid Protease in a Membrane We next sought to reveal the atomic basis of how peptide aldehydes interact with rhomboid as it catalyzes proteolysis in the membrane environment. Prior work established that an inactive mutant of E. coli rhomboid GlpG, whose structure has been studied extensively (Brooks and Lemieux, 2013), can be crystallized from a membrane bicelle (Figure 2A) (Faham and Bowie, 2002; Vinothkumar, 2011). Although such an inactive mutant prohibits studying catalysis, this strategy provided a starting point for crystallizing an active rhomboid in a membrane. We therefore searched our bank of >200 GlpG mutants for those that maintain activity and structural stability (Baker and Urban, 2012) and ultimately succeeded in crystallizing an active GlpG variant (Y205F) in a membrane (Figure 2B). Notably, phenylalanine at position 205 occurs naturally in many rhomboid orthologs (Bondar et al., 2009), had no effect on the structural stability of GlpG (Figure 2C), and slowed proteolytic rate less than 2-fold (Figure 2D). This is a very mild effect, because the natural variation in catalytic rate between rhomboid orthologs ranges 10,000-fold (Dickey et al., 2013). The resulting structure revealed a gate-open conformer with lateral displacement of the entire TM5 gating helix and fully disordered overlying L5 Cap loop, while the rest of the protein was unchanged relative to the closed state (Figure 2B). We next used this open conformer of rhomboid in a membrane environment as an excellent starting point for visualizing substrate binding and catalysis. Structure of Rhomboid in Catalytic Complex with Peptide Aldehydes To form the catalytic complex, we reacted GlpG in the open conformation crystallized from a membrane bicelle with our most potent Gurken peptide aldehydes. After iterative optimization, we ultimately succeeded in collecting four data sets that differed in reaction conditions and ranged from 2.2–2.4 Å in resolution and solved the resulting structures using molecular replacement (Figure 3A; Table 1). Globally, only minor changes in the protease accompany substrate binding and catalysis (Figure 3B). The greatest changes occurred in the extracellular L5 loop to the top of TM6. In the open conformer, the distal part of the L5 Cap became ordered, while TM5 maintained its open form (Figure 3B). The small size of our peptide aldehydes also allowed us to form complexes with gate-closed GlpG, which defined the minimal movements necessary for catalysis (Figure 3C). When we used the gateclosed form as a starting point, residues of the extracellular L5 loop and extending to the top of TM6 were displaced outward by substrate (Figure 3C). Excluding these 13 residues, the root-mean-square deviation (rmsd) along the remaining 158 alpha carbons of GlpG was only 0.65 Å. Moreover, the catalytic residues themselves maintained positions that were very similar Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. 331 Figure 2. Structure of an Active, Open Rhomboid Protease in a Membrane (A) Schematic of E. coli rhomboid GlpG in a DMPC/ CHAPSO membrane bicelle. The short-chain CHAPSO seals the edges of the DMPC membrane. (B) Structure of an open E. coli GlpG Y205F (yellow) in a membrane overlaid with a gate-closed form (red, 2IC8 crystallized from detergent). Left is topdown view (membrane in plane of the page), middle is a lateral view (dashed lines indicate approximate location of the membrane), and right are distance measurements between landmark TM5 and TM2 residue alpha carbons (beads). In the membrane environment, TM5 has shifted up and away from TM2. (C) Thermostability analysis of GlpG with tyrosine versus phenylalanine at position 205. (D) Real-time Michaelis-Menten kinetic analysis of wild-type versus Y205F GlpG reconstituted in liposomes formed from E. coli lipids. Inset compared kinetic parameters of Y205F relative to wild-type. Kinetic parameters are represented as mean ± SEM. to their arrangement in the apoenzyme (Figure 3D). These minor changes reveal that the apoenzyme monomer observed in most structures is in the catalytically active form, although it must be realized that natural substrates are much longer, and greater motions away from the active site undoubtedly are required to accommodate intramembrane proteolysis normally. The Tetrahedral Intermediate At the active site, the electron density of catalytic S201 was continuous with the substrate alanine aldehyde residue, indicating that covalent catalysis indeed took place (Figures 3A and 3E). Interestingly, catalytic H254 moved slightly but still made a hydrogen bond to S201 (at distances of 2.85 and 3.11 Å in the tripeptide and tetrapeptide structures, respectively). The aldehyde carbon was tetrahedral, thus mimicking the transition state and directly revealing the enzyme contacts that stabilize it during catalysis. Nucleophilic attack of the peptide carbonyl by the serine generates a negatively charged oxygen, the oxyanion (Figure 2A), that must be stabilized in the transition state for catalysis to proceed (Henderson, 1970). This key electrophilic catalysis feature of rhomboid proteases has been difficult to define, because prior studies used diverse non-peptidic inhibitors that suggested a series of possible interactions and at various distances (Vinothkumar et al., 2010, 2013; Xue et al., 2012; Xue and Ha, 2012); initial structures with isocoumarins suggested backbone interactions with L200 and S201 and weak (3.4 Å distance) side-chain interactions with H150 and N154, while the most recent b-lactam and chloromethylketone structures have the oxygen moiety positioned outside the oxyanion hole (Vinothkumar et al., 2013; Zoll et al., 2014). Our complex structure with a true peptide oxyanion now resolves these discrepancies (Figure 3E): the oxyanion hole of rhomboid is ultimately tripartite, being formed by the side chain of H150 (2.69 Å distance), amide backbone of catalytic S201 (2.91 Å distance), and the side chain of N154 (3.04 Å distance). Extended Substrate Interactions Beyond the site of catalysis, the substrate makes a network of interactions with the enzyme, some of which explain previously enigmatic observations. We recently engineered and quantified the effect of 200 mutants on both rhomboid structural stability and protease activity (Baker and Urban, 2012). Residues in the third extracellular loop (L3) in particular are generally not conserved and have little effect on enzyme architecture, yet dramatically compromise proteolytic activity (Baker and Urban, 2012). Although the basis for this was unclear, the substrateenzyme structure now reveals that L3 makes a collection of close backbone interactions with the substrate (Figure 4A), the geometry of which is most likely perturbed by even subtle side-chain alterations in L3. In fact, most enzyme-substrate interactions are backbone interactions (Figure 4A), explaining why rhomboid proteases display rather broad sequence selectivity (Akiyama and Maegawa, 2007; Moin and Urban, 2012; Strisovsky et al., 2009). A total of five hydrogen bonds formed between the substrate backbone and two enzyme loops: three with the inner L3 loop and two with the overlying L5 loop. Moving along the substrate from the site of catalysis (in the substrate termed P1 and moving sequentially outward from the aldehyde to P4), the five sites of contact are P1 NH with CO of G198 (L3), P2 CO with NH of A250 (L5), P2 NH with CO of S248 (L5), P3 CO with NH of G198 (L3), and P3 NH with CO of W196 (L3). A number of restrictions have been mapped in substrate side chains that rhomboid proteases can accommodate, the 332 Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. Figure 3. Conformation of Rhomboid Protease GlpG in Catalytic Complex with Substrate Peptide Aldehydes (A) Electron density map (2Fo–Fc at 2s) of peptide aldehydes (in cyan and red) in the active site of GlpG in the open and closed forms. (B) Top view of overlaid open GlpG with (yellow) and without (blue) substrate bound reveals changes in conformation. Note that residues 248–252 in the L5 loop were structured around substrate (arrow). (C) Top view of overlaid gate-closed GlpG with (yellow) and without (blue) substrate bound reveals minimal global changes in conformation. (D) Nucleophilic and electrophilic catalysis residues display minor positional changes between apoenzyme (blue) and substrate-bound (yellow) forms. (E) The substrate oxyanion (red mesh) is stabilized (dashed green lines) by the side-chain nitrogens of H150 (at 2.69 Å) and N154 (at 3.04 Å), as well as the backbone nitrogen of S201 (at 3.3 Å). Shown in mesh is the electron density map (2Fo–Fc at 2s). structural basis of which is now revealed in the substrateenzyme complex. In particular, rhomboid enzymes normally cleave only after small side chains (Akiyama and Maegawa, 2007; Moin and Urban, 2012; Strisovsky et al., 2009). However, the methyl group of the alanine points into a surprisingly large cavity (Figure 4B), part of which we previously proposed forms a water-retention site (Zhou et al., 2012). All three waters were unperturbed upon substrate binding, and the substrate-enzyme interaction at this restricted P1 site was surprisingly loose, again consistent with low affinity for substrates (Dickey et al., 2013). Yet changing the P1 alanine to valine alone severely compromised peptide aldehyde inhibition of GlpG (Figure 4C). Another side-chain preference has been found at P4 (Strisovsky et al., 2009), where some, but not all (Moin and Urban, 2012; Parussini et al., 2012), rhomboid enzymes prefer large hydrophobic residues. The substrate P4 valine formed hydrophobic interactions with F146 and M120 on the L1 loop (Figure 4D). However, this too was a rather shallow interaction, and reacting GlpG with a tripeptide missing the P4 residue entirely produced a nearly identical substrate conformation (Figure 4E). This observation suggests that the P4 residue has limited impact on GlpG catalysis. Indeed, mutation of large substrate residues at both P4 and P20 (I5A+F10A) that dramatically reduce proteolysis by 100-fold with its natural rhomboid protease P. stuartii AarA (Dickey et al., 2013) reduced E. coli GlpG cleavage by only 2-fold (Figure 4F). Even providing a large phenylalanine at the P4 residue did not enhance peptide aldehyde inhibitor potency against GlpG (Figure 4G). Moving further outward, we extended our structural analysis to Gurken hexapeptide aldehydes (Figure 4H). However, the N-terminal P5 and P6 residues were disordered in our structure, and their presence did not alter the position of P1–P4, arguing that GlpG does not contain specific substrate-interacting subsites beyond P4. Unexpected Stabilizing Interactions at P2 and P3 Particularly surprising were interactions made by the P2 and P3 side chains, because these positions were thought to be completely unrestricted in substrates (Akiyama and Maegawa, 2007; Moin and Urban, 2012; Strisovsky et al., 2009). Indeed, both side chains pointed upward and out from the active site (Figure 5A), explaining the apparent lack of restriction at these substrate positions. Nevertheless, the P3 arginine of Gurken in particular made ‘‘extra’’ contacts (Figure 5B); its guanidinium group made a series of interactions with the top surface of the L5 loop, including directly with the CO of the M247 backbone, as well as the side chains of S248, N251, and S193 (on L3), potentially through bridging water molecules. Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. 333 Table 1. Crystallography Statistics GlpG + VRMA GlpG-Y205F Bicelle GlpG-Y205F Bicelle +RMA GlpG-Y205F Bicelle +VRMA GlpG-Y205F Bicelle +RKVRMA R32 C222(1) C222(1) C222(1) C222(1) 110.6 71.2 70.8 70.6 70.3 110.6 98.1 97.5 98.0 96.1 126.7 63.0 62.8 62.6 62.7 44.80–2.00 (2.05–2.00) 42.56–2.20 (2.24–2.20) 57.51–2.20 (2.27–2.20) 62.55–2.30 (2.39–2.30) 56.75–2.30 (2.38–2.30) Data Collection Space Group Unit cell (Å) a, b, c Resolution (Å)a Observations 159,585 89,344 55,382 50,063 48,096 Unique reflectionsa 19,181 10,370 10,422 9,921 9,474 Multiplicity 7.9 (8.0) 6.8 (6.1) 5.3 (5.2) 5.0 (5.1) 5.1 (5.2) I/s(I) 8.2 (1.9) 6.9 (1.9) 6.7 (1.8) 5.7 (1.9) 5.8 (2.4) Completeness (%)a 99.3 (100.0) 98.4 (74.1) 91.3 (94.0) 99.8 (99.7) 97.6 (97.2) Rmergea 0.113 (0.443) 0.135 (0.437) 0.138 (0.739) 0.190 (0.571) 0.169 (0.522) 22.40–2.30 (2.36–2.30) 42.56–2.50 (2.56–2.50) 50.00–2.30 (2.36–2.30) 50.01–2.40 (2.46–2.40) 50.01–2.4 (2.46–2.40) Refinement Resolution (Å)a No. of reflections 12,636 7,905 8,921 8,287 7,947 Rwork/Rfree 0.215/0.263 (0.289/0.332) 0.222/0.287 (0.244/0.397) 0.209/0.250 (0.265/0.332) 0.235/0.263 (0.318/0.343) 0.237 /0.286 (0.357/0.408) No. of atoms 1,526 1,520 1,487 1,504 1,499 Protein 1,482 1,467 1,464 1,475 1,449 Water 44 53 23 29 50 Average B factors (Å2) 54.8 50.6 45.8 39.4 50.6 Protein 54.8 50.3 44.1 37.8 48.4 Water 52.6 55.4 47.5 41.0 52.6 Bonds (Å) 0.018 0.012 0.008 0.012 0.011 Angles ( ) 1.89 1.37 1.10 1.38 1.32 Rmsd Ramachandran plot Outliers (%) 0.6 0.6 1.1 0.52 0.7 Favored (%) 95.5 95.0 95.5 93.1 90.7 a Values in parentheses correspond to highest resolution shell. Rfree was calculated over reflections in a test set not included in the atomic refinement: GlpG-VRMA, 4.9%; Y205F, 5.2%; Y205F-RMA, 4.8%; Y205F-VRMA, 5.1%; and Y205F-RKVRMA, 4.8%. To evaluate the contribution of these side-chain interactions to affinity, we mutated the corresponding residues individually in GlpG to alanine and quantified the ability of the Gurken tetrapeptide aldehyde to inhibit the GlpG variants (Figure 5C). The N251A mutant alone proved to be 3-fold less sensitive to peptide aldehyde inhibition (p = 6.4 3 1011), while S248A conferred a mild (1.5-fold) but significant (p = 0.00015) reduction in inhibition. Even mutating M247, which makes a backbone interaction with the arginine side chain, increased Ki by 2-fold (p = 5.4 3 107), perhaps by altering conformation of the L5 loop. In contrast to these L5 residues, mutating the L3 residue S193 that may also make a distant interaction with the P3 arginine did not affect inhibition (p = 0.51). Consistent with a network of interactions between GlpG and the P3 residue, combining these GlpG loop mutants proved to be synergistic (Figure 5D). The S248A+N251A double mutant became 6-fold less sensitive to inhibition, while further incorporating S193A into a triple mutant decreased inhibition by 7.6-fold. As such, residues within the L5 loop provide multifaceted stabilizing interactions with the P3 side chain, with additional minor contributions offered by other nearby residues, including S193. To evaluate the functional consequences of these interactions in substrate processing in living cells, we made the corresponding R243A mutant in full-length Gurken and found that even this single mutant reduced processing of Gurken in living E. coli cells by 2-fold (Figure 5E). These unanticipated ‘‘extra’’ interactions may thus explain ‘‘sequence contexts’’ that lead to some substrates being cleaved more efficiently than others (Akiyama and Maegawa, 2007; Moin and Urban, 2012; Strisovsky et al., 2009). 334 Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. Figure 4. An Interaction Network Stabilizes the Substrate in the Rhomboid Active Site (A) Backbone interactions between the extended peptide substrate (cyan) and the L5 (left) and L3 (right) loops of GlpG stabilize the catalytic complex. (B) Magnified view of the P1 substrate alanine side chain in the S1 and water-retention site (shaded), showing the three bound water molecules (red spheres). (C) Inhibition kinetics of Gurken tetrapeptide aldehydes with P1 alanine versus valine. (D) The P4 side chain forms hydrophobic interactions with M120 and F146 of the L1 loop as an S4 pocket (shaded). (E) Overlay of substrate conformation in the GlpG active site with (tetrapeptide in cyan) and without (tripeptide in gray) the P4 valine. (F) Kinetics of P. stuartii AarA versus E. coli GlpG on TatA harboring alanines at both P4 and P20 (I5A+F10A) in reconstituted liposomes formed from E. coli lipids. (G) Inhibition kinetics of the indicated P4 Gurken tetrapeptide aldehydes. (H) Overlay of the tetrapeptide (gray) and hexapeptide (orange) substrate conformation in the GlpG active site. Shown in mesh is the experimental electron density map (2Fo–Fc at 2s), revealing that the P5 and P6 residues (hypothetically drawn in ball and stick) were disordered and thus not making specific contacts with GlpG. Finally, because the rhomboid:substrate interactions that we visualized at P3 had an effect even on processing of a fulllength substrate, we examined whether these interactions play a particularly important role in achieving inhibition of rhomboid by small molecules. In agreement with this postulate, we found that a Gurken tetrapeptide aldehyde with just its P3 residue changed to alanine, as occurs naturally in the TatA substrate, was 10-fold less potent at inhibiting GlpG (Figure 5F). As such, our structural and kinetic observations reveal how contacts at ‘‘unrestricted’’ positions can make ‘‘extra’’ stabilizing interactions that dramatically increase inhibitor potency. Peptide Aldehydes Are Potent and Specific Rhomboid Inhibitors in Living Cells Our serendipitous discovery potentially offers a strategy for the rational design of small-molecule rhomboid inhibitors that are effective in living cells. Prior groundbreaking high-throughput screens of >58,000 compounds using detergent-solubilized rhomboid enzymes identified a series of b-lactams that proved to be potent inhibitors in vitro but failed to block endogenous GlpG activity completely in E. coli cells even when pre-incubated with cells prior to inducing substrate expression (Pierrat et al., 2011). We therefore examined whether our peptide aldehydes were effective in vivo by examining their activity against endogenous GlpG in the same E. coli strain. Adding Ac-VRMA-CHO to growing E. coli cells resulted in a complete block to TatA-Flag processing by GlpG, and this block did not require pre-incubating cells with inhibitor prior to inducing substrate expression (Figure 6A). As expected, inhibition relied on the aldehyde moiety, because converting it to an amide in Ac-VRMA-NH2 abolished inhibition completely. Even the apparent half maximal inhibitory concentration (IC50) of AcVRMA-CHO (98 ± 11 mM) was indistinguishable from the Ki (113 ± 4 mM) that we measured using our proteoliposome system (Figure 6B), indicating that proteoliposomes are an effective mimic of GlpG in the natural setting of the E. coli membrane. We next examined whether the ‘‘extra’’ elements we found to be important for inhibition in vitro are required for blocking GlpG activity in living cells (Figure 6C). Importantly, substituting the P3 arginine with alanine essentially abolished inhibitor efficacy, again indicating that, in addition to the much studied P4 and P1 residues, ‘‘extra’’ interaction at neglected residues can be very effective at driving inhibitor potency. Contrary to expectation, however, we discovered that converting the aldehyde moiety to a chloromethylketone warhead in our tetrapeptides actually compromised inhibitor potency by nearly an order of magnitude (Figure 6C). To date, all rhomboid Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. 335 Figure 5. Unanticipated P3 Residue Interactions and Their Implications on Substrate Cleavage and Rhomboid Inhibition (A) The P2 (methionine) and P3 (arginine) residue side chains point out from the active site. Shown (and in B) is Ac-VRMA-CHO bound to GlpG, with dashed lines denoting hydrogen bonds. (B) The guanidinium group of the P3 substrate arginine side chain makes a series of ‘‘extra’’ stabilizing contacts, including hydrogen bonding (dashed line) directly with the backbone CO of M247 at a distance of 3.06 Å, and potentially with the hydroxyl side chains of S193, S248, and/or N251 (perhaps via water-mediated interactions). (C) Inhibition kinetics of Gurken tetrapeptide aldehyde Ac-VRMA-CHO quantified against four single GlpG loop mutants. (D) Bar graph comparing the Ki of Ac-VRMA-CHO with eight different GlpG loop variants (wild-type, single, double, and triple mutants are shown, with p values relative to wild-type above each bar). (E) Mutating the P3 arginine in Gurken (R243A) alone reduced cleavage 2-fold in E. coli cells. (F) Inhibition kinetics of Gurken tetrapeptide aldehydes with arginine versus alanine at the P3 position. inhibitors are irreversible and form covalent adducts with the protease (Nguyen et al., 2015), which is thought to increase inhibitor potency. Instead, our analysis indicates that the key element for achieving effective inhibition of rhomboid in vivo is a reversible warhead. Finally, although we did not set out to develop a potent inhibitor, even our limited series of peptide aldehydes already yielded an inhibitor that was effective at blocking endogenous GlpG activity in living E. coli cells with an IC50 in the low micromolar range: Ac-RKVRMA-CHO displayed an IC50 of 2.9 ± 0.3 mM (Figure 6D). These early observations provide proof of principle for using reversible warheads and cultivating ‘‘extra’’ interactions at nonessential substrate positions as a promising strategy for developing rhomboid inhibitors. DISCUSSION In conclusion, we integrated kinetic analysis of rhomboid inhibition by substrate-mimicking peptide aldehydes with highresolution X-ray crystallography to interrogate the enzymatic mechanism of rhomboid proteases. An important advance is our ability to study rhomboid catalysis with reversible inhibitors and entirely inside the membrane (both kinetically and crystallographically), which is known to affect the catalytic properties of these membrane-immersed enzymes (Bondar et al., 2009; Moin and Urban, 2012; Urban and Moin, 2014; Vinothkumar, 2011). Ultimately, we identified an additional, early step in the kinetic scheme for rhomboid proteolysis, visualized several key features of rhomboid catalysis at atomic resolution, and, unexpectedly, discovered a promising alternative avenue for targeting these enzymes. Inhibition Kinetics Identify Two Distinct SubstrateBound Complexes Particularly unusual but very informative is the non-competitive mode of inhibition by substrate-mimicking peptide aldehydes, because it reveals that rhomboid proteases exist in two distinct substrate-bound forms (Figure 7). To result in non-competitive inhibition, peptide aldehydes must, by definition, be able to bind to the substrate-enzyme Michaelis complex. Importantly, in addition to structural analysis, we further validated the catalytic center as the binding site that leads to this non-competitive 336 Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. Figure 6. Peptide Aldehydes Inhibit Endogenous GlpG in Living E. coli Cells (A) Western analysis (anti-Flag) of TatA-Flag cleavage by endogenous GlpG in cells cultured in the presence or absence of 1 mM tetrapeptides Ac-VRMA-NH2 (NH2) or Ac-VRMA-CHO (CHO). In all panels, D indicates a control strain in which the endogenous GlpG gene was deleted, while ± indicates whether expression of the TatA-Flag substrate was induced or not induced. The first (and in C last) lane of each gel image shows cells in which substrate expression was not induced. (B) Quantification of inhibition by Ac-VRMA-CHO of endogenous GlpG in living cells (blue line) or pure GlpG reconstituted in proteoliposomes (black line). (C) Western analysis (anti-Flag) of TatA-Flag cleavage by endogenous GlpG in cells cultured in the presence or absence of the indicated tetrapeptides (CMK denotes chloromethylketone). (D) Western analysis (top, anti-Flag) and quantification (lower graph) of inhibition by Ac-RKVRMA-CHO of endogenous GlpG in living cells. inhibition by engineering ‘‘resistant’’ mutants in GlpG that compromised peptide aldehyde inhibition. Therefore, in the Michaelis complex, substrate does not contact the catalytic center. Non-competitive inhibition exhibited by substrate peptide aldehydes reveals what is likely to be the organizing feature underlying rhomboid proteolysis. We previously speculated that, by analogy to DNA glycosylases (Friedman and Stivers, 2010), rhomboid proteases could exist in two forms: the interrogation complex and the scission complex (Dickey et al., 2013). Initial binding of a substrate transmembrane segment laterally in the membrane to a gate-open rhomboid would form the interrogation complex, which in kinetic terms is the Michaelis complex. At this point, a ‘‘pause’’ would give those transmembrane segments that are predisposed to unwind an opportunity to extend toward the inner site of catalysis, while those that do not unwind could not be cleaved and are ultimately ejected (Dickey et al., 2013; Moin and Urban, 2012). This has functional implications, because it gives rhomboid a separate step for discriminating substrates from non-substrates, but this step would render the substrate-bound Michaelis complex nevertheless sensitive to peptide aldehyde attack. The non-competitive mode of inhibition now provides direct experimental evidence for two such functionally separate states of the protease. The kinetic scheme for rhomboid proteolysis has, therefore, a step in which a conformation change converts the Michaelis complex to a catalytic complex. This is an important distinction, because it corrects a recent study with peptide chloromethylketones bound at the catalytic center that was assumed to be the Michaelis complex (Zoll et al., 2014). This was an entirely reasonable assumption at the time, because the kinetic mode of inhibition could not be determined in that study because rhomboid had to be pre-incubated with the irreversible inhibitors for 3 hr prior to adding substrate. We next used X-ray crystallography to visualize the catalytic features of the scission complex itself, namely, covalent linkage between substrate and nucleophilic serine, the tetrahedral intermediate formed by substrate, electrophilic stabilization of the oxyanion by rhomboid, and the network of interactions that secure substrate for catalysis. Electrophilic Catalysis and Oxyanion Stabilization The decisive step in serine protease catalysis is formation of the high-energy transition state tetrahedral intermediate. In fact, although most attention is usually paid to the nucleophilic attack by the serine, progressing through this step absolutely relies on enzymatic stabilization of the oxyanion. Our work here finally reveals the true nature of this electrophilic form of catalysis by rhomboid proteases, because it visualizes a peptide with a true tetrahedral carbon and oxyanion. Prior structures observed a variety of oxyanion interactions and distances, which have been confounding to extrapolate to natural proteolysis because the inhibitors that were studied distort the catalytic center by covalently attaching to both the catalytic serine and histidine, and/or were not peptidic, and/or did not produce a carbon-oxyanion (Vinothkumar et al., 2010, 2013; Vosyka et al., 2013; Xue et al., 2012; Xue and Ha, 2012; Zoll et al., 2014). Interestingly, the mechanism of oxyanion stabilization in rhomboid also proved to be unexpected, because it is tripartite, which has not, to our knowledge, been observed previously for any protease. This multifaceted oxyanion stabilizing network might also explain the long-standing puzzle of why mutating the asparagine residue compromises protease activity to different extents in different rhomboid enzymes (Urban et al., 2001); local geometric Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. 337 Figure 7. Kinetic Scheme of Rhomboid Intramembrane Proteolysis and Its Inhibition by Substrate-Mimicking Peptide Aldehydes A rhomboid protease is drawn in blue, with the substrate transmembrane helix in green, and the aldehyde moiety of the substrate-mimicking peptide aldehyde inhibitor depicted as a red star. The white S (scission site) in rhomboid marks the inner site of catalysis, while the I (interrogation site) indicates the peripheral gateopen site to which transmembrane segments first dock. We previously determined experimentally that KM is equal to Kd for rhomboid proteolysis (Dickey et al., 2013). differences in some rhomboid enzymes might allow them to compensate more easily than others for the absence of asparagine during oxyanion stabilization. Distal Rhomboid:Substrate Interactions Beyond the site of catalysis, we also visualized the network of distal rhomboid:substrate interactions. Rhomboid proteases generally display broad sequence selectivity, and indeed the predominant substrate:enzyme interactions were backbonebackbone interactions with the extended substrate chain sandwiched between the L3 and L5 loops. Nevertheless various substrate mutagenesis studies have converged on the importance of small P1 residues for substrate cleavage (Akiyama and Maegawa, 2007; Moin and Urban, 2012; Strisovsky et al., 2009). Contrary to expectation, the P1 methyl side chain points into a surprisingly large cavity that could potentially accommodate valine, yet valine in this position blocks cleavage completely (Dickey et al., 2013; Strisovsky et al., 2009). This large cavity contains three water molecules that we previously identified and postulated form a water-retention site to aid proteolysis (Zhou et al., 2012). The stark requirement for small residues at P1 may thus not be steric but rather may result from a disruptive effect larger side chains may have on the water-retention site. It must be emphasized that further analysis is required to test this hypothesis. Some rhomboid enzymes (Strisovsky et al., 2009), but not others (Moin and Urban, 2012; Parussini et al., 2012), also display a preference for hydrophobic P4 residues for efficient substrate proteolysis. With GlpG, the P4 side chain forms a shallow but specific interaction with the top of the L1 loop. However, because GlpG has only a mild preference for large residues at this position, visualizing a P4-dependent rhomboid like AarA would provide more information. More distant residues appear to have limited effect on proteolysis; both P5 and P6 residues were disordered in our structure, arguing that GlpG does not contain specific substrate-binding subsites beyond P4. The P1 and P4 interactions we observed are consistent with a beautiful study that used peptide chloromethylketones modeled on the TatA substrate (Zoll et al., 2014); despite major differences in warhead chemistry and peptide sequence, the overall distal contacts are similar, providing confidence that these approaches are revealing true characteristics of extended substrate:rhomboid interactions. Unexpected Substrate Interactions Drive Rhomboid Inhibition More surprising and potentially useful are the unexpected P2 and P3 interactions we discovered, because these positions were thought to be unrestricted in rhomboid substrates, and the side chains capable of making these interactions were not studied in other structures (Strisovsky et al., 2009; Zoll et al., 2014). Indeed, although the side chains face up and out of the active site, they make fortuitous yet highly stabilizing interactions. The series of ‘‘extra’’ P3 arginine contacts with the L5 loop alone increase peptide aldehyde potency by 10-fold, yield inhibitors that are effective without requiring any pre-incubation with rhomboid, and are able to block proteolysis inside the membrane (rather than in detergent, which renders the active site more accessible). In fact, these peptide aldehydes were effective at inhibiting endogenous rhomboid activity to completion in living E. coli cells. An even more surprising feature that proved to be critical for effective inhibition in vivo was the reversible aldehyde warhead itself. It is widely (and reasonably) assumed that irreversible inhibitors are more potent, because they trap proteases covalently in dead-end complexes. This, however, turned out to be 338 Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. incorrect with rhomboid, because identical peptides harboring the well-studied irreversible chloromethylketone warhead in place of the aldehyde were 10-fold less effective at inhibiting rhomboid in living cells. Whether this results from the unusual catalytic properties of rhomboid proteases (Dickey et al., 2013) and/or reflects chemical differences imparted by the membrane environment (Moin and Urban, 2012) remains to be determined. Nevertheless, these observations should motivate a re-evaluation of current strategies for rhomboid inhibition, which are centered entirely on irreversible warheads (Nguyen et al., 2015). In simple terms, starting with reversible warheads such as aldehydes and systematically cultivating interactions at neglected ‘‘nonessential’’ positions in substrates may prove to be an untapped but valuable strategy for the rational design of selective rhomboid inhibitors. Finally, although this marks a promising start both to understanding catalysis in the membrane and discovering opportunities for its selective inhibition, additional high-resolution structures are essential to achieving a full understanding of rhomboid proteolysis. We now have structures of the catalytic scission complex but an obvious lingering gap is the Michaelis complex, which remains a mystery. Nevertheless, our current efforts establish a platform for leveraging the unusual features of rhomboid proteases to achieving these goals for this fascinating but incompletely understood class of membrane-immersed enzymes. EXPERIMENTAL PROCEDURES Protein Purification, Crystallization, Data Collection, and Refinement E. coli GlpG (DN-GlpG: residues 87–276) was expressed and purified as described (Dickey et al., 2013). Thermostability analysis was performed using a StarGazer-384 differential static light-scattering system (Harbinger Biotech) as described in detail previously (Baker and Urban, 2012). For structural analysis, all rhomboid enzymes were expressed and purified under identical conditions, indicating that differences in conformation result mainly from their final environment (detergent versus bicelle) during crystallization. The active DN-GlpG Y205F variant was crystallized from a bicelle composed of DMPC/ CHAPSO (2.8:1) that was prepared using a previously developed method as described (Faham and Bowie, 2002). Reservoir buffer contained 0.1 M NaOAc (pH 5.5), 3M NaCl, and 5% ethylene glycol. Crystals of DN-GlpG in the gateclosed form were obtained in hanging drops at room temperature over a reservoir containing 0.1 M Tris (pH 8.5), 3 M NaNO3, and 15% glycerol. Peptide aldehydes were custom synthesized commercially using solid-phase chemistry, purified to >90% purity by reverse-phase high-performance liquid chromatography, and verified by mass spectrometry. Crystals were incubated with 2.5 mM peptide aldehydes in reservoir buffer for 7–24 hr at room temperature to generate the substrate complex, and flash-frozen in a nitrogen stream. X-ray diffraction data were collected at the Cornell High Energy Synchrotron Source, processed with iMosflm 1.0.7, and scaled with Aimless in the CCP4 program suite. Structures were solved by molecular replacement using Molrep. Electron density maps calculated from the solution of molecular replacement clearly revealed density directly above of connected with catalytic S201, and was modeled as the substrate aldehyde. Structures were further refined by refmac5 and PHENIX with iterative manual model building using COOT. In Vitro Rhomboid Proteolysis Assay Real-time kinetic assays were performed as described (Dickey et al., 2013): 2–80 pmoles of rhomboid were mixed with 30 mg of liposomes formed from an E. coli polar lipid extract (Avanti Polar Lipids) and 25–1,600 pmoles of fluorescein isothiocyanate (FITC)-TatA substrate in 50 mM Na-acetate (pH 4.0) and 150 mM NaCl. Proteoliposomes were formed by rapid dilution and collected by ultracentrifugation at 600,000 3 g for 30 min, and proteolysis was initiated by neutralizing proteoliposomes with 50 mM Tris (pH 7.4) and 150 mM NaCl (and simultaneously adding peptides without pre-incubation for inhibition experiments). Because membranes quench FITC-TatA, rhomboid-mediated proteolytic release was monitored at 37 C in real time by quantifying FITC fluorescence in a Synergy H4 Hybrid plate reader (BioTek). For inhibition experiments, initial rates were extracted from progress curves monitored in real time, and reduction in rate was plotted as a function of inhibitor concentration and modeled using the R environment with the equation v0;I Ki ; = v0 Ki + ½I in which v0,I and v0 are initial rates in the presence and absence of inhibitor, respectively. Analysis of Rhomboid Proteolysis and Inhibition in E. coli Cells The GlpG+ E. coli K12 strain NR698 (kind gift from Tom Silhavy, Princeton University) that allows more accurate IC50 determination by rendering the outer membrane more permeable to small molecules (Ruiz et al., 2005) was used to generate a DGlpG strain in a two-step process. First, we introduced GlpG::Kan through P1-mediated transduction and then excised the Kan gene using FRT-mediated recombination. The wild-type and DGlpG strains were transformed with pBAD-PsTatA-Flag (Dickey et al., 2013) and grown in LB + ampicillin to an optical density at 600 nm of 0.4 by shaking cultures at 250 rpm at 37 C. Expression of the PsTatA-Flag substrate was then induced by adding arabinose to 25 mM or 1 mM, the indicated peptides were added to the induced cultures, and the cultures were grown for 2 hr, shaking at 250 rpm at 37 C. Whole-cell lysates were prepared by pelleting 0.8 ml of each culture and resuspending the cell pellets in reducing Tricine-SDS sample buffer (Life Technologies) and lysed by sonication. Proteins were resolved on 16% Tricine-SDS-PAGE gels (Life Technologies), transferred to nitrocellulose using the Trans-Blot Turbo system (Bio-Rad), and probed with the indicated antibodies. Immune complexes were visualized and quantified by infrared laser scanning on an Odyssey imager (Li-Cor Biosciences). ACCESSION NUMBERS Structure coordinates have been deposited into the PDB (http://www.rcsb. org/pdb/home/home.do) under the accession codes PDB: 5F5B (GlpG+ VRMA), PDB: 5F5D (GlpG-Y205F_bicelle), PDB: 5F5G (GlpG-Y205F_bicelle+ RMA), PDB: 5F5J (GlpG-Y205F_bicelle+VRMA), and PDB: 5F5K (GlpGY205F_bicelle+RKVRMA). SUPPLEMENTAL INFORMATION Supplemental Information includes one figure and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2015.12.022. AUTHOR CONTRIBUTIONS S.U. designed the study and oversaw its execution. S.C. solved the structures of GlpG with substrate peptides. S.U. and S.W.D. generated DNA constructs, S.W.D. performed enzyme activity and inhibition analyses, and S.U. conducted the in vivo inhibition experiments. S.U. wrote the paper, and all authors approved the final manuscript. ACKNOWLEDGMENTS We are grateful to members of the Urban lab for helpful discussions, particularly to Rosanna Baker, who purified the mutants analyzed in Figures 5C and 5D. This work was supported by National Institutes of Health (NIH) grants 2R01AI066025 and R01AI110925, the Howard Hughes Medical Institute, and the David and Lucile Packard Foundation. X-ray diffraction data were collected using instruments at the Cornell High Energy Synchrotron Source (beamline supported by National Science Foundation grant DMR-0936384 and NIH grant GM-103485). Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc. 339 Received: July 16, 2015 Revised: November 20, 2015 Accepted: December 15, 2015 Published: January 21, 2016 Nguyen, M.T., Kersavond, T.V., and Verhelst, S.H. (2015). Chemical Tools for the Study of Intramembrane Proteases. ACS Chem. Biol. 10, 2423–2434. REFERENCES Parussini, F., Tang, Q., Moin, S.M., Mital, J., Urban, S., and Ward, G.E. (2012). Intramembrane proteolysis of Toxoplasma apical membrane antigen 1 facilitates host-cell invasion but is dispensable for replication. Proc. Natl. Acad. Sci. U S A 109, 7463–7468. Akiyama, Y., and Maegawa, S. (2007). Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol. Microbiol. 64, 1028–1037. Pierrat, O.A., Strisovsky, K., Christova, Y., Large, J., Ansell, K., Bouloc, N., Smiljanic, E., and Freeman, M. (2011). Monocyclic b-lactams are selective, mechanism-based inhibitors of rhomboid intramembrane proteases. ACS Chem. Biol. 6, 325–335. Baker, R.P., and Urban, S. (2012). Architectural and thermodynamic principles underlying intramembrane protease function. Nat. Chem. Biol. 8, 759–768. Bondar, A.N., del Val, C., and White, S.H. (2009). Rhomboid protease dynamics and lipid interactions. Structure 17, 395–405. Brooks, C.L., and Lemieux, M.J. (2013). Untangling structure-function relationships in the rhomboid family of intramembrane proteases. Biochim. Biophys. Acta 1828, 2862–2872. Brown, M.S., Ye, J., Rawson, R.B., and Goldstein, J.L. (2000). Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–398. Powers, J.C., Asgian, J.L., Ekici, O.D., and James, K.E. (2002). Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 102, 4639– 4750. Ruiz, N., Falcone, B., Kahne, D., and Silhavy, T.J. (2005). Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121, 307–317. Strisovsky, K., Sharpe, H.J., and Freeman, M. (2009). Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol. Cell 36, 1048–1059. Urban, S. (2009). Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat. Rev. Microbiol. 7, 411–423. Chan, E.Y., and McQuibban, G.A. (2013). The mitochondrial rhomboid protease: its rise from obscurity to the pinnacle of disease-relevant genes. Biochim. Biophys. Acta 1828, 2916–2925. Urban, S., and Moin, S.M. (2014). A subset of membrane-altering agents and g-secretase modulators provoke nonsubstrate cleavage by rhomboid proteases. Cell Rep. 8, 1241–1247. De Strooper, B., and Annaert, W. (2010). Novel research horizons for presenilins and g-secretases in cell biology and disease. Annu. Rev. Cell Dev. Biol. 26, 235–260. Urban, S., Lee, J.R., and Freeman, M. (2001). Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107, 173–182. Dickey, S.W., Baker, R.P., Cho, S., and Urban, S. (2013). Proteolysis inside the membrane is a rate-governed reaction not driven by substrate affinity. Cell 155, 1270–1281. Drag, M., and Salvesen, G.S. (2010). Emerging principles in protease-based drug discovery. Nat. Rev. 9, 690–701. Faham, S., and Bowie, J.U. (2002). Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J. Mol. Biol. 316, 1–6. Friedman, J.I., and Stivers, J.T. (2010). Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry 49, 4957–4967. Golde, T.E., Koo, E.H., Felsenstein, K.M., Osborne, B.A., and Miele, L. (2013). g-Secretase inhibitors and modulators. Biochim. Biophys. Acta 1828, 2898– 2907. Hedstrom, L. (2002). Serine protease mechanism and specificity. Chem. Rev. 102, 4501–4524. Urban, S., Schlieper, D., and Freeman, M. (2002). Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr. Biol. 12, 1507–1512. Vinothkumar, K.R. (2011). Structure of rhomboid protease in a lipid environment. J. Mol. Biol. 407, 232–247. Vinothkumar, K.R., Strisovsky, K., Andreeva, A., Christova, Y., Verhelst, S., and Freeman, M. (2010). The structural basis for catalysis and substrate specificity of a rhomboid protease. EMBO J. 29, 3797–3809. Vinothkumar, K.R., Pierrat, O.A., Large, J.M., and Freeman, M. (2013). Structure of rhomboid protease in complex with b-lactam inhibitors defines the S20 cavity. Structure 21, 1051–1058. Vosyka, O., Vinothkumar, K.R., Wolf, E.V., Brouwer, A.J., Liskamp, R.M., and Verhelst, S.H. (2013). Activity-based probes for rhomboid proteases discovered in a mass spectrometry-based assay. Proc. Natl. Acad. Sci. U S A 110, 2472–2477. Wolfe, M.S. (2009). Intramembrane proteolysis. Chem. Rev. 109, 1599–1612. Henderson, R. (1970). Structure of crystalline alpha-chymotrypsin. IV. The structure of indoleacryloyl-alpha-chyotrypsin and its relevance to the hydrolytic mechanism of the enzyme. J. Mol. Biol. 54, 341–354. Xue, Y., and Ha, Y. (2012). Catalytic mechanism of rhomboid protease GlpG probed by 3,4-dichloroisocoumarin and diisopropyl fluorophosphonate. J. Biol. Chem. 287, 3099–3107. Kamp, F., Winkler, E., Trambauer, J., Ebke, A., Fluhrer, R., and Steiner, H. (2015). Intramembrane proteolysis of b-amyloid precursor protein by g-secretase is an unusually slow process. Biophys. J. 108, 1229–1237. Xue, Y., Chowdhury, S., Liu, X., Akiyama, Y., Ellman, J., and Ha, Y. (2012). Conformational change in rhomboid protease GlpG induced by inhibitor binding to its S0 subsites. Biochemistry 51, 3723–3731. Manolaridis, I., Kulkarni, K., Dodd, R.B., Ogasawara, S., Zhang, Z., Bineva, G., O’Reilly, N., Hanrahan, S.J., Thompson, A.J., Cronin, N., et al. (2013). Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature 504, 301–305. Zhou, Y., Moin, S.M., Urban, S., and Zhang, Y. (2012). An internal water-retention site in the rhomboid intramembrane protease GlpG ensures catalytic efficiency. Structure 20, 1255–1263. Moin, S.M., and Urban, S. (2012). Membrane immersion allows rhomboid proteases to achieve specificity by reading transmembrane segment dynamics. eLife 1, e00173. Zoll, S., Stanchev, S., Began, J., Skerle, J., Lepsı́k, M., Peclinovská, L., Majer, P., and Strisovsky, K. (2014). Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures. EMBO J. 33, 2408–2421. 340 Molecular Cell 61, 329–340, February 4, 2016 ª2016 Elsevier Inc.





