1 Modelling Group fMRI Data – The fully general, ugly case
advertisement

Tom Nichols
ST416 - Advanced Topics in Biostatistics - Part 1: fMRI
1
ST416 - Advanced Topics in Biostatistics - Lecture 7
Multi-Subject Modelling
1
Modelling Group fMRI Data – The fully general, ugly case
Most of the lectures (until the end of last week, when I talked about permutation) have focused on modelling single subject’s data. Specifically, fitting time series regression models that
account for the experimental variation in the data Y while accounting for temporal autocorrelation in the errors . Now, we move on to consider modelling fMRI data from a group of
subjects with the goal of making inference on the population from which they were sampled.
For have N subject’s data, let the time series model for the kth subjects be
Yk = Xk βk + k ,
(1)
where Yk is a T -vector of the observed data, Xk is the T ×p predictor matrix, and k the T -vector
of random errors. Xk contains the BOLD HRF-convolved blocks or event that define the fMRI
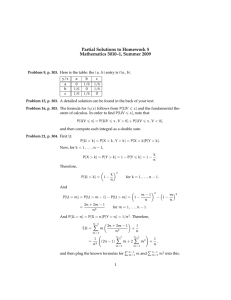
experiment (see “Experimental Predictors” in Fig. 1), as well as possibly drift regressors. In
practice, T may actually differ between subjects, but we leave it homogeneous for simplicity.
The the only thing that is required is that each column of the design matrices Xk means the
same thing in every subject.
Let’s assemble all subjects’ data into a grand multi-subject model:
X1
···
0
1
Y1
.
.
..
..
..
.
.
..
(2)
,
=
Y = Yk , X = ..
,
Xk
.
.k
.
..
..
...
N
YN
0
···
XN
where Y and are length-N T column vectors, and X is N T × N T block diagonal matrix.
Then we can write all these N “first level” models at once as
V1 σ12
···
0
..
.
.
..
2
Y = Xβ + , ∼ N (0, V ), V = ..
(3)
Vk σk
.
..
.
2
0
···
VN σN
where β is a length N p vector. Recall that the Vk model temporal autocorrelation; e.g. if an
|i−j|
AR(1) model with parameter ρk is used, ((Vk ))ij = ρk .
Tom Nichols
ST416 - Advanced Topics in Biostatistics - Part 1: fMRI
2
Experimental
Stimuli
0
20
40
60
80
100
120
Experimental
Predictors
0
20
40
60
80
100
120
0
20
40
60
80
100
120
+
+
...
+
Figure 1: The experimental stimuli and predictors associated with the BOLD response from a
singe voxel over time. The top color bar indicates when the subject was cued to tap their fingers
randomly (red), sequentially (green), only the index (yellow), or not at all (cyan). The associated experimental stimuli are shown, as well as the experimental predictors that are created by
convolving the stimuli with a HRF. Finally, the original BOLD response (black) is shown with
the predicted model fit (blue) based on the model formed with the experimental predictors.
Tom Nichols
ST416 - Advanced Topics in Biostatistics - Part 1: fMRI
3
So far, we have regarded the β as fixed, representing the properties of each individual subject. If we instead consider that we have sampled our N subjects at random from a population,
then we can pose a “second level” or group model and consider the β’s as random. Moreover, we can try to model the β’s and make inferences on properties of the group as a whole.
Specifically,
β = XG βG + G
(4)
where XG is the N p × pG group level design matrix, and G are the N p-vector of group-level
errors. For the experimental design illustrated in Figure 1, with 4 predictors, a XG matrix could
be just average each of the predictors,
I4
.
..
XG = I4
(5)
.
..
I4
where I4 is a 4 × 4 identity matrix. The interpretation of each element of βG would be the
average effect of the first three BOLD predictors in the population. Or we could model all of
the men and women separately; e.g. if all the women were listed first and then men,
I4
..
.
I
4
(6)
XG =
I4
..
.
I4
the first 4 elements of βG would represent the women’s population responses, and the second 4
elements would represent the men’s.
Subject responses are not independent. In particular, we expect that a subject that has the
largest BOLD response for one stimuli type is likely to have the largest BOLD response for
another stimuli type. It can also occur that subjects subjects with the largest response on one
type may have the smallest on others, exhibiting a correlations. Thus Var(G ) is not identity
Tom Nichols
but rather
ST416 - Advanced Topics in Biostatistics - Part 1: fMRI
···
VG
.
Var(G ) = ..
0
..
.
VG
···
0
4
.. 2
σ ,
.
G
..
.
VG
(7)
2
where pG × pG VG is the correlation in group effects, and σG
is the group-level variance.
Combining the first level (3) and second level (4) models into one, we get
Y = XXG βG + XG + .
(8)
This is sometimes referred to a variance components model, because we are modelling multiple sources of variation: Intrasubject measurement error variation in and between-subject
variation in XG . If you estimate these variance components, you can use Generalized Least
Squares (GLS) to ‘whiten’ the data and model with the matrix square root inverse (Cholesky
decomposition) of
Var(Y ) = Var(XG ) + Var()
(9)
and then find estimates of βG using OLS.
This model, however, is a disaster for fMRI. It says that if you want to make inference
on population parameters βG you need to keep all N subjects length-T time series around.
But each subject has 10’s of 1,000’s of voxels, and each voxel’s time series length T = 100
to T = 1, 000. This is feasible on modern computer hardware, but still much slower and
cumbersome than it needs to be.
2
Modelling Group fMRI Data – The summary statistics approach
While the models fit to each subject are involved (Fig. 1 is a quite a simple design) in practice
all investigators want to make inference on is one contrast at a time. I will show how, if there is
only 1 measure per subject taken to the second level model, the maths simplify tremendously,
we can see better what is actually happening (and, crucially, it is computationally very fast).
To restate notation once more, for data on N subjects, the kth subject is modelled as
Yk = Xk βk + k
where Yk is a length T vector, Xk is N × p matrix of BOLD predictors, and k is the length
T vector of random errors. To keep things simple, I will forget about autocorrelation, and let
Tom Nichols
ST416 - Advanced Topics in Biostatistics - Part 1: fMRI
5
k ∼ N (0, Iσk2 ); this would correspond to the setting where there is neglible autocorrelation
(rare) or when we have already decorrelated data and model by some means.
We are interested in just one particular contrast c of the parameters, cβk , estimated with
cβ̂k = c(Xk0 Xk )−1 Xk0 Yk , E(cβ̂k ) = cβk , Var(cβ̂k ) = c(Xk0 Xk )−1 c0 σk2 .
It will be helpful to have this result in hand
cβ̂k = cβk + c(Xk0 Xk )−1 Xk0 k
clearly showing how cβ̂k is just a peturbed version of cβk (can you show this!?).
Again, we are not just intersted in subject k, but want to make inference on the population
from which these N subjects were drawn. Thus the cβk are random, as they will change with
each new draw of N random subjects. Assemble these random variables of interest into γ =
[cβ1 , . . . , cβN ]0 , and then our model for these unobservable quantities is
γ = XG βG + G ,
2
G ∼ N (0, IσG
)
where XG is a N × pG “second level” design matrix, and G is the N -vector of econd level
2
.
errors, with “pure between subject” variance σG
Of course we don’t observe γ, but only γ̂ = [cβ̂1 , . . . , cβ̂N ]0 . A little manipulation shows
the model for γ̂ is
γ̂ = γ + γ̂ − γ
= XG βG + G + (γ̂ − γ)
= XG βG + G̃ .
2
where G̃ is the “mixed effects” error. Var(G̃ ) is the sum IσG
and Var(γ̂ − γ), but what is this
second term? Of course Var(γ̂ + a) = Var(γ̂) for any constant a, but γ is not a constant but a
random variable. Due to the helpful result above, though, it works out as if γ were a constant:
For the kth element of (γ̂ − γ)
Var ((γ̂ − γ)k ) = Var(cβ̂k − cβk )
= Var(cβk + c(Xk0 Xk )−1 Xk0 k − cβk )
= Var(c(Xk0 Xk )−1 Xk0 k )
= c(Xk0 Xk )−1 Xk0 Var(k )Xk (Xk0 Xk )−1 c0
= c(Xk0 Xk )−1 c0 σk2
= Var(cβ̂k )
where I have used the independence of k , but in fact you can get the equivalent result when
Var(k ) = Vk σk2 and whitening has been used.
Tom Nichols
2.1
ST416 - Advanced Topics in Biostatistics - Part 1: fMRI
6
Estimating the Group fMRI Model - Summary Statistics Approach
So, to review, for group fMRI data, we fit the estimated contrast data γ̂ with the model
γ̂ = XG βG + G̃
(10)
Var(γ̂) = Var(G̃ )
2
= IσG
+ diag({Var(cβ̂k )}).
This shows that this is a “components of variance” model: The variance in the group contrast
estimates γ̂ (when considered as samples from a population and not just as individual subjects)
is a sum of a pure between-subject variance (same for all subjects) and a within-subject term
(possibly different for each subject k).
Once again we can use Generalised Least Squares (GLS) to estimate this. As a reminder,
for a vanilla General Linear Model
Y = Xβ + ,
the GLS estimate is obtained after “prewhitening” data and model:
Var()−1/2 Y = Var()−1/2 X β + Var()−1/2 and finding estimates with
β̂GLS =
=
−1
(Var()−1/2 X)0 (Var()−1/2 X)
(Var()−1/2 X)0 Var()−1/2 Y
−1
X 0 Var()−1 X
X 0 Var()−1 Y.
(11)
Note that, while the whitening matrix is an inverse square-root of variance, it appears in the
final expression for the estimate (11) as inverse variance.
For our model (10) on γ̂, the prewhitening matrix Var(G̃ )−1/2 is diagonal, with elements
1
((Var(G̃ )−1/2 ))k,k = q
2
σG
+ Var(cβ̂k )
2
Thus each of the N contrast estimates γ̂k is weighted according to the balance of σG
+Var(cβ̂k ).
“Bad” subjects, those with large Var(cβ̂k ) will be shrunk more than “good” subject with
2
smaller Var(cβ̂k ). But of course, the exact weight depends on σG
+ Var(cβ̂k ). In particu2
lar, if σG Var(cβ̂k ) it won’t matter what the relative values of Var(cβ̂k ) are for different k as
2
2
σG
+ Var(cβ̂k ) ≈ σG
for all k.
Tom Nichols
2.2
ST416 - Advanced Topics in Biostatistics - Part 1: fMRI
7
Summary Statistics Approach – Special case of one-sample group
model
To see more exactly how this works, consider the special case of a one-sample group model,
when pG = 1 and XG = [1, . . . , 1]0 . This is the case when we’re just trying to infer on the population mean of the bold response described by cβk . Using the expression for the whitened
P 2
−1
estimate (11), note that the term corresponding to (X 0 Var()−1 X) reduces to k (σG
+
Var(cβ̂k ))−1 , and so we get
, N
N
X
X
1
cβ̂k
.
γ̂G =
2
2
k=1 σG + Var(cβ̂k )
k=1 σG + Var(cβ̂k )
2.3
Summary Statistics Approach – Special case of homogeneous subjects
Consider the case when all subjects have the same intrasubject variance, i.e. Var(cβ̂k ) =
Var(cβ̂1 ) for all k = 2, . . . , N . Then the weighting term is a constant and can be brought
out
, N
N
X σ 2 + Var(cβ̂1 )
X
G
cβ̂k
γ̂G =
2
σ
k=1 G + Var(cβ̂1 )
k=1
,
N
X
cβ̂k
N.
=
k=1
This shows that when there is no hetereogeneity over subjects, the weighting vanishes and the
calculation reduces to the usual one-sample estimate, i.e. the average. This results will hold
2
.
approximately when the variation in Var(cβ̂k ) over subjects is small relative to σG
In general, for model 11, you can show that if the whitening matrix is a multiple of a identity
matrix, the GLS estimates are identical to the OLS estimates.
2.4
Estimating the Variance Components
So far I’ve blithly said that GLS is easy, just prewhiten by the square root inverse of the errors.
2
In practice, this can be diifficult. For example, how do we obtain estimates of σG
and Var(cβ̂k )
to perform the whitening of (10)? The standard answer is Restricted Maximum Likelihood
(REML). REML consists of writing down the log likelihood of the residuals eG = γ̂ − β̂G ,
e0G (Var(G̃ ))−1 eG + log |Var(G̃ )| + log |XG0 (Var(G̃ ))−1 XG |,
(12)
2
and maximizing with respect to the variance parameters in Var(G̃ )) = IσG
+diag({Var(cβ̂k )}).
2
Specifically, the variance parameters include σG and all parameters inside Var(cβ̂k ) (which includes σk2 and, if we were modelling serial autocorrelation, any parameters in Vk ).
Tom Nichols
ST416 - Advanced Topics in Biostatistics - Part 1: fMRI
8
In fMRI, however, we can take a wonderful short cut: Because we have hundreds of observations for each subject’s first level model, Var(cβ̂k ) = c(Xk0 Xk )−1 σk2 is very well estimated
by
d β̂k ) = c(X 0 Xk )−1 σ̂ 2 .
Var(c
k
k
Hence we can take Var(cβ̂k ) as fixed and known, and then there is but one variance parameter,
2
σG
.
This simplifies the iterative optimization that is needed to maximize (12) and makes it quite
fast, so much so that it can be done voxel-wise no problem. I won’t go into any more details of
2
REML, but for intution, it is handy to see how a methods of moments estimator of σG
works
2.5
2
with Methods of Moments
Estimating σG
Consider the usual sample variance of the N group level observations
1 X
(γ̂k − γ̂· )2
S 2 (γ̂) =
N −1 k
where γ̂· is the sample mean of the N observations.
For the one-sample group model, when XG = [1, . . . , 1]0 , you can show that
2
E S 2 (γ̂) = σG
+ Var(c0 β̂· )
where Var(c0 β̂· ) is the sample mean of the N intrasubect variance estimates. The method of
2
2
moments estimator of σG
is obtained by setting S 2 (γ̂) equal to E (S 2 (γ̂)) and solving for σG
:
2
σ̃G
= max{0, S 2 (γ̂) − Var(c0 β̂· )},
where I have used the maximum operator to prevent negative variance estimates from occuring.
Acknowledgments
These notes are based in part on Mumford & Nichols (2006).
References
Beckmann, C. F., Jenkinson, M., & Smith, S. M. (2003). General multilevel linear modeling
for group analysis in FMRI. NeuroImage, 20(2), 1052-1063.
Mumford, J. A., & Nichols, T. E. (2006). Modeling and inference of multisubject fMRI
data. IEEE Engineering in Medicine and Biology Magazine, 25(2), 42-51.