Document 12071403
Reconstructing long-term limnological trends in phytoplankton and toxin production using paleogenomics and next generation sequencing
a Toxicology Center, University of Saskatchewan, Saskatoon, Saskatchewan, Canada. b Department of Veterinary Biomedical Sciences, University of Saskatchewan, Saskatoon, Saskatchewan, Canada. c Global Institute for Water Security, University of Saskatchewan, Saskatoon, Saskatchewan, Canada. c School of Environment and Sustainability, University of Saskatchewan, Saskatoon, Saskatchewan, Canada.
INTRODUCTION
Tse, T. J.
a , Hill, A.
a , Wiseman, S.
a , Hecker, M.
a , Giesy J.P.
ab , Doig, L. E.
a , Wheater H.
c , and Jones, P. D.
ad
MATERIALS & METHODS PRELIMINARY RESULTS AND DISCUSSION
•
Lake Diefenbaker (LD):
• Large multi-purpose reservoir (225-km long), formed in 1967, located in southern Saskatchewan, Canada.
• Provides water for 45% of Saskatchewan residences (WSA 2012).
• Local reports and concerns regarding increasing frequency and abundance of algal blooms and their potential impacts on water quality and usability.
• Moderately eutrophic 1 up-reservoir to oligo-mesotrophic downreservoir 2 .
•
Buffalo Pound Lake (BPL):
• Provides water for 25% of Saskatchewan residences (WSA 2014).
• Hepato- and neurotoxins were observed in water samples collected in the summer of 2014.
• Naturally rich in nutrients (e.g. phosphate and nitrogen and dissolved organic carbon) which encourage the growth of phytoplankton (City of Regina Annual Report 2012).
• Eutrophic 3 .
•
Objectives:
• Reconstruct historical trends of the phytoplankton community in both reservoirs.
• Identify potential invasive and harmful cyanobacterial species.
• Identify toxin producing genes if known toxin-producers are present.
MATERIALS
& METHODS
•
Sediment Collection and Sectioning –
Sediment cores were collected from the Gardiner arm (red), and Qu’Appelle arm (yellow) from LD (summer of
2011) and BPL (green; fall of 2013) (Figure 1). Samples were sectioned at 1cm increments.
Figure 1: Map of sediment core locations from Lake Diefenbaker Gardiner arm (red),
Qu’Appelle arm (yellow) and Buffalo Pound Lake (green) in Saskatchewan, Canada.
Created by Peter Downing – Educational Media Access and Production © 2011
Sediment core
…….. .………
Bio-Rad Thermocycler MiSeq Desktop Sequencer
• Sequencing was completed using a • Genomic DNA (gDNA) was
MiSeq Desktop Sequencer. extracted from 1-g of sediment
• Trimming and selection of high (wet wt.) from each 1-cm quality sequences were performed increment using EZNA Soil using the Mothur software package DNA kits.
(v 1.33.3). Only sequences between • PCR amplification of the 23S
440 and 460 bp in length with no rDNA was accomplished using modified primer pairs
P23SrV_f1 and P23SrV_r1 4 ambiguous bases were retained.
These sequences were then identified using Blast2GO (v 2.7.2) in the nonwith Illumina recommended overhangs. redundant database.
PRELIMINARY RESULTS AND DISCUSSION
DNA Purity DNA Concentration
Figure 2. Genomic DNA concentrations (left) and purity
(right) from sediment cores collected from Buffalo Pound
Lake and Lake Diefenbaker,
Saskatchewan, Canada.
• gDNA was successfully extracted throughout each sediment core.
• Increasing concentration of total extracted gDNA in more recent sediments.
• DNA purity was relatively conserved in all sediment increments with average
260/280 absorbances of:
• BPL ~ 1.85
• LD: Gardiner arm ~ 1.79
• LD: Qu’Appelle arm ~ 1.65
16
18
20
22
9
10
12
14
Sediment depth
(cm)
1
2
3
4
5
6
7
8
24
26
28
30
Chlorella sp.
✔
✔
✔
✔
✔
✔
✔
✔
✔
✔
Green Algae
Chrysodidymus sp.
Cryptomonas sp.
✔
✔
✔
✔
✔
✔
✔
✔
✔
✔
Nannochloropsis sp.
✔
✔
✔
✔
✔
✔
✔
✔
✔
✔
✔
✔
Synechococcus sp.
✔
✔
✔
✔
✔
✔
✔
✔
✔
✔
✔
Cyanobacteria
Synechocystis sp.
Cyanobium sp.
✔
✔
✔
✔
Anabaena sp.
✔
✔
✔
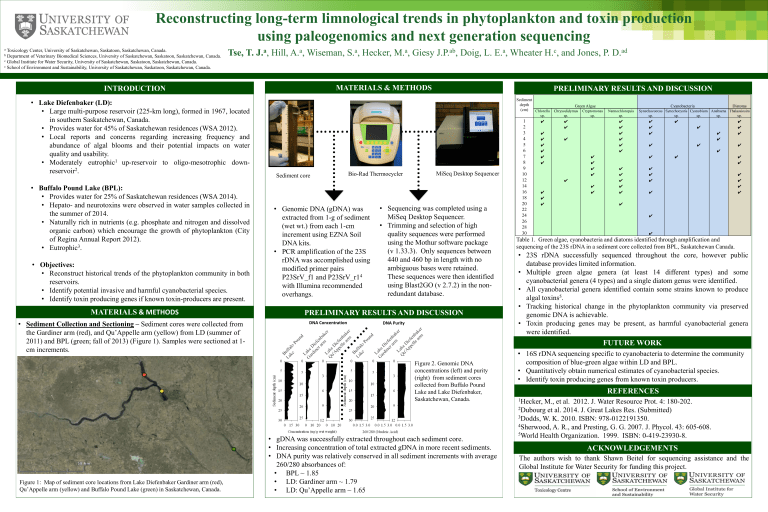
Table 1. Green algae, cyanobacteria and diatoms identified through amplification and sequencing of the 23S rDNA in a sediment core collected from BPL, Saskatchewan Canada.
Diatoms
Thalassiosira sp.
✔
✔
✔
✔
✔
✔
✔
✔
✔
✔
• 23S rDNA successfully sequenced throughout the core, however public database provides limited information.
• Multiple green algae genera (at least 14 different types) and some cyanobacterial genera (4 types) and a single diatom genus were identified.
• All cyanobacterial genera identified contain some strains known to produce algal toxins 5 .
• Tracking historical change in the phytoplankton community via preserved genomic DNA is achievable.
• Toxin producing genes may be present, as harmful cyanobacterial genera were identified.
FUTURE WORK
• 16S rDNA sequencing specific to cyanobacteria to determine the community composition of blue-green algae within LD and BPL.
• Quantitatively obtain numerical estimates of cyanobacterial species.
• Identify toxin producing genes from known toxin producers.
REFERENCES
1 Hecker, M., et al. 2012. J. Water Resource Prot. 4: 180-202.
2 Dubourg et al. 2014. J. Great Lakes Res. (Submitted)
3 Dodds, W. K. 2010. ISBN: 978-0122191350.
4 Sherwood, A. R., and Presting, G. G. 2007. J. Phycol. 43: 605-608.
5 World Health Organization. 1999. ISBN: 0-419-23930-8.
ACKNOWLEDGEMENTS
The authors wish to thank Shawn Beitel for sequencing assistance and the
Global Institute for Water Security for funding this project.




