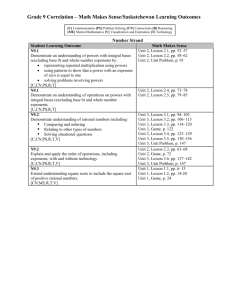
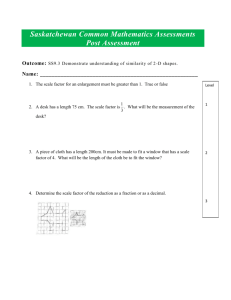
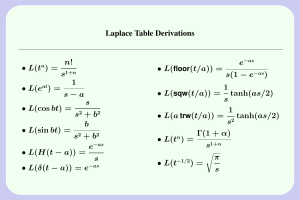
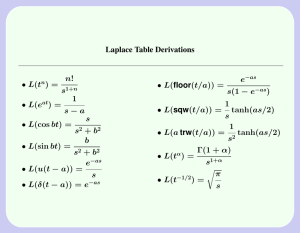
Single-cell enabled comparative genomics of a deep ocean SAR11 bathytype
advertisement

Supplementary Information for Single-cell enabled comparative genomics of a deep ocean SAR11 bathytype J. Cameron Thrash1,2,*, Ben Temperton1, Brandon K. Swan3, Zachary C. Landry1, Tanja Woyke4, Edward F. DeLong5, Ramunas Stepanauskas3 and Stephan J. Giovannoni1 1. Department of Microbiology, Oregon State University, Corvallis, OR 97331 2. Department of Biological Sciences, Louisiana State University, Baton Rouge, LA, 70803 3. Bigelow Laboratory for Ocean Sciences, East Boothbay, ME 04544 4. DOE Joint Genome Institute, Walnut Creek, CA 94598 5. Departments of Civil & Environmental Engineering and Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139 *To whom correspondence should be addressed: thrashc@lsu.edu Supplementary information contains: Supplementary Text Supplementary Methods Supplementary Figures S1-15 Supplementary Table_S1_F.xlsx Additional supporting text files, including scripts, supporting text files and fasta files, and SAG genome data, are available on the Giovannoni Lab website at: http://giovannonilab.science.oregonstate.edu/publications Page 1 of 21 Supplementary Text Basic comparative genomics results Table S1 details the results of our comparative genomics analysis using categorization of the 1764 orthologous clusters (OCs) found in the SAGs according to other SAR11 membership: those unique to a given SAG (unique), those shared by 2-4 SAGs but not by any other SAR11 genomes (shared Ic), those shared by all isolates and 1-3 SAGs (possible core), and those shared in all SAGs and all isolates (true core). All other distributions were not categorized. Each gene categorization and OC membership is cataloged in Table S1 and displayed in order along each scaffold in Figure S15. The incomplete nature of the SAGs makes it impossible to rule out the presence of an ortholog in the source cell and given the high percentage of conserved core genes among distantly related pure culture strains (Grote et al., 2012), possible core orthologs were so termed because their presence in all isolate genomes and at least one SAG made these the most likely candidates for missing orthologs in the remaining SAGs. There were 172 true core (10%) and 480 possible core (27%) OCs. The 544 SAG-specific clusters contained 400 unique (23%) and 144 shared Ic (8%) clusters, and 568 OCs (32%) had some other shared distribution among SAGs and pure-culture genomes. Negative data with regards to previously reported deep-ocean characteristics Many characteristics previously associated with deep ocean microorganisms, whether obtained from cultivated strains (Lauro and Bartlett, 2008, Nagata et al., 2010, Simonato et al., 2006)(and references therein) or from cultivation-independent analyses (Konstantinidis et al., 2009), were either not observed in the subclade Ic genomes, or were present, but shared with surface genomes. We examined amino acid substitution patterns among orthologs shared between the SAGs and the surface genomes (Konstantinidis et al., 2009) (Figs. 6, S10). While the overall magnitude of the substitution differences was small, the general trend was for relative increases in cysteine, isoleucine, lysine, asparagine, arginine and tryptophan in the predicted subclade Ic protein sequences at the expense of alanine, aspartatic acid, glutamic acid, methionine, glutamine, threonine and valine. These trends mostly held true when comparing amino acid substitutions with biochemically similar or different residues (+ and - results in blast output, respectively). The exceptions were in the cases of cysteine and asparagine, which showed relative increases only when examining different and similar amino acid substitutions, respectively (Fig. S12). Increased arginine and tryptophan residues were reported in a 4000 m sample from station ALOHA (Konstantinidis et al., 2009), yet many of the other depthassociated trends observed in that whole-community metagenome study were opposite our Page 2 of 21 more targeted SAR11 taxon analysis, for example, histidine and alanine were more frequently substituted in the surface strains compared to the SAGs, and the reverse was true for isoleucine and lysine. We identified no unique genes for polyunsaturated fatty acid synthesis, no unique duplications in ATPases or cytochrome c oxidases, and no unique genes for complex polymer utilization, such as pullulan, chitin, or cellulose. AAA240-E13 had a unique predicted xylanase/chitin deacetylase, however an OC with the same annotation was found in the isolate genomes (Table S1). In spite of their isolation from the ALOHA OMZ and considerable recruitment of metagenomic sequences from the heart of the ETSP OMZ at 200 m, the SAGs did not encode a high affinity cbb3-type cytochrome c oxidase, nor any genes for alternative terminal electron accepting processes, such as nitrate reductases. The SAG 16S rRNA genes didn’t have insertions associated with some piezophilic strains (Lauro et al., 2007) (Fig. S16). There were no unique tonB-type transporters, or ompH/L genes, and none of the SAGs had genes for the Strickland reaction other than thioredoxin, thioredoxin reductase, and a translation elongation factor (these three genes were not unique to the SAGs). The SAGs had putative cold shock protein genes and copies of the groE/L genes, but these were not unique to the Ic clade. AAA240-E13 and AAA288-E13 had a unique pulJ type II secretion component, but many of the surface strains had pillin and type II secretion genes. AAA288-N07 contained an intact aerobic carbon monoxide dehydrogenase (CODH) gene cluster, but this was shared with HIMB5 and HIMB114. Unlike the SAR324 and Arctic96-BD19 SAGs reported by (Swan et al., 2011), none of the SAR11 SAGs contained unique autotrophy pathways. Finally, unlike the results of (Konstantinidis et al., 2009), the SAGs showed no significant increases transposable elements compared to the surface SAR11s (Table S1). Additional observations based on relative abundance of genes in metagenomic datasets Qualitative assessment of additional clusters showed several additional genes of interest that, although were not statistically more abundant in deep samples compared to surface samples, nevertheless had considerable recruitment at depth compared to surface samples (Table S1). Among the additional genes highly abundant at depth that were also conserved among SAR11 genomes outside of the Ic subclade were predicted ABC-type amino acid transporters, TRAP transporters, glutamate and glutamine synthases, FeS cluster assembly genes, sec and twin arginine protein translocases, the coxL CODH subunit, subunits of the hetertetrameric sarcosine oxidase (Yilmaz et al., 2011), thioredoxin reductase, and ferrodoxin (Table S1). The unique xylanase/chitin deacetylase in AAA240-E13 (2236673789) was more abundant in deep water Page 3 of 21 samples, however, the copy of this gene shared with other SAGs and the isolate genomes (2236673495) was also among the most abundant genes in surface samples (Table S1). Ribosomal proteins and respiratory proteins such as cytochromes, NADPH:quinone oxidoreductase, and F0F1-type ATP synthases were also highly abundant in deep samples, usually with much lower abundance in surface samples, indicating that these highly conserved SAR11 genes probably have enough nucleotide divergence to serve as clade-specific markers. There were also a number of highly conserved (possible or true core) hypothetical genes that showed high abundance with depth. Additional phage information The SAGs shared genes with other SAR11 genomes that had at least partial homology to Pelagiphage genes. Whole genome BLAST of the four recently sequenced Pelagiphage genomes (Zhao et al., 2013) against the SAGs indicated significant hits > 100bp from two of the Pelagiphage, HTVC008M and HTVC019P, to true core and possible core genes such as NAD synthetase, peroxiredoxin, and ribonuclotide reductase, as well as flexible genome genes such as that encoding a small heat shock protein, ribosomal protein S21, and hypothetical proteins (Table S1). Supplementary methods Single-cell separation/ MDA Water samples for single cell analyses were collected from the mesopelagic (770m) using Niskin bottles during Hawaii Ocean Time-series (HOT) Cruise 215, station ALOHA (22°45ʹ′N, 158°00ʹ′W; 9 September 2009). Replicate, 1 mL aliquots of water were cryopreserved with 6% glycine betaine (Sigma) and stored at −80ºC (Cleland et al., 2004). Prior to cell sorting, samples with prokaryote cell abundances above 5x105 mL-1 were diluted 10x with filter-sterilized field samples and pre-screened through a 70 µm mesh-size cell strainer (BD). For heterotrophic prokaryote detection, diluted subsamples (1-3 mL) were incubated for 10-120 min with SYTO-9 DNA stain (5 µM; Invitrogen). Cell sorting was performed with a MoFlo™ (Beckman Coulter) flow cytometer using a 488 nm argon laser for excitation, a 70 µm nozzle orifice and a CyClone™ robotic arm for droplet deposition into microplates. The cytometer was triggered on side scatter. The “purify 1 drop” mode was used for maximal sort purity. Prokaryote cells were separated from eukaryotes, viruses, and detritus based on SYTO-9 fluorescence (proxy to nucleic acid content) and light side scatter (proxy to particle size)(del Giorgio et al., 1996). Synechococcus cells were excluded, based on their autofluorescence Page 4 of 21 signal. Target cells were deposited into 384-well plates containing 600 nL per well of 1x TE and stored at −80ºC until further processing. Of the 384 wells, 315 were dedicated for single cells, 66 were used as negative controls (no droplet deposited) and 3 received 10 cells each (positive controls). Cells were lysed and their DNA was denatured using cold KOH (Raghunathan et al., 2005). Genomic DNA from the lysed cells was amplified using multiple displacement amplification (MDA) (Dean et al., 2002, Raghunathan et al., 2005) in 10 µL final volume. The MDA reactions contained 2 U/µL Repliphi polymerase (Epicentre), 1x reaction buffer (Epicentre), 0.4 mM each dNTP (Epicentre), 2 mM DTT (Epicentre), 50 mM phosphorylated random hexamers (IDT) and 1 µM SYTO-9 (Invitrogen) (all final concentration). The MDA reactions were run at 30°C for 1216 h, then inactivated by a 15 min incubation at 65°C. Amplified genomic DNA was stored at 80°C until further processing. We refer to the MDA products originating from individual cells as single amplified genomes (SAGs). Prior to cell sorting, the instrument and the workspace were decontaminated for DNA as previously described (Stepanauskas and Sieracki, 2007). High molecular weight DNA contaminants were removed from all MDA reagents by a UV treatment in Stratalinker (Stratagene). An empirical optimization of the UV exposure was performed to remove all detectable contaminants without inactivating the reaction. Cell sorting and MDA setup were performed in a HEPA-filtered environment. As a quality control, the kinetics of all MDA reactions was monitored by measuring the SYTO-9 fluorescence using FLUOstar Omega (BMG). The critical point (Cp) was determined for each MDA reaction as the time required to produce half of the maximal fluorescence. The Cp is inversely correlated to the amount of DNA template (Zhang et al., 2006). The Cp values were significantly lower in 1-cell wells compared to 0-cell wells in all microplates for which MDA kinetics was monitored (p<0.001; Wilcoxon Two Sample Test). MDA products were diluted 50-fold in TE buffer and 500 nL aliquots of diluted MDA product served as the template DNA in 5 µL final volume real-time PCR screens. All PCR reactions were performed using LightCycler 480 SYBR Green I Master Mix (Roche) and the Roche LightCycler® 480 II real-time thermal cycler. PCR amplification of SSU rRNA and metabolic genes from SAGs was done using primers and conditions listed in Table S1. Forward (5´−GTAAAACGACGGCCAGT−3´) and reverse (5´−CAGGAAACAGCTATGACC−3´) M13 sequencing primers were added to the 5´ ends of each target primer pair to aid direct sequencing of PCR products. All PCR reactions were run for 40 cycles at the appropriate annealing temperature, followed by melting curve analysis performed as follows: 95°C for 5 s, Page 5 of 21 52°C for 1 min, and a continuous temperature ramp (0.11°C/s) from 52 to 97°C. Real-time PCR kinetics and amplicon melting curves served as proxies for detecting SAGs positive for target genes. New, 20 µL PCR reactions were set up for all PCR-positive SAGs and amplicons were sequenced from both ends using Sanger technology by Beckman Coulter Genomics. Single cell sorting, whole genome amplification, real-time PCR screens and PCR product sequence analyses were performed at the Bigelow Single Cell Genomics Center (www.bigelow.org/scgc). Selection based on MDA reaction kinetics Four SAGs in the Ic subclade were picked to cover the total depth of branching observed in the 16S phylogeny, and selected based on MDA reaction kinetics to improve the possibility of more complete genome recovery. The kinetics of all MDA reactions was monitored by measuring the SYTO-9 fluorescence using a FLUOstar Omega (BMG) plate reader. The MDA critical point (Cp) was determined for each reaction as the time required to produce half of the maximal fluorescence. The Cp value is inversely correlated to the amount of DNA template (Zhang et al., 2006). Sequencing/Assembly/Annotation Draft genomes were generated at the DOE Joint genome Institute (JGI) using the Illumina technology (Béjà et al., 2002). Illumina standard shotgun libraries were constructed and sequenced using the Illumina HiSeq 2000 platform. All general aspects of library construction and sequencing performed at the JGI can be found at http://www.jgi.doe.gov. All raw Illumina sequence data was passed through DUK, a filtering program developed at JGI, which removes known Illumina sequencing and library preparation artifacts (unpublished). The following steps were then performed for assembly: 1) filtered Illumina reads were assembled using Velvet v. 1.1.04 (Stein et al., 1996), 2) 1–3 kbp simulated paired end reads were created from Velvet contigs using wgsim (http://github.com/lh3/wgsim), 3) Illumina reads were assembled with simulated read pairs using Allpaths–LG v. r41043 (Hsiao et al., 2005). Parameters for assembly steps were: 1) Velvet: 63 -shortPaired and velvetg: -very clean yes -export -Filtered yes -min contig lgth 500 -scaffolding no -cov cutoff 10, 2) wgsim: -e 0 -1 100 -2 100 -r 0 -R 0 -X 0, 3) Allpaths: -LG PrepareAllpathsInputs: PHRED 64=1 PLOIDY=1 FRAG COVERAGE=125 JUMP COVERAGE=25 LONG JUMP COV=50, RunAllpathsLG: THREADS=8 RUN=std shredpairs TARGETS=standard VAPI WARN ONLY=True OVERWRITE=True. Page 6 of 21 Each raw sequence data set was screened against all finished bacterial and archaeal genome sequences (downloaded from NCBI) and the human genome to identify potential contamination in the sample. Reads were mapped against reference genomes with bwa version 0.5.9 (Philippot, 2002) using default parameters (96% identity threshold). None of the libraries showed significant contamination. Additionally, gene sequences of the final assemblies were compared against the GenBank nr database by BLASTX and taxonomically classified using MEGAN (Beaumont et al., 2002). Genes were identified using Prodigal (Hyatt et al., 2010). The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database (nr), UniProt, TIGRFam, Pfam, KEGG, COG, and InterPro databases. The tRNAScan-SE tool (Hacker and Kaper, 2000) was used to find tRNA genes, whereas ribosomal RNA genes were found by searches against models of the ribosomal RNA genes built from SILVA (Pruesse et al., 2007). Other non–coding RNAs such as the RNA components of the protein secretion complex and the RNase P were identified by searching genomes for the corresponding Rfam profiles using INFERNAL (Makarova et al., 1999). Additional gene prediction analysis and manual functional annotation was performed within the Integrated Microbial Genomes (IMG) (Markowitz et al., 2008) platform developed by the Joint Genome Institute, Walnut Creek, CA, USA (http://img.jgi.doe.gov). Orthology with other SAR11s SAG orthologs were determined using the automated phylogenomics pipeline Hal (Robbertse et al., 2011) combined with several curation steps. Protein-coding fastas for 8 pure-culture SAR11 genomes were obtained from IMG (https://img.jgi.doe.gov). Initial orthologous clusters were established by all vs. all BLASTP and subsequent Markov clustering (MCL) at the inflation parameter (I) 1.5, consistently with our previous work (Grote et al., 2012). These clusters were then filtered for genes with ≥ 30% difference to the median gene length of the cluster, similarly to (Grote et al., 2012), using cluster_size_filter.pl iteratively until clusters no longer split. Because of the fragmentary nature of the SAGs, genes from these genomes were not subjected to the same size filter, as it has been shown that highly incomplete genomes contain large numbers of partial open reading frames (ORFs) (Klassen and Currie, 2012). A separate evaluation of orthologs, based on synteny, was conducted using the methods of (Yelton et al., 2011). Syntenic genes from all pairwise comparisons were coalesced into clusters using synteny_clustering.py, and these clusters were compared with those from the Hal pipeline using cluster_comparison.py. In cases where the syntenic clusters contained additional genes to those in the Hal clusters, the cluster assignment based on synteny was used. Hypothetical Page 7 of 21 proteins associated with extraneous, unused scaffolds from HTCC1002 (scaffolds 3-5) (Grote et al., 2012) were manually removed. Finally, orthologous clusters for single-copy housekeeping genes were manually inspected to ensure no erroneous mis-assignment during these automated steps. As part of the Hal pipeline, cluster data is output in the form of a heatmap. This heatmap was manually curated to reflect the changes to the original clustering based on our filters. This heat map can be queried to establish numbers of shared orthologs between different genomes. Table S1 and Figure S15 display five gene categories: 1) orthologs unique to an individual SAG (unique), 2) orthologs shared among SAGs but not other SAR11 strains (shared Ic), 3) orthologs shared among all pure culture SAR11 genomes and between one and three SAGs (potential core), 4) orthologs shared among all SAR11 pure culture genomes and all SAGs (true core), 5) everything else. Note that many clusters contain more than one gene from a given genome. These may be considered paralogs for the purposes of this analysis, the same as in (Grote et al., 2012), and paralogs were ignored for the purposes of counting shared orthologs across clusters. However, caution should be used in interpreting duplicate genes within clusters for two reasons: 1) due to the incomplete nature of the SAGs and the potential abundance of partial or split ORFs across multiple scaffolds, duplicate genes from SAGs in a given cluster may not reflect “real” duplicates, and 2) given the clustering is the result of a partially automated pipeline and not hand-curated for each genome, the possibility remains that clusters may be artificially concatenated due to the inflation parameter selected for Markov Clustering. Post-assembly SAG scaffold QC Amplification of genomes from single cells increases the risk of contaminant DNA being included in the downstream genome sequencing and subsequent assembly compared to traditional sequencing of microbial genomes using billions-trillions of chromosomal copies. Although pre- and post-amplification QC has very high success with decreasing such contamination, sequenced SAG scaffolds were evaluated for possible cases of contamination using BLASTN, tetramer analysis, and manual inspection all scaffolds smaller than 1000bp for regions of low predicted ORF density. BLASTN of each scaffold was performed against the nr database with default settings. Scaffolds were flagged for further evaluation if they contained BLASTN hits with > 500bp alignment length to non-alphaproteobacteria, or if all BLASTN hits were to non-alphaproteobacteria. Tetramer analysis of each scaffold was carried out using a sliding window of 2000bp and a 200bp step. Principal Components Analysis (PCA) was carried out on the results to identify outlying scaffolds/regions of scaffolds, and these were flagged for Page 8 of 21 further evaluation. Those scaffolds flagged from the BLASTN and tetramer analyses were manually inspected for gene conservation among the other SAR11 strains. Those containing genes conserved in other strains were ruled out as contaminants. Finally all scaffolds smaller than 1000bp were manually inspected for predicted ORF density. The streamlined nature of SAR11 genomes results in very small non-coding regions throughout the genome, so scaffolds were visually screened based on coding density. All flagged scaffolds resulting from these analyses are identified in Table S1. However, it should be noted that a scaffold with BLASTN hits to non-alphaproteobacteria or outlying tetramer scores is not sufficient to identify contamination as these may simply reflect hypervariable regions within the SAG and/or instances of horizontal gene transfer. Therefore, scaffolds flagged as possible contaminants are to be treated with caution. Circos plot Circos plots were generated using Circos version 0.62-1. Scaffolds for each SAG were placed in order of size from largest to smallest. IMG annotations and OC designations were used as the base categorizations for the plot. GC content for each gene is represented as displayed in IMG. Links files were produced using a custom python script to organize segment duplications by OC designations using previously created tab-delimited files. The python script, Circos configuration files, and input data are available upon request. Estimated genome completion SAG genome completion was evaluated based on 599 single-copy genes present in all eight pure-culture SAR11 genomes. Overall SAG genome completion percentage was based on the percentage of these genes found in the SAGs (Table S1). AAI vs. 16S identity Average amino acid identity was calculated with the scripts/methods of (Yelton et al., 2011). Pairwise 16S rRNA gene identity was calculated with megablast using default settings. All data is available in Table S1. The heatplot of AAI vs. 16S was made in R. COG distribution The barplot (Fig. 4) and boxplot (Fig. S9) of the COG distribution among SAR11 genomes was built using data supplied by IMG (Table S1) with R. Page 9 of 21 Amino acid distribution Amino acid substitution pattern analysis was performed as described in Konstantinidis et al. 2009 (Konstantinidis et al., 2009). Briefly, the fasta file for each HAL cluster was used as a BLASTP query against a database containing non-redundant proteins from 8 surface SAR11 strains and 4 SAGs from 770m at ALOHA. Alignments with an expect value of less than 1 x 1020 were discarded. For remaining alignments, the number of similar amino acid substitutions, different amino acid substitutions and gaps was calculated between all surface strain proteins and SAG proteins. The fold-change in abundance of each amino acid between surface strain proteins and SAG proteins was calculated. All analyses are available in the IPython notebook ‘Calculating amino acid changes.ipynb’. Intergenic spacer analysis Intergenic spacer regions are provided as part of the IMG annotation process. Sizes and statistics for each set of intergenic regions were calculated using the fasta_length_counter.pl script. Distribution of intergenic regions was examined in R with histograms and box plots, and R was also used to run the Wilcoxon rank sum analysis: > wilcox.test(hist$surface,histr$Ic,paired=FALSE,conf.int=TRUE,conf.level=0.95) All analyses only included non-zero intergenic regions calculated by IMG. Transposable elements Transposable elements were assessed using the sequences collected by Brian Haas of the Broad Institute for the program TransposonPSI (http://transposonpsi.sourceforge.net). Sequences from this library (1537) were searched against a database of SAR11 genomes (see metagenomic reciprocal best blast, below) using tblastn on default settings. Deep water adaptation genes Genes identified as potential deep ocean adaptations were searched for by inspection of the SAG annotations, inspection of KEGG pathways on IMG, and BLASTP of homologs for the various features against the SAR11 genomes. The fasta file containing these homologs is available in Supplemental Information. The presence of 16S rRNA gene insertions was tested by aligning all SAR11 16S sequences with those of known piezophiles carrying the insertions (Lauro et al., 2007) using MUSCLE (Edgar, 2004), and manually inspecting the alignment. CRISPR region analysis and cas gene search Page 10 of 21 CRISPRs are annotated by IMG using the CRISPR recognition tool and PILER (see http://img.jgi.doe.gov for more details). The designated AAA240-E13 CRISPR region was further analyzed by examining reciprocal best BLAST hits (see “Metagenomics,” below) to those coordinates (scaffold 14, bases 24207-26268). Sequences were identified as having recruited to the corresponding coordinates of the single-scaffold AA240-E13 genome (Table S1) by use of metagenome_index.pl. To build the local recruitment plot in Figure 7, these sequences were aligned to the intergenic/CRISPR region with BLASTN with low-complexity filtering disabled. Additionally, the entire region was searched against the IMG v400 custom protein database with BLASTX on default settings to identify potential protein-coding regions. HMMs developed for a wide range of cas protein families were developed by Haft et al. (Haft et al., 2005) and Makarova et al. (Makarova et al., 2011), and made available through TIGRFAM (http://www.jcvi.org/cgi-bin/tigrfams/index.cgi). Each of the 46 HMMs from Haft et al. Table 1 and the 32 additional HMMs from Makarova et al. Table S4 were searched against the proteincoding sequences from all four SAGs using hmmsearch (part of the HMMER3 package (Eddy, 2011)) on default settings. Only 8 HMMs returned any hits to the SAGs. Of those, only three had hits in AAA240-E13. TIGR00372 had hits to hypothetical proteins in orthologous cluster 15001317. TIGR01587 had hits to type II DNA helicases conserved in all sequenced SAR11 genomes. TIGR03117 had hits to a RecG-like helicase, also conserved in all SAR11 genomes. The remaining HMMs with hits in SAR11 had low scores and small alignment regions. All positive search results are provided with our additional files. Phylogenetics SAG 16S rRNA gene sequences were combined with those of SAR11 strains in previously established subclades (Grote et al., 2012, Morris et al., 2005, Vergin et al., 2013) (SAG_reference.fasta). Sequences were aligned with MUSCLE (Edgar, 2004) using default settings and the tree was created with RAxML (Stamatakis et al., 2008). Thirty seven alphaproteobacterial and two outgroup 16S rRNA sequences were aligned with the subclade Ic clone sequence from (Vergin et al., 2013) and those of the sequenced Ic SAGs. AAA240-E13 had two partial 16S rRNA gene fragments after assembly, and the larger of the two was used. Alignments were performed with MUSCLE using default settings (Edgar, 2004), poorly aligned positions were removed with Gblocks (Castresana, 2000) using the following settings: –b1=(seqs/2)+1 –b2=(seqs/2)+1 –b3=seqs/2 –b4=2 –b5=h The final tree was computed using RAxML (Stamatakis, 2006) using the following settings: raxmlHPC-PTHREADS -x 1234 -T 4 -f a -m GTRGAMMA -\# 1000 Page 11 of 21 The phylogenomic tree of SAR11 strains was calculated as part of the Hal pipeline (Robbertse et al., 2011, Thrash et al., 2011), in this case, using liberal Gblocks settings, maximumlikelihood (RAxML), and 10% allowed missing data, which resulted in a concatenated alignment of 322 protein-coding genes (84,355 sites). Because of the small length of the proteorhodopsin gene and difficulty with the alignments using MUSCLE (data not shown), the combined, iterative alignment/phylogeny program HandAlign was utilized for the proteorhodopsin phylogeny (Westesson et al., 2012). Homologs were selected based on a BLASTP of the proteorhodopsin orthologs (cluster 1500536) in SAR11 against the IMG v350 protein gene database using default settings. Large gene homologs were removed using the cluster_size_filter.pl script: $ perl ~/scripts/cluster_size_filter.pl rhodopsin.tab 70 130 Eukaryotic rhodopsin homologs were removed manually as well as those from HTCC2255/HTCC2999 as these constituted potential contaminants. The tree was calculated using the following commands: $ handalign clean_rhodopsins3.faa --hmmoc-root hmmoc -l 255 -ts 2000 -af clean_rhodopsins3.trace.stk -ub > clean_rhodopsins3.MAP.stk $ stocktree.pl clean_rhodopsins3.MAP.stk > clean_rhodopsins3.MAP.newick Based on the results of (Grote et al., 2012), we knew distinction of the sarcosine dehydrogenase (sardh) and dimethylglycine dehydrogenase (dmgdh) from within the same OC (cluster 150013) required phylogenetic analysis. This was done using the bash script protPipeline3, which automates a phylogenetic pipeline including alignment with MUSCLE (Edgar, 2004), Gblocks editing of poorly aligned sites (Castresana, 2000), ProtTest amino acid substitution modeling (Abascal et al., 2005), and maximum-likelihood tree construction using RAxML (Stamatakis, 2006). The clusters formed based on the tree were designated 1500013.f.ok (dmgdh), 150013.f1.ok, 1500013.f2.ok, and 1500013.f3.ok (sardh). All unaligned fasta files for each of the single gene trees and the super alignment and model file for the concatenated protein tree provided in Supplemental Information. All tree labels were manipulated using the Newick utilities (Junier and Zdobnov, 2010). Metagenomics Seven samples were collected at the BATS site (31° 40’ N, 64° 10’ W) during August 19, 2002 (0, 40, 80, 120, 160, 200, and 250 m). To collect the microbial biomass, ~ 70 L of seawater was filtered through 0.2-µm polyethersulfone membranes (Supor, Pall, East Hills, NY, USA) using McLane Research Laboratories, Inc. (East Falmouth, MA) in situ water transfer systems and Page 12 of 21 membranes were stored at -80°C until further processing. Nucleic acids were extracted and purified as described (Morris et al., 2005). Library construction and shotgun sequencing were performed using a first-generation GS-FLX protocol (454 Life Sciences), yielding average read lengths of 226 bp. BATS metagenomes and corresponding geochemical metadata are available on CAMERA (http://camera.calit2.net) under the project ID CAM_PROJ_BATS. Data was also analyzed from 454 metagenomic sequences collected from Station ALOHA (Shi et al., 2011), the ETSP OMZ (Stewart et al., 2012), Puerto Rico Trench (Eloe et al., 2011a), the Sea of Marmara (Quaiser et al., 2010), and the Matapan-Vavilov Deep in the Mediterranean Sea (Smedile et al., 2013). All 454 metagenomic datasets were quality trimmed using lucy v1.2 (http://sourceforge.net/projects/lucy/) using a minimum good sequence length of 100bp and default error settings. Trimmed reads were dereplicated using CD-HIT-454 v4.6.1 (https://code.google.com/p/cdhit/) with the following parameters: (-c 1.00 –n 8). Sanger sequences from ALOHA (Konstantinidis et al., 2009), and the GOS (Rusch et al., 2007, Venter et al., 2004), were also subjected to the reciprocal best blast protocol, below, but without normalization due to their difference in read size. GOS sequences were first separated according to sample, and individual databases were created for each; samples with FILTER_MIN > 0.22 µm were excluded, as well as a single sample MOVE858 where FILTER_MAX was 0.22 µm. GOS surface sequences from Antarctica and the Southern Ocean (Brown et al., 2012) (Table S1) were dereplicated as above, but without quality trimming (no fastq files were available). For these samples, all available size fractions were combined for each sample. These were also excluded from gene centric normalization. Comparative recruitment of metagenomic sequences was completed using a reciprocal best BLAST (rbb). A total of 12 SAR11 genomes were included in the rbb- eight pure culture genomes (HTCC1062, HTCC1002, HTCC9565, HTCC7211, HIMB5, HIMB114, IMCC9063, HIMB59), and the four SAR11 SAGs. All incomplete SAR11 genome sequences were artificially concatenated in the order presented by IMG, with a string of nine “N” at each scaffold break for later identification of boundaries. The concatenated SAR11 genome sequences were searched against each metagenome database with BLASTN on default settings. All hits to SAR11 genomes were then searched against the entire IMG database (v400), which had been customized to contain only these 12 SAR11 genome sequences (others have been completed and deposited since this analysis began) using BLASTN on default settings except – max_target_seqs, set to 100000000, to allow for inclusion of all possible hits. Amino acid and nucleotide versions of this database, IMG v400 custom, were used in the average genome size and relative abundance calculations, respectively, below. Best hits from the second BLASTN Page 13 of 21 (reciprocal best hits- rbh) to the 12 SAR11 sequences were plotted with fragment_recruiter.r. Localization of each rbh to each gene in the SAR11 genomes was completed using the script metagenome_index.pl. Gene coordinates were established for each genome fasta using the gene coordinate workflow available in Supplemental Information. The gene coordinates for the concatenated SAGs is recorded in Table S1 under the tabs Start Coord (single) and End Coord (single). For relative abundance estimates of SAR11 in a given dataset, each set of 454 metagenomic sequences by sample was searched against the entire IMG v400 custom genome database using megablast with default settings. Best hits were then counted as either SAR11like or non SAR11-like, based on whether they recruited to any of the 12 SAR11 genomes or not, respectively. Investigating enrichment of SAR11 gene clusters at depth in metagenomic samples A complete description of methods can be found in the supplementary file ‘Statistical_analyses.pdf’. This document was generated with a runnable Sweave script, which is available on the Giovannoni GitHub repository (git@github.com:giovannoni-lab/bathytype.git). Briefly, metagenomes were classified as ‘surface’ (<200m) or ‘deep’ (≥200m) and the abundance of gene clusters as determined by reciprocal best-BLAST (see above) in each dataset was calculated. To identify statistically significant differences in gene cluster abundance between deep and surface samples, the R package DESeq (Anders & Huber, 2010) was used to correctly account for over-dispersion typically associated with gene abundances in high throughput sequencing studies. To validate the a posteriori classification of ‘deep’ and ‘surface’ samples, blind estimation of dispersion followed by clustering analysis of samples was performed. This analysis showed the MATA, MARM, PRT and CHOMZ.800 samples did not cluster correctly with other ‘deep’ samples. Therefore, DESeq analysis was performed both against the complete dataset and a subset with samples MATA, MARM, PRT and CHOMZ.800 removed. Investigating SAR11 Ic subclade abundance as a function of temperature A complete description of methods can be found in the supplementary file ‘Statistical_analyses.pdf’. This document was generated with a runnable Sweave script, which is available on the Giovannoni GitHub repository (git@github.com:giovannoni-lab/bathytype.git). To determine if increased SAR11 Ic abundance at depth was a result of an adaptation to lower temperatures, SAR11 Ic abundance and total SAR11 abundance was calculated in 91 GOS samples by reciprocal best-BLAST (see above). Negative binomial regression GLM analysis Page 14 of 21 was then applied to the data, using the total SAR11 counts as an offset. As the data consisted of different sequencing types (Sanger and 454), the sequencing type was included as a factor in the model. Model terms were sequentially dropped to test each term for significance. Sequencing type was shown to not be a significant regression factor and so was dropped from the model. The resultant model was then compared to the null model and the Nagelkerke coefficient of determination (Nagelkerke, 1991) was calculated. References Abascal F, Zardoya R, Posada D (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104-2105. Beaumont HJE, Hommes NG, Sayavedra-Soto LA, Arp DJ, Arciero DM, Hooper AB et al. (2002). Nitrite reductase of Nitrosomonas europaea is not essential for production of gaseous nitrogen oxides and confers tolerance to nitrite. J Bacteriol 184: 2557–2560. Béjà O, Koonin EV, Aravind L, Taylor LT, Seitz H, Stein JL et al. (2002). Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl Environ Microb 68: 335–345. Brown MV, Lauro FM, DeMaere MZ, Les M, Wilkins D, Thomas T et al. (2012). Global biogeography of SAR11 marine bacteria. Mol Sys Biol 8: 1-13. Castresana J (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540-552. Cleland D, Krader P, McCree C, Tang J, Emerson D (2004). Glycine betaine as a cryoprotectant for prokaryotes. J Microbiol Methods 58: 31-38. Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P et al. (2002). Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA 99: 5261-5266. del Giorgio PA, Bird DF, Prairie YT, Planas D (1996). Flow cytometric determination of bacterial abundance in lake plankton with the green nucleic acid stain SYTO 13. Limnol Oceanogr 41. Dobbek H, Gremer L, Meyer O, Huber R (1999). Crystal structure and mechanism of CO dehydrogenase, a molybdo iron-sulfur flavoprotein containing S-selanylcysteine. Proc Natl Acad Sci USA 96: 8884-8889. Eddy SR (2011). Accelerated Profile HMM Searches. PLoS Comput Biol 7: e1002195. Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792-1797. Page 15 of 21 Eloe EA, Fadrosh DW, Novotny M, Zeigler Allen L, Kim M, Lombardo M-J et al. (2011a). Going Deeper: Metagenome of a Hadopelagic Microbial Community. PLOS ONE 6: e20388. Eloe EA, Malfatti F, Gutierrez J, Hardy K, Schmidt WE, Pogliano K et al. (2011b). Isolation and characterization of a psychropiezophilic alphaproteobacterium. Appl Environ Microbiol 77: 81458153. Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ et al. (2012). Streamlining and Core Genome Conservation among Highly Divergent Members of the SAR11 Clade. mBio 3: e00252-00212. Hacker J, Kaper JB (2000). Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54: 641–679. Haft DH, Selengut J, Mongodin EF, Nelson KE (2005). A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLOS Comput Biol 1: e60. Hsiao WWL, Ung K, Aeschliman D, Bryan J, Finlay BB, Brinkman FSL (2005). Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLOS Genet 1: e62. Hyatt D, Chen G-L, LoCascio P, Land M, Larimer F, Hauser L (2010). Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. Junier T, Zdobnov EM (2010). The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26: 1669-1670. Klassen JL, Currie CR (2012). Gene fragmentation in bacterial draft genomes: extent, consequences and mitigation. BMC Genomics 13: 14. Konstantinidis KT, Braff J, Karl DM, DeLong EF (2009). Comparative Metagenomic Analysis of a Microbial Community Residing at a Depth of 4,000 Meters at Station ALOHA in the North Pacific Subtropical Gyre. Appl Environ Microbiol 75: 5345-5355. Lauro FM, Chastain RA, Blankenship LE, Yayanos AA, Bartlett DH (2007). The unique 16S rRNA genes of piezophiles reflect both phylogeny and adaptation. Appl Environ Microbiol 73: 838-845. Lauro FM, Bartlett DH (2008). Prokaryotic lifestyles in deep sea habitats. Extremophiles 12: 1525. Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI et al. (1999). Comparative genomics of the Archaea (Euryarchaeota): Evolution of conserved protein families, the stable core, and the variable shell. Gen Res 9: 608–628. Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P et al. (2011). Evolution and classification of the CRISPR/Cas systems. Nat Rev Micro 9: 467-477. Markowitz VM, Ivanova NN, Szeto E, Palaniappan K, Chu K, Dalevi D et al. (2008). IMG/M: a data management and analysis system for metagenomes. Nucl Acids Res 36: D534-538. Page 16 of 21 Morris R, Vergin K, Cho J, Rappe M, Carlson C, Giovannoni S (2005). Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr 50: 1687-1696. Nagata T, Tamburini C, Arístegui J, Baltar F, Bochdansky AB, Fonda-Umani S et al. (2010). Emerging concepts on microbial processes in the bathypelagic ocean – ecology, biogeochemistry, and genomics. Deep-Sea Res II 57: 1519-1536. Nagelkerke NJD (1991). A note on a general definition of the coefficient of determination. Biometrika 78: 691-692. Philippot L (2002). Denitrifying genes in bacterial and Archaeal genomes. BBA-Gene Struct Expr 1577: 355–376. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res 35: 7188–7196. Quaiser A, Zivanovic Y, Moreira D, López-García P (2010). Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J 5: 285-304. Raghunathan A, Ferguson Jr HR, Bornarth CJ, Song W, Driscoll M, Lasken RS (2005). Genomic DNA amplification from a single bacterium. Appl Environ Microbiol 71: 3342-3347. Robbertse B, Yoder RJ, Boyd A, Reeves J, Spatafora JW (2011). Hal: an Automated Pipeline for Phylogenetic Analyses of Genomic Data. PLOS Curr ToL 3: RRN1213. Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLOS Biol 5: e77. Shi Y, Tyson GW, Eppley JM, DeLong EF (2011). Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J 5: 9991013. Simonato F, Campanaro S, Lauro FM, Vezzi A, D&apos;Angelo M, Vitulo N et al. (2006). Piezophilic adaptation: a genomic point of view. J Biotechnol 126: 11-25. Smedile F, Messina E, La Cono V, Tsoy O, Monticelli LS, Borghini M et al. (2013). Metagenomic analysis of hadopelagic microbial assemblages thriving at the deepest part of Mediterranean Sea, Matapan-Vavilov Deep. Environ Microbiol 15: 167-182. Stamatakis A (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688-2690. Stamatakis A, Hoover P, Rougemont J (2008). A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol 57: 758-771. Stein JL, Marsh TL, Wu KY, Shizuya H, DeLong EF (1996). Characterization of uncultivated prokaryotes: Isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol 178: 591–599. Page 17 of 21 Stepanauskas R, Sieracki ME (2007). Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc Natl Acad Sci USA 104: 9052–9057. Stewart FJ, Ulloa O, DeLong EF (2012). Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol 14: 23-40. Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for Chemolithoautotrophy Among Ubiquitous Bacteria Lineages in the Dark Ocean. Science 333: 1296-1300. Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ et al. (2011). Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep 1: 1-9. Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66-74. Vergin KL, Beszteri B, Monier A, Thrash JC, Temperton B, Treusch AH et al. (2013). Highresolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-series Study site by phylogenetic placement of pyrosequences. ISME J: 1-11. Westesson O, Barquist L, Holmes I (2012). HandAlign: Bayesian multiple sequence alignment, phylogeny and ancestral reconstruction. Bioinformatics 28: 1170-1171. Yelton AP, Thomas BC, Simmons SL, Wilmes P, Zemla A, Thelen MP et al. (2011). A SemiQuantitative, Synteny-Based Method to Improve Functional Predictions for Hypothetical and Poorly Annotated Bacterial and Archaeal Genes. PLOS Comput Biol 7: e1002230. Yilmaz P, Gilbert JA, Knight R, Amaral-Zettler L, Karsch-Mizrachi I, Cochrane G et al. (2011). The genomic standards consortium: bringing standards to life for microbial ecology. ISME J 5: 1565-1567. Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW et al. (2006). Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol 24: 680-686. Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC et al. (2013). Abundant SAR11 viruses in the ocean. Nature 494: 357-360. Page 18 of 21 Supplementary Figures Figure S1. Maximum-likelihood tree of 16S rRNA genes for the SAR11-type SAGs used for initial selection of SAGs for genome sequencing. Outgroup is R. prowazekii; bootstrap values (n=100) are indicated at the nodes; scale bar indicates 0.3 changes per position. Figure S2. Recruitment plots of reciprocal best hit metagenomic sequences for Station ALOHA, by genome. Colors indicate depth. Y-axes: % identity; x-axes, genome length. Figure S3. Recruitment plots of reciprocal best hit metagenomic sequences for the Eastern Tropical South Pacific Oxygen Minimum Zone, by genome. Colors indicate depth. Y-axes: % identity; x-axes, genome length. Figure S4. Recruitment plots of reciprocal best hit metagenomic sequences for BATS, by genome. Colors indicate depth. Y-axes: % identity; x-axes, genome length. Figure S5. Recruitment plots of reciprocal best hit metagenomic sequences for the Puerto Rico Trench, by genome. Y-axes: % identity; x-axes, genome length. Figure S6. Recruitment plots of reciprocal best hit metagenomic sequences for the Sea of Marmara, by genome. Y-axes: % identity; x-axes, genome length. Figure S7. Recruitment plots of reciprocal best hit metagenomic sequences for the MatapanVavilov Deep, by genome. Y-axes: % identity; x-axes, genome length. Figure S8. Recruitment plots of reciprocal best hit Sanger sequenced metagenomic sequences for Station ALOHA at 4000 m, by genome. Y-axes: % identity; x-axes, genome length. Figure S9. Boxplot of COG distributions between surface (red) and subclade Ic genomes (“sag”, blue). Boxes indicate the upper and lower quartile, with the median as a black bar, error bars indicate adjacent values, and open circles indicate outside values. Orange boxes highlight COG categories with non-overlapping distributions. Figure S10. Relative abundance of amino acids based on their classification as similar or different amino acid substitutions. Page 19 of 21 Figure S11. Analysis of intergenic space. Distributions of intergenic spaces are plotted in histograms for A) all surface genomes and B) all Ic genomes, with an x-axis limit of 1000. Box plots of the same data, C) including outliers and D) excluding outliers to more easily view the median values. Wilcoxon rank sum results are reported in C. Figure S12. Putative partial pathway for purine degradation in subclade Ic (taken from KEGG). Enzymes for each transition are labeled, along with the corresponding OC from this analysis, and colored according to the key. The uncolored S-allantoin amidohydrolase indicates no matching annotations in any of the genomes. Gene organization on AAA240-E13 is depicted in the upper right. Note that the xanthine dehydrogenase clustered with the aerobic carbon monoxide dehydrogenase large subunit genes (Table S1), but we expect this was because they are part of a larger molybdenum hydroxylase family (Dobbek et al., 1999), and not true orthologs in this case. Although present in AAA288-E13 and AAA288-N07, the adenosine deaminase was missing in AAA240-E13 and therefore not included in the gene neighborhoods. Figure S13. Relative abundance of orthologous clusters (OCs) by depth and location according to number of reciprocal best blast hits in that sample. Each heat plot contains all OCs found in the pan-genome (x-axis) of the 12 genomes analyzed (y-axis). Colors (blue to white to red) indicate increasing relative abundance, according to each plot-specific scale. Relative abundance of OCs for each genome was used to calculate Bray-Curtis dissimilarities and hierarchically clustered using unweighted pair group method with arithmetic mean (UPGMA). All descendent links below a cluster node k share a color if k is the first node below the cut threshold of 0.7 × maximum cluster difference (default matplotlib/MATLAB behavior for dendrograms). Red boxes highlight samples where the sublclade Ic SAGs grouped together hierarchically based on gene recruitment patterns. ALOHA- Station ALOHA, ESTP OMZEastern Subtropical Pacific Oxygen Minimum Zone, BATS- Bermuda Atlantic Time-series Study site, MARM- Sea of Marmara, PRT- Puerto Rico Trench, MATA- Matapan-Vavilov Deep. Figure S14. Bayesian phylogenetic analysis of proteorhodopsin sequences using HandAlign. SAR11 proteorhodopsins are highlighted in red, along with subclade designations. Scale bar indicates changes per position. Page 20 of 21 Figure S15. Circos plot of SAG genomes, arranged by scaffold length in descending order. Outer ring: GC content. Center ring: scaffolds proportionate to size. Inner ring: predicted genes colored according to OC category: blue- true core; green- potential core; orange- shared Ic; redunique Ic; black- rRNA/tRNA; uncolored- remaining distributions. Additionally, shared Ic clusters are connected by lines of the same color. Figure S16. MUSCLE alignment of the 16S rRNA gene for genome-sequenced SAR11 strains, including the SAGs, as well as sequences with known insertions from (Eloe et al., 2011b, Lauro et al., 2007) for comparison. Supplementary Table Table S1. Excel spreadsheet (Table_S1_F.xlsx) of the comparative genomics metagenomics data for the SAR11 subclade Ic SAGs. Page 21 of 21 and Figure S1 ref. sequences w/out cultured representatives genome-sequenced representatives SAGs selected for sequencing Maine Mediteranean Surface Mesopelagic Subclade Ia Subclade Ib “Subclade Ic” Subclade II Subclade IIIa 0.3 Subclade V Figure S2 Figure S3 Figure S4 Figure S5 Figure S6 Figure S7 Station ALOHA 4000m Sanger shotgun sequences 90 60 70 80 70 80 90 90 80 70 AAA288−N07 100 AAA288−G21 100 100 AAA288−E13 60 60 70 80 90 100 AAA240−E13 60 Figure S8 955384 637000375_HTCC1062 638371090_HTCC1002 2503290285_HTCC9565 2503290180_HTCC7211 80 70 60 60 60 70 70 80 90 100 90 90 80 90 80 70 60 100 911028 100 813817 100 1402975 1456888 2503985354_HIMB5b 2503290182_HIMB114 2505679264_IMCC9063 2503985355_HIMB59b 80 60 60 1237371 4000m rev 70 80 70 70 60 4000m fwd 90 100 90 90 80 90 80 70 60 1343202 100 1279701 100 1323004 100 1308759 1284727 1410127 E.sag E.surface G.sag G.surface D.sag D.surface N.sag N.surface M.sag M.surface B.sag B.surface H.sag H.surface Z.sag Z.surface V.sag V.surface C.sag C.surface S.sag S.surface R.sag R.surface P.sag P.surface U.sag U.surface I.sag I.surface F.sag F.surface O.sag O.surface L.sag L.surface Q.sag Q.surface T.sag T.surface K.sag K.surface J.sag J.surface 0 5 10 Percentage of genes 15 Figure S9 B Ala Cys Asp Glu Phe Gly His Ile Lys Leu Met Asn Pro Gln Arg Ser Thr Val Trp X Tyr gap More in the surface Similar a.a. substitutions More in the SAGs More in the surface A Ala Cys Asp Glu Phe Gly His Ile Lys Leu Met Asn Pro Gln Arg Ser Thr Val Trp X Tyr gap More in the SAGs Figure S10 Different a.a. substitutions Figure S11 A B SAR11 Ic intergenic sizes 100 80 0 0 20 50 40 60 intergenic length 150 100 intergenic length 200 120 250 140 surface SAR11 intergenic sizes 0 200 400 600 800 0 200 Frequency 600 800 Frequency D SAR11 intergenic spacer distribution SAR11 intergenic spacer distribution (no outliers) 250 C 400 200 150 100 length 2000 50 1000 0 length 3000 Wilcoxon rank sum results W = 4905338 p-value = 4.506e-06 95% confidence interval: -7.000018 -2.999993 difference in location (surface vs. Ic): -5.000034 surface Ic surface Ic Figure S12 Allantoicase Xanthine Xanthine dehydrogenase 1500887 1500965 Putative urate catabolism protein Ureidoglycolate hydrolase AAA240-E13 Scaffold 7 OHCU decarboxylase Urate Putative urate catabolism protein ? 15001376 Cytosine/uracil/thiamine/allantoin permeases Xanthine dehydrogenase SSU Xanthine dehydrogenase Cytosine/adenosine deaminase AAA240-E13 Scaffold 13 5-hydroxyisourate OHCU Hydroxyisourate hydrolase 15001369 subclade Ic only subclade Ic and other SAR11 OHCU 15001217 S-allantoin Allantoate 2-oxo-4-hydroxy-4-carboxy5-ureidoimidazoline OR 5-hydroxy-2-oxo-4-ureido2,5-dihydro-1H-imidazole5-carboxylate OHCU decarboxylase S-allantoin amidohydrolase Hydroxyisourate hydrolase Allantoicase 15001375 Ureidoglycolate hydrolase Ureidoglycolate Urea 15001374 NH3 + CO2 _41246 _DSM M_7 _H-43 _DS tuosa linguale a_trac _ arivirg irosoma psin_M _Sp rhodo opsin 2783_ rhod Ftrac_ 643_ 7451_ 64978 lin_1 37_S 952 6464 Figure S14 rio ac ter rio rh _r m do ps _s ho iu er 35 ib e_ in Ra _E xig da lston nti uo i_E ia_e ch ut IIIa ba c 70 rop H 3 ha I_0 teriu _H 2ip m 16 Zr _s S p_ da AT acte rioop 1b sin_ Gloe 8_Ro obac seRS _296 ter_ 6_rho 4325_rho dopsin violaceu dopsin_T s_P _Ros CC_ hermus 637844493_SR e _a U_1500_xanthorh 7 21 quaticusiflexus_sp odopsin_Salinib _R4 S-1 acter_ruber_M31__Y51MC23 DSM_13855 649911837_Isop_3505_rhodopsin_Isosphaera_pallida_IS1B_ATCC_43644 645187 730_Ta qDRAFT_ s_2 0738 us_3 bacter_antarcticu opsin_Octadecaacter_antarctic CCK2B11831 _178_xanthorhod ecab _H_Tsp_ L1979 _sp ily_Octad m _BA B1 leste am u bf ila ri su h p L sin_ thylo teobac ea_spp_E iorhodop s in_Me 5_Bacter prote beta_pro _Nisacter_ 07_151 etical_ sin oba 316_OA3 3 sin_ ypoth p h dop o 647596 _ d 5 96 rho Marin rho o 0094 o 44 _ h 1 th t _ 8 1 M_ xan xan otein _MB2 632_ r DS 77_ 6000 _ p 4 8 _ 3 9 4 1 9 1 l_ B 63 _1 7_K tica 1-2 199 B 2608 the SP BAL 17 6476 ypo SO 27_ D- 04 4_h 322 er_ 8 f SS 122 4 i 6 1 6 m 64 e_ __774 0 l e i _ c t CC 7 c B1 _ra tra PCC EL ter on p_ P ac 3_M _c _s sp_ ob 542 on ma toc e_ 3 d s 6 s te 640 pla No hec _K _ ot alo ein sin n _H rot op op Cya sin d m p o _ do hro rh in _c ho rio ops _r _a te 95 sin ac hod op 15 b d 0 _ 5_ 7_r rho 16 2 CO 7_ 46 lr3 _39 LP _3 _a _H 4 ac 2 4 r 2 4 5 4 9 5 _K 7 1 3 9 4 3 C 09 16 72 10 PC 65 86 63 2_ 64 15 2 8 34 64 16 -1 1 AA _B C HT HHH HU_J_0 W 18 P _03rv V kw 4 Po8 3 664IMH_I_00 _0022nOC VFfbG 4104 182zMACS2fA u8b 9204 uesmr _01m 71662 d R aD 41 47_A _VND IB 4_ H 0 AR 20 _0 73_ 21 ba 60 cte _B rio ac rh te odo rio p rh sin od _ op Vib ri si n_ o_c am Vi br pb io el _h lii_ ar AN ve D4 yi _B B1 20 _A TC v2A cte ba _b 07 640632429_FBBAL38_10567_bacteriorhodopsin_Flavobact eria_sp_BAL38 _4H-3-77555 00 ter_diaphorus _7 _rhodopsin_Krokinobac uis_ATCC 650834282_Krodi_2220 xus_torq sychrofle C 4 44-3-D215-AP23 odopsin_P 02 1 SS 02E bacteriorh _MM 5_00947_ _Mi_D23 i teriaia_ is ME 0_P70075 bac cter ensp_ns 63911976 o s a v e a _Fla vob ngh er_irg psin Fla do ct r_ 90 in_ a_ ba te odo 72 _rh dops doni olariibac o 805 _1 k r h M T_0 Do n_P la 2_r o _ i RAF DS 146 sin ps _P _ CD p o 5_ FT do od sin lav3 A -1 5_F rho orh op DR i 55 od _2 m cu iri _Fla 9_ b_ 667 10 644 579 cte 9655 ba 6405 to_b 44 ilar_ 28 _sim as 11 0198 07 0_gll a_ 104 5757 ey me T1 ct 6374 74 EA er ick 0_ M_ _D l_p _1 ein ke 4_ rot nic 0_ ba l_p e_ 21 ca tiv 06 uo eti uta 52 _p oth 13 63 yp adio to _Sp leran s iros om _SRS a_li ngu3021 ale_ 6 DS psin _h 69 xig 63 38 G0 07 21 s_r 64 PH D1 5_ P_ odo 49 3_ ME _rh ccu 27 70 _E oco Dd 23 ine SAR11 d o rh m am _g in s op an s_ tu a did b eo ot pr a_ M _I m iu er t ac Pu SAR116 lum iril isp e nic _I um rin a _m 22 13 C MC 4 A3 SK sp_ _ ium 4 _S1 2 -212 BAA CC_ _AT o s e t h u ac 2143 thic _P 9 t tob TCC ben sin 25 othe sp_H arib op Pho 8_ p um_ od in_ io_c acteri 08 _hy Vibr orh odops b i _ o r 3 in te e 6 s pro 2207 dop act iorh CC 212 ma_ _HTCC _b acter riorho M _gam rium_sp 14 _I 16_ arine acte 71 0_b obacte 22 R1Va sin_M _008776334_b _prote a p 5 4 m o d m A 3 _ rho e_ga 27 S KA 14 0_0 _Marin cterio 14 50_E _SVASC201 dopsin 8_ba IMCC2047 6517I5wid 20932824__VIB eriorho 0328 bacterium_ 713 143__10311_bact 703 990205219 2 ma_proteo P 6 am _ G _g 6663530 122585__GB2207 odopsin 64K orh H ote Pr 1 dm_2278_ 66430901017 540_imjuxnf 651417AD_03 Ic Ia Ic IIIa 7_ o te o Pr l_ ica o pr te C in_ r cte a tob _ rium ang um ust 0.3 238 647608769_OA 48 _C ba tJ_ 63 98 01 68 01 94 00 0_ 00 64 FP 44 16 25 96 40 06 _b _0 85 ac 22 8_ te 50 64 M rio _h 68 44 op 58 yp 6_ 35 s o in_ the 7_ 05 Tra tic Ci 37 a d_ l_p trom 651 650 _r 2 94 633 rot ho 832 i c 4 045 e _rh 538 do in_ rob _Me o _ ps do Kro Fu ium talD p in lvi di_ RAF sin _b _M m 0 T _ 510 6468 _13 ari ath Tru et 5766 _rh 90_ n ep hy a_ yo 7_Tra hyp odo e m pe a lo othe d_22 psin ra_r l ba ag rin tica 52_rh ad _K i ct l_ u i _ o rok odop pro HT m er tein ino victr sin_ _ iu CCJL _Me bac ix_ True m 23 pera R _s _rad thylom ter_dia Q-2 50564 p_ icro iovic 4 p _ h b trix_ 4RQ-2ium_a orusD_SM 46 4_D lbum 4H _17 SM_ _B -3- 0 63803 9 7 1709G8 -5 3 6824_ 3 Rxyl_ 2037_ rhodo psin_R 64358641 3_Cyan74 ubrob acter_ 25_3441_ xylano rhodopsin philus _Cyanoth 638050316_HQ1017A_ _DSM ece_sp_P bacteriorhodopsin_pre _9941 CC_7425 cursor_Squarebop_II_H aloquadratum_walsbyi _HBSQ001_DSM_1679 0 72 13 3 63816 64150387_VNG1 sensor 63784 66928_OE33660G_ 48F_sensoryy_rhodopsin_I_Halobacterium_sp_NRC-1 645 66454196715 5500_ _rhodopsin SRU_ 021 411120_H _I_Halobac 2511_ 6_Huta_0 63 025 terium_sali S e L 8 4 n 6 so _ 4 R 18 Hm narum_R1_ ry_rho TI_1 8_rho 64 34815 17 uk dopsin DSM_671 763 dopsin 37 1667 79 _2 _ S 5 R _ 0 I_Salin _ _rr 680 Halo 07 40607 rhod ib n rh a _ ct abdu ops AC 14 4__O er_rub r in_H s_uta er_M3 00 hodop 6_ VNE3 1_DSM 14 hens alor Hl G 48 _se sin_H is_A hab _1385 ac 17 0R X-2_ 5 dus alo nso _0 64 _s DSM _tia mic ry_ _129 mate 08 G_ ens rob r 4 h 0 ium a_S 5_ se ory od A _ n o _ RL4 rh mu psi od sory rhod koh B n_ 1_ ata _ o op Ha ei_ sin rhod psin arg loa op _II_ _H r 2 c _D ula Ha sin al SM _m lob _I or _12 ari I_ ub 286 sm Ha acte ru ort riu lob m u m_ i_A _l ac ac s TC te ali us riu C_ na 43 pr m_ rum 04 of sp _R 9 un _N 1_ di RC DS _A M_ -1 TC 67 C_ 1 49 23 9 49 30 _4 C C AT i_ 90 tu 67 or _1 m 0 is SM 16 ar _2 _D m 01 SM a_ D Q0 ul rc a_ BS ar oa i_H al ab by H G als n_ is_ _w tio on um ra t nc 54 ra ha fu d 68 _p n_ ua _1 as ow loq 9 SM on 04 kn Ha _D m 43 23 un _I_ C_ no C C f_ op tro yi_ AT b _o re lsb Na tui_ a r in a II_ qu mo _w ps _ S 40 aris in _o r_ tum 129 s m a r so 59 SM_ la_ op ad ur rcu -2_D 05 od qu ec loa _AX rh pr alo AC nsis Ha L4B y_ in_ I_H tahe AR or_ s rrn u _ or _ urs a_S op sin dus ns 4_ c e t d op rhab ma pre se 27 ho d lo _ ia a A_ ior rho s_t sin 82 r in_H u p o 49 34 ops do abd 81 cte eri o 8 h _430 r rhod ba act iorh 63 P4 ATCC Halo _0315_ N A_ _b ter ortui_ sin_ uta 14 016 bac 4_ arism dop 374_H 10 _1 30_ ula_m 96 rho 1 loarc a 22 HQqrw 20 15_ 64497 sin_H p 3__H nAC 77 -1 141 o p_NRC TI_ 3139 _rr orhod 63 ri te rium_s 56068657 HLR _bac obacte 19_ M_671 1802183 3161 sin_Hal rum_R1_DS 665338 16404 4_rrnAC rhodop erium_salina acterio 65 8469 psin_Halobact 67G_b 6381 bacteriorhodo VNG14 0_OE3106F_ 786_ 8160 6623 6315 64 m_walsbyi_C23_DSM_16854 651266842_Hqrw_1019_bacteriorhodopsin_II_Haloquadratu 64 515 9_ sin _K n_6 23 op sin _Sli 36 od 071 dop rh 500 rho 04 9_ 95_ 646 64 41 _17 48 _1 64 ig rad 4_ PI Ex _K ED _M 23 5_ 7_ 457 60 29 16 01 825 644 6 24 89 63 89 63 05 90 63 87 16 64 640 88 30 CC Figure S15 Figure S16 Alignment: /Users/jozenn/Documents/Giovannoni Lab/Projects/SAGs/Results/16S_tree_for_ms/piezo16s_cln_al.fna Seaview [blocks=10 fontsize=6 LETTER] on Tue Jan 8 09:20:25 2013 1 ---------------------------------------------------------------------------------------CCCT ------CCCT ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTTTAATTG ----------------------------------------------------------------------------------------------------------------------------------TAACTT ------------------------------------------------------------------------- ---------------------------------------------------------------------------------TAGAGTTTGA TAGAGTTTGA ----------------------GTTTGA -------AAA -----------GAGTTTGA ----GTTTGA --GAGTTTGA --GAGTTTGA --GAGTTTGA ----GTTTGA ----GTTTGA --GAGTTTGA ----GTTTGA --GAGTTTGA --GAGTTTGA --GAGTTTGA --GAGTTTGA --GAGTTTGA -------------------------------------------TGA AAGAGTTTGA ----------TGAGTTTGA ----------------------------AGAGTTTGA -------------------------------------------------------AGAGTTTGA -AGAGTTTGA GAGAGTTTGA -AGAGTTTGA ----------AGAGTTTGA -------------------AGAGTTTGA ----------AGAGTTTGA ---------------------------------------------------------------------------------TCATGGCTCA TCATGGCTCA ------------------TCATGGCTCA TCATGGCTCA ---------TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA ------------------------------------TCCTGGCTCA TCATGGCTCA ---------TCCTGGCTCA ----GGCTCA -----------ATGGCTCA TCCTGGCTCA -----------ATGGCTCA --ATGGCTCA --ATGGCTCA -----------ATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA TCATGGCTCA -----GCTCA TCATGGCTCA ------------------TCATGGCTCA --ATGGCTCA TCATGGCTCA --TTGAACGC --TTGAACGC --TTGAACGC --TTGAACGC --TTGAACGC --TTGAACGC --TTGAACGC --TTGAACGC --TTGAACGC GATTGAACGC GATTGAACGC ----GAACGC ---------GATTGAACGC GATTGAACGC ---------GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC GATTGAACGC ------------------------------------GATTGAACGC GATTGAACGC ---------C GGACGAACGC GAACGAACGC --ACGAACGC GAACGAACGC GAACGTACGC -AACGTACGC GAACGTACGC GAACGTACGC GAACGTACGC ---------GAACGTACGC GAACGTACGC GAACGTACGC GAACGTACGC GAACGTACGC GGACGTACGC GAACGTACGC -AACGTACGC -AACGTACGC GAATGTACGC GAACGTACGC GAACGTACGC TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG ----GGCAGG TGGCGGCAGG TGGCGGCAGG ---CGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG TGGCGGCAGG ------------------------------------TGGCGGCAGG TGGCGGCAGG TGGCGGCATG TGGCGGCATG TGGCGGCACG TGGCGGCAGG TTGCTGCATG TGGCGGCACG TGGCGGCACG TGGCGGCACG TGGCGGCACG TGGCGGCACG ---------TGGCGGCACG TGGCGGCACG TGGCGGCACG TGGCGGCACG TGGCGGCACG TGGCGGCACTGGCGGCACG TGGCGGCACG TGGCGGCACG TGGCGGCACG TGGCGGCACG TGGCGGCACG CTTAACACAT CTTAACACAT CTTAACACAT CTTAACACAT CTTAACACAT CTTAACACAT CTTAACACAT CTTAACACAT CTTAACACAT CTTAACACAT CTTAACACAT CTTAACACAT CTTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT --TTACACAT ---------------------------CCTAACACAT CCTAACACAT C-TAATACAT CCTAATACAT CCTAACACAT CCTAACACAT CTTTATACAT CTT-ATACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT ---------CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CGTTATACAT CCTTATACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT CCTAACACAT GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG -------------------------GAG GCAAGTCGAG GCAAGTCGAG GCAAGTCGAA GCAAGTCGAA GCAAGTCGAG GCAAGTCGAG GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA ---------GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAA GCAAGTCGAG GCAAGTCGAG GCAAGTCGAG CGGTAACA-CGGTAACA-CGGTAACA-CGGTAACA-CGGTAACA-CGGTAACA-CGGTAACA-CGGTAACA-CGGTAACA-CGGTAACATCGGTAACA-CGGTAACA-CGGTAACA-CGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGTAACAGCGGAAACG-------------------CGGAAACAGCGGAAACAGCGGAAACAGCGCTTTAATT CGCTTTAATT C--------CG-------CGAAGGCCCC--------CG----CAGC--------C--------C-----------------C--------C--------C--------C--------C--------C--------C--------C--------C--------C--------C--------C--------- ------GAGA ------GAGA ------GAGA ------GAGA ------GAGA ------GAGA ------GAGA ------GAGA ------GAGA ------GAAC ------TAAA ------GAGA ------GAGA ------GAAT ------GAAT ------GAAT ------GGAT ------GAAT ------GAAG ------GAAT ------GAAG ------GAAT ------GAAT ------GAAT ------GAAT ------GAAT ------GAAT ------GAAT ------GAAT ------GAAT ------AAGA ------------------------GAAG ------GAAG ------GAAG TCACCGGGTG TCACCGGGTG ---------------------------------GCAG ---------------GAAG ------GAAG ------GAAG ---------------GAAG ------GAAG ------GAAG ------GAAG ------GAAG ------GAAG ------GAAG ------GCAG ------GCAG ------GAGG ------GGGG ------GAGG -TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGCCTAGCTTGCGTAGCTTGCGTAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGCGAAGCTTGCT -TAGCTTGCAAAGCTTGCT -TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGC-TAGCTTGCATAGCTTGC-------------------GTGCTTGC-GTGCTTGC-GTGCTTGCCTTGCACCCCTTGCACCC---GCTT---CGCCTT---TAGCTTGCT -TAGCAA---TAGCAA---TAGCAA---TAGCAA---TAGCAA------------TAGCAA---TAGCAA---TAGCAA---TAGCAA---TAGCAA---TAGCAA---TAGCAA---TAGCAA---TAGCAA---TACTTG---TAGCAA---TAGCAA--- TA-TCTGCTG TA-TCTGCTG TA-TCTGCTG TA-TCTGCTG TA-TCTGCTG TA-TCTGCTG TA-TCTGCTG TA-TCTGCTG TA-TCTGCTG TAGTTTGATG TAGTTTGATG TACTTTGCTG TACTTTGCTG TAATTCGCTG TAATTTGCTG TAATTCGCTG TAATTCGCTG TAATTCGCTG TTCTTTGCTG TAATTTGCTG TTCTTTGCTG TAATTTGCTG TAATTTGCTG TAATTCGCTG TAATTCGCTG TAATTTGCTG TAATTTGCTG TAATTTGCTG TAATTTGCTG TAATTCGCTG TATTCTGGCG ------------------ACCTTTGCTG ACCTTTGCTG ACCTTTGCTG ATCGAAGTTA ATCGAAGTTA TCCTTCGGGA -------CGG AGGTGTCCT--------TA --------TA --------TA --------TA --------TA -----------------TA --------TA --------TA --------TA --------TA --------TA --------TA --------TA --------TA --------TA --------TA --------TA 111 ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGTGGCG ACGAGTGGCG ACGAGTGGCG ACGAGTGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG ACGAGCGGCG TCGAGCGGCG ------------------TCGAGCGGCG TCGAGCGGCG TCGAGCGGCG AGGAGTGGCG AGGAGTGGCG AGGAGCGGCG GCGAGCGGCG --TAGTGGCG CTGAGTGGCA CTGAGTGGCA CTTAGTGGCA CTTAGTGGCA CTTAGTGGCA ---------CTTAGTGGCA CTTAGTGGCA CTTAGTGGCA CTTAGTGGCA CTTAGTGGCA CTTAGTGGCA CTTAGTGGCA CTGAGTGGCA CTGAGTGGCA CCTAGCGGCA CCTAGCGGCA CCTAGCGGCA GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG ----GGTGAG -ACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTTAG GACGGGTTAG CACGGGTGAG GACGGGTGAG AACGGGTGAG AACGGGTGAG AACGGGTGAG GACGGGTGAG ---------GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGAG GACGGGTGCG GACGGGTGCG GACGGGTGCG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCTTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCCTGG TAATGCTTGG TAATGCCTAG TAATGCCTAG TAATGCCTAG TAATGCCTAG TAATGCCTAG TAACACGTGG TAACACGTGG TAACGCGTGG TAACGCGTGG TAACACGTGG TATAATGTGG TATAATGTGG TAATATGTGG TAATATGTGG TAATATGTGG ---------TAACATGTAG TAACATGTAG TAACATGTGG TAACATGTGG TAACATGTGG TAACATGTAG TAACATGTGG TAACATGTGG TAACATGTGG TAACATGTGG TAACATGTGG TAACATGTGG G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATATGCC G-AATCTGCC G-GAACTGCC G-GAACTGCC G-GAACTGCC G-GAACTGCC G-GAACTGCC GTAACCTGCC GTAACCTGCC G-AACATACC G-AACGTGCC G-AACATACC G-AATCTGCC G-AATCTGCC G-AATCTGCC G-AATCTGCC G-TATCTACC ---------G-TATCTGCC G-TATCTGCC G-TATCTACC G-TATCTACC G-TATCTACC G-TATCTTCC G-TATCTTCC G-AATCTGCC G-AATCTGCC G-AATCTGCC G-AATCTGCC G-AATCTGCC TTATGGTGGG TTATGGTGGG TTATGGTGGG TTATGGTGGG TTATGGTGGG TTATGGTGGG TTATGGTGGG TTATGGTGGG TTATGGTGGG TTGGAGTGGG TTGGAGTGGG TTAACGTGGG TTAACGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TTAGTGTGGG TAGTCGAGGG CAGTCGAAGG CAGTCGAGGG CAGTCGAGGG CAGTCGAGGG CAGTCGAGGG CATAAGAGGG CATAAGAGGG CATCACTAAG CAGATCTACG TCATAGTACG TTTTGGTTTG TTTTGGTTTG CTTCGGTCTG CTTCGGTCTG TTTCGGTCTG ---------CTTTGGCCTG CTTTGGCCTG CTTTGGCCTG CTTTGGCCTG CTTTGGCCTG CTTTGGTGAG CTTTGGTGAG TTGTGGTTCA TTGTGGTTCA TTTTGGTTCG TTTTGGTTCG TTTTGGTTCG GGACAACAGT GGACAACAGT GGACAACAGT GGACAACAGT GGACAACAGT GGACAACAGT GGACAACAGT GGACAACAGT GGACAACAGT GGACAACAGT GGACAACAGT GGACAACAGT GGACAACAGT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGATAACTAT GGACAACAGT GGATAACAGT GGATAACAGT GGATAACAGT GGATAACAGT GGATAACAGT GGATAACATT GGATAACATT GAATAGCCTC GAATAGCCAC GAATAACTGC GAATAACTCG GAATAACACG GAATAACACG GAATAACATG GAATAACATG ---------GAATAACACG GAATAACACG GAATAACACG GAATAACACG GAATAACACG GAATGACACG GAATAACACG GGACAACATT GGACAACATT GAACAACATG GAACAACATG GAATAACACG TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAT TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC TGGAAACGAC CGGAAACGGA CGGAAACGGA GGGAAACTGA TGGAAACGGT GGGAAACTGT GGGAAACTTG GGGAAACTTG AGGAAACTTG AGGAAACTTA GGGAAACTTA ---------AGGAAACTTG AGGAAACTTG AGGAAACTTG AGGAAACTTG AGGAAACTTG AGGAAACTTG AGGAAACTTG GGGAAACCGA GGGAAACCGA GGGAAACCTG GGGAAACCTG GGGAAACCTG TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC AGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC GAGTAATACC GAGTAATACC AGCTAATACC AGCTAATACC TGCTAATACC CGCTAATACC TGCTAATACC TGCTAATACC ---------TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACC TGCTAATACT TGCTAATACT TGCTAATACC TGCTAATACC TGCTAATACC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTG G-ATAACGTG GCATAATGTC GCATAATGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAATGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATAACGTC GCATACGACC GCATACGCTGCATACGCCC GCATACGCCC GCATACGCCC GCATACGCCC GCATATTTCT GCATATTTCT TTATACGCCC GTATACGCCC GTATATT--GGATAAGCCC GAATAAGCCC GGATAAGCCT GGATAAGCCT GGATAAGCCT ---------AGATAAGTCT GGATAAGTCT GGATAAGTCT GGATAAGTCT GGATAAGTCT TCATAAGTCT TCATAAGTCT GGATAAGCCC GGATAAGCCC GGATAAGTCC GGATAAGTCC GGATAAGTCC T--------T--------T--------T--------T--------T--------T--------T--------T--------CTT--TTTGT CTT--TTTGT T--------T--------TTTG-TTCAT T--------T--------CTTG-TTCAT T--------CTTG-TTCAT CTTG-TTCAT CTTG-TTCAT CTTG-TTCAT CTTG-TTCAT CTTG-TTCAT CTTG-TTCAT CTTG-TTCAT CTTG-TTCAT CTTG-TTCAT T--------T--------T-----------------T--------T--------T--------T--------AATCGTCTCC AATCGTCTCC T--------T-----------------TTAC-----TTAC-----TTAC-----TTAC-----TTAC--------------TTAC-----TTAC-----TTAC-----TTAC-----TTAC-----TTAC-----TTAC-----TTAC-----TTAC-----TTAC-----T--------TTAC------ ------ACGG ------ACGG ------ACGG ------ACGG ------ACGG ------ACGG ------ACGG ------ACGG ------ACGG AGTGAAAAGT AGTGAAAAGT ------ACGG ------ACGG TACGAGCGGG ------TCGG ------TCGG TACGAGCGGG ------TCGG TACGAGCGGG TACGAGCGGG TACGAGCGGG TACGAGCGGG TACGAGCGGG TACGAGCGGG TACGAGCGGG TACGAGCGGG TACGAGCGGG TACGAGCGGG ------TCGG ------TCGG ------ACGG ------ACGG ------ACGG ------ACGG ------ACGG ------ACGG TGACGAATGG TGACGAATGG ------TCGG ------TCGG ------------------G ---------G ---------A ---------A ---------A ------------------G ---------G ---------G ---------G ---------G ---------G ---------G ---------G ---------G ---------G ------AACG ---------G 640698396_C_psychrerythraea_34H 640698404_C_psychrerythraea_34H 640698412_C_psychrerythraea_34H 640698493_C_psychrerythraea_34H 640698426_C_psychrerythraea_34H 640698400_C_psychrerythraea_34H 640698408_C_psychrerythraea_34H 640698416_C_psychrerythraea_34H 640698390_C_psychrerythraea_34H DQ027051.1_Colwellia_sp_MT41 DQ027052.1_Colwellia_sp_KT27 DQ027056.1_Psychromonas_sp_CNPT3 DQ027057.1_Psychromonas_sp_HS11 640705772_P_profundum_SS9 DQ027054.1_P_profundum_3TCK DQ027055.1_P_profundum_1230sf1 640705946_P_profundum_SS9 640705754_P_profundum_SS9 640705856_P_profundum_SS9 640705924_P_profundum_SS9 640705919_P_profundum_SS9 640705763_P_profundum_SS9 640705741_P_profundum_SS9 640705850_P_profundum_SS9 640705738_P_profundum_SS9 640705907_P_profundum_SS9 640705913_P_profundum_SS9 640705910_P_profundum_SS9 640705881_P_profundum_SS9 640705884_P_profundum_SS9 DQ027053.1_Moritella_sp_PE36 DQ027059.1_Shewanella_sp_PT48 DQ027060.1_Shewanella_sp_PT64 641466167_P_profundum_SS9 DQ027058.1_Shewanella_sp_KT99 641463346_P_profundum_SS9 DQ027061.1_Carnobacterium_sp_AT7 DQ027062.1_Carnobacterium_sp_AT12 JF303756.1_Rhodobacterales_PRT1 639101207_P_profundum_3TCK 2504111167_HIMB59b U75256.1_SAR211g2 AF353214.1_Arctic95B-1g2 2236674967_AAA288-E13 2236446051_AAA288-G21 2236452142_AAA288-N07 2236674298_AAA240-E13 2504109332_HIMB5b 2503352260_HTCC7211 2503364240_HTCC9565 640708248_HTCC1062 642513240_HTCC1002 X52172.1_SAR11g1b U75649.1_SAR193g1b Z99997.1_LD12 AB154304.1_S9D-28fresh 2503356127_HIMB114 2505687060_IMCC9063 U70686.1_OM155g3 640698396_C_psychrerythraea_34H 640698404_C_psychrerythraea_34H 640698412_C_psychrerythraea_34H 640698493_C_psychrerythraea_34H 640698426_C_psychrerythraea_34H 640698400_C_psychrerythraea_34H 640698408_C_psychrerythraea_34H 640698416_C_psychrerythraea_34H 640698390_C_psychrerythraea_34H DQ027051.1_Colwellia_sp_MT41 DQ027052.1_Colwellia_sp_KT27 DQ027056.1_Psychromonas_sp_CNPT3 DQ027057.1_Psychromonas_sp_HS11 640705772_P_profundum_SS9 DQ027054.1_P_profundum_3TCK DQ027055.1_P_profundum_1230sf1 640705946_P_profundum_SS9 640705754_P_profundum_SS9 640705856_P_profundum_SS9 640705924_P_profundum_SS9 640705919_P_profundum_SS9 640705763_P_profundum_SS9 640705741_P_profundum_SS9 640705850_P_profundum_SS9 640705738_P_profundum_SS9 640705907_P_profundum_SS9 640705913_P_profundum_SS9 640705910_P_profundum_SS9 640705881_P_profundum_SS9 640705884_P_profundum_SS9 DQ027053.1_Moritella_sp_PE36 DQ027059.1_Shewanella_sp_PT48 DQ027060.1_Shewanella_sp_PT64 641466167_P_profundum_SS9 DQ027058.1_Shewanella_sp_KT99 641463346_P_profundum_SS9 DQ027061.1_Carnobacterium_sp_AT7 DQ027062.1_Carnobacterium_sp_AT12 JF303756.1_Rhodobacterales_PRT1 639101207_P_profundum_3TCK 2504111167_HIMB59b U75256.1_SAR211g2 AF353214.1_Arctic95B-1g2 2236674967_AAA288-E13 2236446051_AAA288-G21 2236452142_AAA288-N07 2236674298_AAA240-E13 2504109332_HIMB5b 2503352260_HTCC7211 2503364240_HTCC9565 640708248_HTCC1062 642513240_HTCC1002 X52172.1_SAR11g1b U75649.1_SAR193g1b Z99997.1_LD12 AB154304.1_S9D-28fresh 2503356127_HIMB114 2505687060_IMCC9063 U70686.1_OM155g3 221 ACCAAAGGGG ACCAAAGGGG ACCAAAGGGG ACCAAAGGGG ACCAAAGGGG ACCAAAGGGG ACCAAAGGGG ACCAAAGGGG ACCAAAGGGG ACCAAAGGGG ACCAAAGGGG ACCAAAGCAG ACCAAAGCAG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG ACCAAAGAGG GTGAAAGGGG GGGAAAGGAG GGGAAAGGAG GGGAAAGGAG GGGAAAGGAG GGGAAAGGAG AAGAAAGGTG AAGAAAGGTG GGGAAAG--GGGAAAG---CCAGAAATG GGGAAAG--GGGAAAG--GGGAAAG--GGGAAAG--GGGAAAG-----------GAGAAAG--GAGAAAG--GAGAAAG--GAGAAAG--GAGAAAG--GAGAAAG--GAGAAAG--GGGAAAG--GGGAAAG--GAGAAAG--GAGAAAG--GAGAAAG--- GGGA-----T GGGA-----T GGGA-----T GGGA-----T GGGA-----T GGGA----TT GGGA----TT GGGA----TT GGGA----TT GGGA-----T GGGA-----T GGGA-----C GGGA-----C GGGA-----A GGGA-----C GGGA-----C GGGA-----A GGGA-----C GGGA-----C GGGA-----C GGGA-----C GGGA-----C GGGA-----C GGGA-----C GGGA-----C GGGA-----C GGGA-----C GGGA-----C GGGA-----C GGGA-----C GCC------T GGGA-----GGGACGCGCT GGGACGCGTT GGGACGCGCT GGGACGCGTT G--------G--------------------------GGAA-----------------------T ---------C ---------C ---------C ---------------------------------------------C ------------------C ---------C ------------------T ---------------------------T CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----TCTTGTTTTT CTTC-----CTTC-----CCTTGTTTTT CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----CTTC-----TTTC-----TTTC-----TTTT-----TTTC-----CTTC-----CTTC--------------------------------CTTT-----TTTA-----TTTA-----TTTA-----TTTA--------------CTTT-----CTTT-----CTTT-----TTTA-----CTTT-----TTTA-----TTTA-----TTTT-----TTTA--------------TTTT-----TTTA------ --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG CGAAACAGGG --------GG --------GG CGAAATGGGG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG --------GG ------TTGA ---------G ---CAATGTG ----GAAGTG ---CGAAGTG ----GAAGTG --------------------------------------------AG ------------------------------------------------------------------------------------------------------------------------------------------------------------------- -ACCTCTCGC -ACCTCTCGC -ACCTCTCGC -ACCTCTCGC -ACCTCTCGC AACCTCTCGC AACCTCTCGC AACCTCTCGC AACCTCTCGC -ACCTCCCGC -ACCTCTCGC -GCCTTGCGC -GCCTTGCGC -TCCTCTCGC -GCCTCTCGC -GCCTCTCGC -TCCTCTCGC -GCCTCTCGC -GCCTCTCGC -GCCTCTCGC -GCCTTTCGC -GCCTCTCGC -GCCTCTCGC -GCCTCTCGC -GCCTCTCGC -GCCTCTCGC -GCCTCTCGC -GCCTCTCGC -GCCTCTCGC -GCCTCTCGC AAGCTCTCGC CGCCTTTCGC CGCCTTTCGC CGCCTTTCGC CGCCTTTCGC CGCCTTTCGC -GGCTACCGC -GGCTACCGC -ATTTATCGG -ATTTATCGG -ATTTATCGC ----ATGCGC ------ACGC -----TGCTC -----TGCTC -----TGCTC -------------ATGCAC ----ATGCAC ----ATGCAC -----TGCAC ----ATGCAC -----TGCGC -----TGCGC -----AATGC ------ATGC -ATTTATCGC -----AATGC ------ATGC CATTTGATTA CATTTGATTA CATTTGATTA CATTTGATTA CATTTGATTA CATTTGATTA CATTTGATTA CATTTGATTA CATTTGATTA TTCTTGATTA CTCTTGATCA GTTTAGAGTA GTTTAGAGTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GCTAAGATTA GACTAGATGA GATTGGATGT GATTGGATGT GATTGGATGT GATTGGATGT GATTGGATGT TTATGGATGG TTATGGATGG TGATGGATTG ATTTGGATCG TATGAGATTG CGAAAGATGA CGAAAGATGA CGATGGATGA CGATTGATGA CGATAGATGA ---------CATTGGATGA CATTGGATGA CAATGGATGA CATTGGATGA CATTGGATGA CGAAGGATGA CAATGGATGA CATAAGATGA CATAAGATGA CGAAAGATGA CAAAAGATGA CAAAAGATGA GCCCAAGTGA GCCCAAGTGA GCCCAAGTGA GCCCAAGTGA GCCCAAGTGA GCCCAAGTGA GCCCAAGTGA GCCCAAGTGA GCCCAAGTGA GCCCAAGTGA GCCCAAGTGA GCCCAAGTGG GCCCAAGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAGGTGG GCCCAAGTGG ACCTAGGTGG ACCTAGGTGG ACCTAGGTGG ACCTAGGTGG ACCTAGGTGG ACCCGCGGCG ACCCGCGGCG GCCCGCGTAA GCCCGCGTTA GCCCGCGCAA GCCCGCACTT GCCCGCACTT GTCCACACTT GCCCACACTT GTCCACACTT ---------GCCTGCACTT GCCTGCACTT GCCCGCACTT GCCCGCACTT GCCCGCACTT GCCTGCACTT GCCCGCACTT GCCCGCATTT GCCCGCATTT GCCCGCACTA GCCCGCATTT GCCCGCATTT GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTTG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG GATTAGCTAG TATTAGCTAG TATTAGCTAG GATTAGATAG GATTAGGTAG GATTAGCTTG GATTAGCTCG GATTAGCTAG GATTAGTTAG GATTAGTTAG GATTAGTTAG ---------GATTAGTTTG GATTAGTTTG GATTAGTTTG GATTAGTTTG GATTAGTTTG GATTAGTTTG GATTAGTTTG GATTAGTTAG GATTAGTTAG GATTAGCTTG GATTAGCTTG GATTAGTTTG TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTAAGGT TTGGTAAGGT TTGGTAAGGT TTGGTAAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT TTGGTAAGGT TTGGTAAGGT TTGGTAAGGT TTGGTAAGGT TTGGTAAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGGGGT TTGGTGGGGT TTGGTAAGGT TTGGTGAGGT TTGGTAAGGT TTGGTGAGGT TTGGTGAGGT TTGGTGAGGT ---------TTGGTGGGGT TTGGTGGGGT TTGGTAGGGT TTGGTAGGGT TTGGTAGGGT TTGGTGGGGT TTGGTGGGGT TTGGTGAGGT TTGGTGAGGT TTGGTAAGGT TTGGTGAGGT TTGGTGAGGT AATGGCTCAC AATGGCTCAC AATGGCTCAC AATGGCTCAC AATGGCTCAC AATGGCTCAC AATGGCTCAC AATGGCTCAC AATGGCTCAC AAAGGCTTAC AAAGGCTTAC AATGGCTTAC AATGGCTTAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAAGGCTCAC AAGAGCTCAC AATGGCTTAC AATGGCTTAC AATGGCTTAC AATGGCTTAC AATGGCTTAC AATGGCTCAC AATGGCTCAC AACGGCCTAC AACGGCCTAC AACGGCTTAC AATGGCTCAC AAAAGCTTAC AAAAGCTCAC AATAGCTCAC AATAGCTCAC ---------AATAGCCTAC AATGGCCTAC AATGGCCTAC AATGGCCTAC AATGGCCTAC AATGGCCTAC AATGGCCTGC AATGGCTCAC AATGGCTCAC AAAAGCTTAC AAAAGCTCAC AATGGCTCAC CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCAACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGACG CAAGGCGATG CAAGGCGATG CAAGTCGACG CAAGCCTACG CAAGGCTCCG CAAGGCAACA CAAGGCAATG CAAGACAATG CAAGACAATG CAAGACAATG ---------CAAGACTATG CAAGACTGTG CAAGACTATG CAAGACTATG CAAGACTATG CAAGACTTTG CAAGACTTTG CAAGACAATG CAAGACAATG CAAGGCTACG CAAGGCTATA CAAGACTACA 331 ATCTCTAGCT ATCTCTAGCT ATCTCTAGCT ATCTCTAGCT ATCTCTAGCT ATCTCTAGCT ATCTCTAGCT ATCTCTAGCT ATCTCTAGCT ATCTCTAGCT ATCTCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATCCCTAGCT ATACGTAGCC ATACGTAGCC ATCTTTAGCT ATCTATAGCT ATCTTTAGCT ATCAATAGCT ATCAATAGCT ATCAATAGCT ATCAATAGCT ATCAATAGCT ---------ATCAATAGCT ATCAATAGCT ATCAATAGCT ATCAATAGCT ATCAATAGCT ATCTATAGCT ATCAATAGCT ATCAATAGCT ATCAATAGCT ATCTATAGCT ATCAATAGCT ATCAATAGCT GG-TTTGAGA GG-TTTGAGA GG-TTTGAGA GG-TTTGAGA GG-TTTGAGA GG-TTTGAGA GG-TTTGAGA GG-TTTGAGA GG-TTTGAGA GG-TTTGAGA GG-TTTGAGA GG-TCTTAGA GG-TCTTAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GG-TCTGAGA GA-CCTGAGA GACCCTGAGA GG-TTTGAGA GG-TTTTAGA GG-TTTGAGA GT-TCTTAGA GT-TCTTAGA GA-TTTGAGA GA-TTTGAGA GA-TTTGAGA ---------GA-TTTGAGA GA-TTTGAGA GA-TTTGAGA GA-TTTGAGA GA-TTTGAGA GA-TCTGAGA GA-TCTGAGA GA-TCTGAGA GA-TCTGAGA GA-TTTGAGA GA-TTTGAGA GA-TTTGAGA GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGACCAG GGATGACCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGGTGATCGG GGGTGATCGG GGATGATCAG GGATGATCAG GGATGATCAG GGAAGACCAT GGAAGACCAG GGATGATCAG GGATGATCAG GGATGATCAG ---------GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG GGATGATCAG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGA CCACACTGGG CCACACTGGG CAACACTGGG CAACACTGGG CCACACTGGG CCACATTGGG CCACATTGGG CCACATTGGG CCACATTGGG CCACATTGGG ---------CCACATTGGG CCACATTGGG CCACATTGGG CCACATTGGG CCACATTGGG CCACATTGGG CCACATTGGG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGG CCACACTGGG ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ---------ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGACAC ACTGAGATAC ACTGAGATAC GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGTCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAAACT GGCCCAAACT GGCCCAAACT ---------GGCCCAAACT GGCCCAAACT GGCCCAAACT GGCCCAAACT GGCCCAAACT GGCCCAAACT GGCCCAAACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT GGCCCAGACT CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGGG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCCAGGGGGG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG ---------CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG CCTACGGGAG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTAG GCAGCAGTAG GCAGCAGTGG GCAGCAGTGG GCAGCAGTAG GCAGCAGTAA GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG ---------GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTAG GCAGCAGTAG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GCAGCAGTGG GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATATTGC GGAATCTTCC GGAATCTTCC GGAATCTTAG GGAATCTTGG GGAATATTGG GGAATCTTTC GGAATCTTGC GGAATCTTGC GGAATCTTGC GGAATCTTGC ---------GGAATCTTGC GGAATCTTGC GGAATCTTGC GGAATCTTGC GGAATCTTGC GAAACATTGC GGAATATTGC GGAATCTTGC GGAATCTTGC GGAATCTTGC GGAATCTTGC GGAATCTTGC ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGAGG ACAATGGAGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGAGG ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGGCG ACAATGGGCG GCAATGGACG GCAATGGACG ACAATGGGCG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGGG ACAATGGGAG ACAATGGGGG ACAATGGGGG ---------ACAATGGAGG ACAATGGAGG ACAATGGAGG ACAATGGAGG ACAATGGAGG ACAATGGAGG GCAATGGAGG ACAATGGAGG ACAATGGAGG ACAATGGAGG ACAATGGGCG ACAATGGGCG AAAGCCTGAT AAAGCCTGAT AAAGCCTGAT AAAGCCTGAT AAAGCCTGAT AAAGCCTGAT AAAGCCTGAT AAAGCCTGAT AAAGCCTGAT AAAGCCTGAT AAAGCCTGAT AAACTCTGAT AAACTCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT AAACTCTGAT CAAGCCTGAT CAAGCCTGAT CAAGCCTGAT CAAGCCTGAT CAAGCCTGAT AAAGTCTGAC AAAGTCTGAC CAAGCCTGAT CAACCCTGAT AAACCCTGAT AAACCCTGAT AAACCCTGAT CAATCCTGAT AAACCCTGAT AAACCCTGAT ---------AAACTCTGAT AAACTCTGAT AAACTCTGAT AAACTCTGAT AAACTCTGAT AAACTCTGAT AAACTCTGAT AAACTCTGAT AAACTCTGAT AAACTCTGAT AAAGCCTGAT AAAGCCTGAT 640698396_C_psychrerythraea_34H 640698404_C_psychrerythraea_34H 640698412_C_psychrerythraea_34H 640698493_C_psychrerythraea_34H 640698426_C_psychrerythraea_34H 640698400_C_psychrerythraea_34H 640698408_C_psychrerythraea_34H 640698416_C_psychrerythraea_34H 640698390_C_psychrerythraea_34H DQ027051.1_Colwellia_sp_MT41 DQ027052.1_Colwellia_sp_KT27 DQ027056.1_Psychromonas_sp_CNPT3 DQ027057.1_Psychromonas_sp_HS11 640705772_P_profundum_SS9 DQ027054.1_P_profundum_3TCK DQ027055.1_P_profundum_1230sf1 640705946_P_profundum_SS9 640705754_P_profundum_SS9 640705856_P_profundum_SS9 640705924_P_profundum_SS9 640705919_P_profundum_SS9 640705763_P_profundum_SS9 640705741_P_profundum_SS9 640705850_P_profundum_SS9 640705738_P_profundum_SS9 640705907_P_profundum_SS9 640705913_P_profundum_SS9 640705910_P_profundum_SS9 640705881_P_profundum_SS9 640705884_P_profundum_SS9 DQ027053.1_Moritella_sp_PE36 DQ027059.1_Shewanella_sp_PT48 DQ027060.1_Shewanella_sp_PT64 641466167_P_profundum_SS9 DQ027058.1_Shewanella_sp_KT99 641463346_P_profundum_SS9 DQ027061.1_Carnobacterium_sp_AT7 DQ027062.1_Carnobacterium_sp_AT12 JF303756.1_Rhodobacterales_PRT1 639101207_P_profundum_3TCK 2504111167_HIMB59b U75256.1_SAR211g2 AF353214.1_Arctic95B-1g2 2236674967_AAA288-E13 2236446051_AAA288-G21 2236452142_AAA288-N07 2236674298_AAA240-E13 2504109332_HIMB5b 2503352260_HTCC7211 2503364240_HTCC9565 640708248_HTCC1062 642513240_HTCC1002 X52172.1_SAR11g1b U75649.1_SAR193g1b Z99997.1_LD12 AB154304.1_S9D-28fresh 2503356127_HIMB114 2505687060_IMCC9063 U70686.1_OM155g3 640698396_C_psychrerythraea_34H 640698404_C_psychrerythraea_34H 640698412_C_psychrerythraea_34H 640698493_C_psychrerythraea_34H 640698426_C_psychrerythraea_34H 640698400_C_psychrerythraea_34H 640698408_C_psychrerythraea_34H 640698416_C_psychrerythraea_34H 640698390_C_psychrerythraea_34H DQ027051.1_Colwellia_sp_MT41 DQ027052.1_Colwellia_sp_KT27 DQ027056.1_Psychromonas_sp_CNPT3 DQ027057.1_Psychromonas_sp_HS11 640705772_P_profundum_SS9 DQ027054.1_P_profundum_3TCK DQ027055.1_P_profundum_1230sf1 640705946_P_profundum_SS9 640705754_P_profundum_SS9 640705856_P_profundum_SS9 640705924_P_profundum_SS9 640705919_P_profundum_SS9 640705763_P_profundum_SS9 640705741_P_profundum_SS9 640705850_P_profundum_SS9 640705738_P_profundum_SS9 640705907_P_profundum_SS9 640705913_P_profundum_SS9 640705910_P_profundum_SS9 640705881_P_profundum_SS9 640705884_P_profundum_SS9 DQ027053.1_Moritella_sp_PE36 DQ027059.1_Shewanella_sp_PT48 DQ027060.1_Shewanella_sp_PT64 641466167_P_profundum_SS9 DQ027058.1_Shewanella_sp_KT99 641463346_P_profundum_SS9 DQ027061.1_Carnobacterium_sp_AT7 DQ027062.1_Carnobacterium_sp_AT12 JF303756.1_Rhodobacterales_PRT1 639101207_P_profundum_3TCK 2504111167_HIMB59b U75256.1_SAR211g2 AF353214.1_Arctic95B-1g2 2236674967_AAA288-E13 2236446051_AAA288-G21 2236452142_AAA288-N07 2236674298_AAA240-E13 2504109332_HIMB5b 2503352260_HTCC7211 2503364240_HTCC9565 640708248_HTCC1062 642513240_HTCC1002 X52172.1_SAR11g1b U75649.1_SAR193g1b Z99997.1_LD12 AB154304.1_S9D-28fresh 2503356127_HIMB114 2505687060_IMCC9063 U70686.1_OM155g3 441 GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGT GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATAC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GCAGCCATGC GGAGCAATGC GGAGCAATGC CTAGCGATGC CCAGCCATGC CCAGCAATGC GCAGCGATGC GCAGCGATGC GCAGCGATGC GCAGCGATGC GCAGCGATGC ---------GCAGCGATGC GCAGCGATGC GCAGCGATGC GCAGCGATGC GCAGCGATGC GCAGGGATGC GCAGCGATGC GCAGCGATGC GCAGCGATGC GCAGCGATGC GCAGCGATGC GCAGCGATGC CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTATG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGTGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG AGCGTGAGTG CCCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG ---------CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG CGCGTGAGTG AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCT AA-GAAGGCT AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGTT AA-GAAGGTT AT-GAAGGTC AA-GAAGGCC AC-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC ---------AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AAGGAAGGCC AA-GAAGGAC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC AA-GAAGGCC TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTAGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGGTTGT TTCGGATCGT TTCGGATCGT TTAGGATCGT CTAGGGTCGT TTAGGGTTGT TTTGGGTTGT TTTGGGTTGT CTTGGGTTGT CTTGGGTTGT TTTGGGTTGT ---------CTTGGGTTGT CTTGGGTTGT CTTGGGTTGT TTTGGGTTGT TTTGGGTTGT TTTGGGTTGT TTTGGGTTGT TTTGGGTTGT TTCGGGTTGT CTTGGGTTGT TTTGGGTTGT TTTGGGTTGT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGTACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAGCACTTT AAAACTCTGT AAAACTCTGT AAAGCTCTTT AAAACTCTTT AAAACTCTTT AAAACTCTTT AAAACTCTTT AAAGCTCTTT AAAGCTCTTT AAAACTCTTT ---------AAAGCTCTTT AAAGCTCTTT AAAGCTCTTT AAAGCTCTTT AAAGCTCTTT AAAGCTCTTT AAAGCTCTTT AAAGCTCTTT AAAGCTCTTT AAAGCTCTTT AAAGCTCTTT AAAGCTCTTT CAGTTGTGAG CAGTTGTGAG CAGTTGTGAG CAGTTGTGAG CAGTTGTGAG CAGTTGTGAG CAGTTGTGAG CAGTTGTGAG CAGTTGTGAG CAGTTGTGAG CAGTTGTGAG CAGCGAGGAG CAGCGAGGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGTCGTGAG CAGCGAGGAG CAGCGAGGAG CAGCGAGGAG CAGCGAGGAG CAGCGAGGAG CAGCGAGGAG TGTTAGAGAA TGTTAGAGAA CGCCAGGGAT CGCCTGTGAT CATCGGTGAG CGTGGGGGAA CGTCGGGGAA CGTCGGGGAA CGTCGGGGAA CGTCGAGGAA ---------CGTCGGGGAA CGTCGGGGAA CGTCGGGGAA CGTCGGGGAA CGTCGGGGAA CGTCGGGGAA CGTCGGGGAA TGTAGGGGAA TGTAGGGGAA TGTCGGGAAA TGTCGGGAAA TGTCGGGAAA GAA-AGGAGT GAA-AGGAGT GAA-AGGAGT GAA-AGGAGT GAA-AGGAGT GAA-AGGGGT GAA-AGGGGT GAA-AGGGGT GAA-AGGGGT GAA-AGGTGG GAA-AGGTGG GAA-AGGGTA GAA-AGGGTA GAA-GGGTAT GAA-GGGTGT GAA-GGGTGT GAA-GACATT GAA-GACATT GAA-GGGTGT GAA-GATTAT GAA-GATTAT GAA-GATTAT GAA-GATTAT GAA-GATTAT GAA-GATTAT GAA-GATTAT GAA-GATTAT GAA-GATTAT GAA-GATTAT GAA-GATTAT GAA-AGGTAG GAA-AGGTTG GAA-AGGTTG GAA-AGGTTG GAA-AGGTTG GAA-AGGTTG GAACAAGGAT GAACAAGGAT GAT-AA---GAT-AA---GAT-AA---GAA-AA---GAA-AA---GAA-AA---GAA-AA---GAA-AA------------GAA-AA---GAA-AA---GAA-AA---GAA-AA---GAA-AA---GAA-AA---GAA-AA---GAT-AA---GAT-AA---GAT-AA---GAT-AA---GAT-AA---- GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA TGATTTAATA ACGTTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA GTAGTTAATA ACAGTTAATA ACAGTTAATA ACAGTTAATA ACAGTTAATA ACAGTTAATA ACAGTTAATA ACAGTTAATA ACAGTTAATA ACAGTTAATA ACAGTTAATA ACAGTTAATA TAAATTAATA TATGTTAATA TAGTTTAATA TAGTTTAATA TAGTTTAATA TAGTTTAATA GAGAGTAACT GAGAGTAACT ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCTGCATTCT GCTGCATTCT GCTGCATTCT GCTGCATTCT GCTGCATTCT GCTGCATTCT GCTGCATTCT GCTGCATTCT GCTGCATTCT AATCATCACT AACGTTCACT ACTGCTATCT ACTGCTATCT GCTGCATGCC GCTGCATGCC GCTGCATGCC GCTGCAGTGT GCTGCAGTGT GCTGCATGCC GCTGTGTGAT GCTGTGTGAT GCTGTGTGAT GCTGTGTGAT GCTGTGTGAT GCTGTGTGAT GCTGTGTGAT GCTGTGTGAT GCTGTGTGAT GCTGTGTGAT GCTGTGTGAT CTTTGCTACT AACTACAGCT AACTACAGCT AACTACAGCT AACTATAGCT AACTACAGCT GCT-CATCCC GCT-CATCCC ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTGACGTTAA GTGACGTTAA GTGACGTTAA GTGACGTTAA GTGACGTTAA GTGACGTTAA GTGACGTTAA GTGACGTTAA GTGACGTTAA GTGACGTTAG GTGACGTTAG GTGACGTTAC GTGACGTTAC TTGACGTTAG TTGACGTTAG TTGACGTTCG TTGACGTTAG TTGACGTTAG TTGACGTTAG TTGACGTTAG TTGACGTTAG TTGACGTTAG TTGACGTTAG TTGACGTTAG TTGACGTTAG TTGACGTTAG TTGACGTTAG TTGACGTTAG TTGACGTTAA TTGACGTTAG GTGACGTTAC GTGACGTTAC GTGACG-TAC ATGACGTTAC GTGACGTTAC GTGACGTTAC CTGACGGTAT CTGACGGTAT -TGACAGTAC -TGACAGTAG -TGACAGTAG -TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC ----------TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC -TGACTGTAC CAACAGAAGA CAACAGAAGA CAACAGAAGA CAACAGAAGA CAACAGAAGA CAACAGAAGA CAACAGAAGA CAACAGAAGA CAACAGAAGA CAACAGAAGA CAACAGAAGA TCGCAGAAGA TCGCAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA CGACAGAAGA TCGCAGAAGA TCGCAGAAGA TCGCAGAAGA TCGCATAAGA TCGCAGAAGA TCGCAGAAGA CTAACCAAAA CTAACCAGAA CTGGTAAAGA CAGGTAAAGA CCGAAGAAGA CCCAATAAGA CCGAATAAGA CCGAATAAGA CCGAATAAGA TCGAATAAGA ---------CCGAATAAGA CCGAATAAGA CCGAATAAGA CCGAATAAGA CCGAATAAGA CCGAATAAGA CCCAATAAGA CCTAAGAATA CCTAAGAATA CCGAAGAATA CCGAAGAATA CCGAAGAATA 551 AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT AGCACCGGCT G---CCGGAT AGCCACGGCT AGCCCCGGCT AACCCCGGCT AGGACCGGCT AGGTCCGGCT AGGTCCGGCT AGGTCCGGCT AGGTCCGGCT AGGTCCGGCT ---------AGGTCCGGCT AGGTCCGGCT AGGTCCGGCT AGGTCCGGCT AGGTCCGGCT AGGTCCGGCT AGGTCCGGYT AGGTCCGGCT AGGTCCGGCT AGGTCCGGCT AGGTCCGGCT AGGTCCGGCT AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTCCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTACGTGC AACTACGTGC AACTCCGTGC AACTCCGTGC AATTCCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC ---------AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC AACTTCGTGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC ---------CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC CAGCAGCCGC GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGT GGTAATACGT GGTAATACGG GGTAATACGG GGTAATACGG GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA ---------GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGTAATACGA GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCGAG AGGGTGCAAG GGGGTGCHAG GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG GGGGTGCAAG AGGTGGCAAG AGGTGGCAAG AGGGGGCTAG AGGGGGTTAG AAGGTCCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG ---------AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG AGGGACCTAG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTACATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGTTA-ATCG CGT---GCCG CGTTG-TCCG CGTTG-TTCG CGTTG-TTCG CGTTG-TTCG CGTAG-TTCG CGTAG-TTCG CGTAG-TTCG CGTAG-TTCG CGTAG-TTCG ---------CGTAG-TTCG CGTAG-TTCG CGTAG-TTCG CGTAG-TTCG CGTAG-TTCG CGTAG-TTCG CGTAG-TTCG CGTAG-TTCG CGTAG-TTCG CGTAA-TTCG CGTAG-TTCG CGTAG-TTCG GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACTGG GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAAT--CT-G GAATTACT-G GAATTACT-G GAATTACT-G GA-TTATT-G GATTTAT--G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G ---------GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GAATTACT-G GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAACAG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTA--AG GGCGTA--AG GGCGTAA-AG GGCGTA--AG GGCGTAA-AG GGCGTA--AG GGCGTA--AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCTTAA-AG GGCTTAA-AG GGCTTAA-AG GGCTTAA-AG GGCTTAA-AG ---------GGCTTAA-AG GGCTTAA-AG GGCTTAA-AG GGCTTAA-AG GGCTTAA-AG GGCTTAA-AG GGCTTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG GGCGTAA-AG CGTTCGTAGG CGTTCGTAGG CGTTCGTAGG CGTTCGTAGG CGTTCGTAGG CGTTCGTAGG CGTTCGTAGG CGTTCGTAGG CGTTCGTAGG CGTTCGTAGG CGTTCGTAGG CGCGCGTA-G CGCGCGTAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGCATGCAGG CGTACGCAGG CGTACGCAGG CGTACGCAGG CGTACGCAGG CGTACGCAGG CGAGCGCAGG CGAGCGCAGG CGTACGTAGG CGCACGTAGG CGCATGTAGG AGCTCGTAGG AGCTCGTAGG AGCTCGTAGG AGCACGTAGG AGCACGTAGG ---------AGTTCGTAGG AGTTCGTAGG AGTTCGTAGG AGTTCGTAGG AGTTCGTAGG AGTTCGTAGG AGTTCGTAGG CGCGCGTAGG CGCGCGTAGG CGCGCGTAGG AGTATGTAGG AGTATGTAGG CGGTCTATTA CGGTCTATTA CGGTCTATTA CGGTCTATTA CGGTCTATTA CGGTCTATTA CGGTCTATTA CGGTCTATTA CGGTCTATTA CGGTTATTTA CGGTTATTTA TGGTTAATTA TGGTT--ATT CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTCTGTTA CGGTTTGTTA CGGTTTGTTCGGTTTGTTCGGTTTGTTA CGGTTTGTTA CGGTTTGTTA CGGTTCTTTA CGGTTCTTTA CCGATTGGAA CGGATTAGAA CGGAACAAAA TGGTTAAAAA TGGTTAAAAA TGGTTAGAAA TGGTTAAAAA TGGTTAAAAA ---------TGGTTGAAAA TGGTTGAAAA TGGTTGAAAA TGGTTGAAAA TGGTTGAAAA TGGTTAAAAA TGGTTAAAAA CCGTTGAGTT CCGTTGAGTT CGGTTAAGTA CGGCTAGATA TGGTTAGATA AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGTCAGATGT AGTCAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCGAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGCAAGATGT AGTCTGATGT AGTCTGATGT AGTAGGGGGT AGTTAGAGGT AGTTAGAAGT AGTTGATGGT AGTTGATGGT AGTTGGTGGT AGTTGGTGGT AGTTGGTGGT ---------AGTTGGTGGT AGTTAGTGGT AGTTGGTGGT AGTTGGTGGT AGTTGGTGGT AGTTGGTGGT AGTTGGTGGT AGTTAATTGT AGTTAATTGT AGTTAATTGT AGTTAATTGT AGTTAATTGT 640698396_C_psychrerythraea_34H 640698404_C_psychrerythraea_34H 640698412_C_psychrerythraea_34H 640698493_C_psychrerythraea_34H 640698426_C_psychrerythraea_34H 640698400_C_psychrerythraea_34H 640698408_C_psychrerythraea_34H 640698416_C_psychrerythraea_34H 640698390_C_psychrerythraea_34H DQ027051.1_Colwellia_sp_MT41 DQ027052.1_Colwellia_sp_KT27 DQ027056.1_Psychromonas_sp_CNPT3 DQ027057.1_Psychromonas_sp_HS11 640705772_P_profundum_SS9 DQ027054.1_P_profundum_3TCK DQ027055.1_P_profundum_1230sf1 640705946_P_profundum_SS9 640705754_P_profundum_SS9 640705856_P_profundum_SS9 640705924_P_profundum_SS9 640705919_P_profundum_SS9 640705763_P_profundum_SS9 640705741_P_profundum_SS9 640705850_P_profundum_SS9 640705738_P_profundum_SS9 640705907_P_profundum_SS9 640705913_P_profundum_SS9 640705910_P_profundum_SS9 640705881_P_profundum_SS9 640705884_P_profundum_SS9 DQ027053.1_Moritella_sp_PE36 DQ027059.1_Shewanella_sp_PT48 DQ027060.1_Shewanella_sp_PT64 641466167_P_profundum_SS9 DQ027058.1_Shewanella_sp_KT99 641463346_P_profundum_SS9 DQ027061.1_Carnobacterium_sp_AT7 DQ027062.1_Carnobacterium_sp_AT12 JF303756.1_Rhodobacterales_PRT1 639101207_P_profundum_3TCK 2504111167_HIMB59b U75256.1_SAR211g2 AF353214.1_Arctic95B-1g2 2236674967_AAA288-E13 2236446051_AAA288-G21 2236452142_AAA288-N07 2236674298_AAA240-E13 2504109332_HIMB5b 2503352260_HTCC7211 2503364240_HTCC9565 640708248_HTCC1062 642513240_HTCC1002 X52172.1_SAR11g1b U75649.1_SAR193g1b Z99997.1_LD12 AB154304.1_S9D-28fresh 2503356127_HIMB114 2505687060_IMCC9063 U70686.1_OM155g3 640698396_C_psychrerythraea_34H 640698404_C_psychrerythraea_34H 640698412_C_psychrerythraea_34H 640698493_C_psychrerythraea_34H 640698426_C_psychrerythraea_34H 640698400_C_psychrerythraea_34H 640698408_C_psychrerythraea_34H 640698416_C_psychrerythraea_34H 640698390_C_psychrerythraea_34H DQ027051.1_Colwellia_sp_MT41 DQ027052.1_Colwellia_sp_KT27 DQ027056.1_Psychromonas_sp_CNPT3 DQ027057.1_Psychromonas_sp_HS11 640705772_P_profundum_SS9 DQ027054.1_P_profundum_3TCK DQ027055.1_P_profundum_1230sf1 640705946_P_profundum_SS9 640705754_P_profundum_SS9 640705856_P_profundum_SS9 640705924_P_profundum_SS9 640705919_P_profundum_SS9 640705763_P_profundum_SS9 640705741_P_profundum_SS9 640705850_P_profundum_SS9 640705738_P_profundum_SS9 640705907_P_profundum_SS9 640705913_P_profundum_SS9 640705910_P_profundum_SS9 640705881_P_profundum_SS9 640705884_P_profundum_SS9 DQ027053.1_Moritella_sp_PE36 DQ027059.1_Shewanella_sp_PT48 DQ027060.1_Shewanella_sp_PT64 641466167_P_profundum_SS9 DQ027058.1_Shewanella_sp_KT99 641463346_P_profundum_SS9 DQ027061.1_Carnobacterium_sp_AT7 DQ027062.1_Carnobacterium_sp_AT12 JF303756.1_Rhodobacterales_PRT1 639101207_P_profundum_3TCK 2504111167_HIMB59b U75256.1_SAR211g2 AF353214.1_Arctic95B-1g2 2236674967_AAA288-E13 2236446051_AAA288-G21 2236452142_AAA288-N07 2236674298_AAA240-E13 2504109332_HIMB5b 2503352260_HTCC7211 2503364240_HTCC9565 640708248_HTCC1062 642513240_HTCC1002 X52172.1_SAR11g1b U75649.1_SAR193g1b Z99997.1_LD12 AB154304.1_S9D-28fresh 2503356127_HIMB114 2505687060_IMCC9063 U70686.1_OM155g3 661 GAAAGCCCAG GAAAGCCCAG GAAAGCCCAG GAAAGCCCAG GAAAGCCCAG GAAAGCCCAG GAAAGCCCAG GAAAGCCCAG GAAAGCCCAG GAAAGCCAAG GAAAGCCCAG GAAAGCCCAG GAAAGCCCAG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCGG GAAAGCCCCG GAAAGCCCCG GAAAGCCCCG GAAAGCCCCG GAAAGCCCCG GAAAGCCCCG GAAAGCCCCC GAAAGCCCCC GAAATCCCAG GAAATCCCGG GAAATCCCTG GAAATCCCAA GAAATCCCAA GAAATCCCAA GAAATCCCAG GAAATCCCAG ---------GAAATCCCAG GAAATCCCAG GAAATCCCAG GAAATCCCAG GAAATCCCAG GAAATCCCAG GAAATCCCAG GAAATCCCAA GAAATCCCAA GAAAGCCCAA GAAAGCCCAA GAAAGCCCAA GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT -GCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTCAACCG GGCTCAACCG GGCTTAACCC GGCTCAACCC GGCTCAACCT GGCTCAACCT GGCTCAACCT GGCTTAACCT AGCTTAACTC AGCTTAACTC ---------AGCTTAACTC AGCTTAACTC AGCTTAACTC AGCTTAACTC AGCTTAACTC AGCTTAACTC AGCTTAACTC AGCTTAACTT AGCTTAACTT AGCTCAACTT AGCTTAACTT AGCTTAACTT TGGAACTGCA TGGAACTGCA TGGAACTGCA TGGAACTGCA TGGAACTGCA TGGAACTGCA TGGAACTGCA TGGAACTGCA TGGAACTGCA TGGAACTGCA TGGAACTGCA TGGAACTGCA TGGAACTGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA CGGAACCGCA GGGAACTGCA GGGAATTGCA GGGAATTGCA GGGAATTGCA GGGAATTGCA GGGAATTGCA GGGAGGGTCA GGGAGGGTCA TGGAACTGCC CGGAACTGCC AGGAATTGCT TGGAACTGCC TGGAACTGCC TGGAACTGCC TGGAACTGCC TGGAACTGCC ---------TGGAACTGCC TGGAACTGCC TGGAACTGCC TGGAACTGCC TGGAACTGCC TGGAACTGCC TGGAACTGCC TGGAACTGCA TGGAACTGCA TGGAATTGCA TGGAACTGCA TGGAATTGCA TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTGAAACTG TTTGAAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTCGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTTTGAACTG TTGGAAACTG TTGGAAACTG TCCTAAACTA TTTAAAACTA TTTAAAACTT ATCAAAACTG ATCAAAACTT GCCAAAACTT ATCAAAACTT ATCAAAACTT ---------ATCAAAACTT ATTAAAACTT ATCAAAACTT ATCAAAACTT ATCAAAACTT ATCAAAACTT ATCAAAACTT ATTAAAACTG ATTAAAACTG ATTAAAACTA ATTAAAACTA ATTAAAACTA GTAGACTAGA GTAGACTAGA GTAGACTAGA GTAGACTAGA GTAGACTAGA GTAGACTAGA GTAGACTAGA GTAGACTAGA GTAGACTAGA GGTAACTAGA GGTAACTAGA GTTGACTAGA GTTGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAGACTAGA GCAAACTAGA GCAAACTAGA GCAAACTAGA GCAAACTAGA GCAAACTAGA GCAAACTAGA GAGAACTTGA GAGAACTTGA ACAGTCTAGA CTAGTCTAGA TTGTTCTGGA TTTAGCTAGA TTTAGCTAGA TTTGACTAGA TTTAGCTAGA TTTAGCTAGA ---------TTCAGCTGGA TTCAGCTAGA TTCAGCTAGA TTCAGCTAGA TTCAGCTAGA TTTAGCTAGA TTTAGCTAGA CTCGACTAGA CTCGACTAGA TTTAGCTAGA TTTAGCTAGA TTTAGCTAGA GTACTGTAGA GTACTGTAGA GTACTGTAGA GTACTGTAGA GTACTGTAGA GTACTGTAGA GTACTGTAGA GTACTGTAGA GTACTGTAGA GTACTGTA-A GTACTGTA-A GT-TTGTAGA GTTTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTTCTTGAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTCTTGTAGA GTGCAGAAGA GTGCAGAAGA GTTCGAGAGA GTTCGAGAGA ATTCAGGAGA GTGTGATAGA GTGTGATAGA GTATGATAGA GTATGATAGA GTGTGATAGA --------GA GTTTGATAGA GTATGATAGA GTATGATAGA GTATGATAGA GTATGATAGA GTATGATAGA GTATGATAGA GTTTGATAGA GTTTGATAGA GTTTATCAGA GTATGATAGA GTATGATAGA GGGTGGTGGA GGGTGGTGGA GGGTGGTGGA GGGTGGTGGA GGGTGGTGGA GGGTGGTGGA GGGTGGTGGA GGGTGGTGGA GGGTGGTGGA GGGTGGTGGA GGGTGGTGGA GGGTGGTAGA GGGTGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGTGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGGGGGTAGA GGAGAGTGGA GGAGAGTGGA GGTGAGTGGA GGTGAGTGGA GGAAAATGGA GGAAAGTGGA GGTAAGCGGA GGAAAGTGGA GGAAAGTAGA GGAAAGTAGA GGAAAGTAGA GGAAAGCAGA GGAAAGCAGA GGAAAGCAGA GGAAAGCAGA GGAAAGCAGA GGAAAGCAGA GGAAAGCAGA GGAAAGCGGA GGAAAGCGGA GGAAAGCGGA GGAAAGCGGA GGAAAGCGGA ATTTCCAGTG ATTTCCAGTG ATTTCCAGTG ATTTCCAGTG ATTTCCAGTG ATTTCCAGTG ATTTCCAGTG ATTTCCAGTG ATTTCCAGTG AT-TCCAGTG ATTTCCAGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTTCAGGTG ATTCCACGTG ATTCCACGTG ATTCCAAGTG ATTCCGAGTG ATTTCCAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATTTCTAGTG ATACATAGTG ATACATAGTG ATACATAGTG ATACATAGTG ATACATAGTG TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGCGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA TAKAGGTGAA TAGAGGTGAA TAGAGGTGAA TAGAGGTGAA ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAG ATGCGTAGAT ATGCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTCGTAGAT ATTGGAAGGA ATTGGAAGGA ATTGGAAGGA ATTGGAAGGA ATTGGAAGGA ATTGGAAGGA ATTGGAAGGA ATTGGAAGGA ATTGGAAGGA ATTGGAWGGA ATTGGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATCTGAAGGA ATGTGGAGGA ATGTGGAGGA ATTTGGAGGA ATTCGGAGGA ATTGGAAGGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTAGAAAGA ATTATGTAGA ATTATGTAGA ATTATGTAGA ATTATGTAGA ATTATGTAGA 771 ACATCAGTGG ACATCAGTGG ACATCAGTGG ACATCAGTGG ACATCAGTGG ACATCAGTGG ACATCAGTGG ACATCAGTGG ACATCAGTGG NCATCAGTGG ACATCAGTGG ATACCAGTGG ATACCAGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCAGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ATACCGGTGG ACACCAGTGG ACACCAGTGG ACACCAGTGG ACACCAGTGG ACACCAGTGG ACACCAAAAG ACACCAAATG ATACCAATTG ATACCAATTG ATACCAATTG ATACCAATTG ATACCAATTG ATACCAATTG ATACCAATTG ATACCAATTG ATACCAATTG ATACCAATTG GTACCAATTG ACACCAGTTG ACACCAGTTG ACACCAGTTG ACACCAGTTG ACACCAGTTG CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAA-GCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAA-GCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGAC CGAAGGCGAC CGAAGGCGGC CGAAGGCGGC CGAAGGCGAT CGAAGGCAAC CGAAGGCAAC CGAAGGCAGC CGAAGGCAGC CGAAGGCAGC CGAAGGCAGC CGAAGGCAGC CGAAGGCAGC CGAAGGCAGC CGAAGGCAGC CGAAGGCAGC CGAAGGCAGC CGAAGGCAGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CGAAGGCGGC CACCTGGACA CACCTGGACA CACCTGGACA CACCTGGACA CACCTGGACA CACCTGGACA CACCTGGACA CACCTGGACA CACCTGGACA CACCTGGACA CACCTGGACA CACCTGGACA CACCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CACCTGGCAA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA CCCCTGGACA TCTCTGGTCT TCTCTGGTCT TCACTGGCTC TCACTGGCTC TTTCTGGACT TTTCTGGATC TTACTGGGTC TTTCTGGATC TTTCTGGATC TTTCTAGATC TTTCTGGATC TTTCTGGATC TTTCTGGATC TTTCTGGATC TTTCTGGATC TTTCTGGATC TTTCTGGATC TTTCTGGATC TTTCTGGATC TTTCTGGATC TTTCTGGGAT TTTCTGGATC TTTCTGGATC GATACTGACG GATACTGACG GATACTGACG GATACTGACG GATACTGACG GATACTGACG GATACTGACG GATACTGACG GATACTGACG GATACTGACG GATACTGACG AAGACTGACA AAGACTGACA AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG GTAACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG AAGACTGACG GTAACTGACG GTAACTGACG GATACTGACG GATACTGACG GATATTGACG ACTACTGACA ACTACTGACA ATTACTGACA ATTACTGACA ATTACTGACA ATTACTGACA AATACTGACA ATTACTGACA ATTACTGACA ATTACTGACA ATTACTGACA ATTACTGACG ATNACTGACG AACACTGACG AACACTGACG AACACTGACG ATTACTGACG ATTACTGACG CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CTGGAGGAAC CT-GAGGCGC CT-GAGGCGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-CAGATGC CT-GAGGTAC CT-GAGGTAC CT-GAGGTAC CT-GAGGTAC CT-GAGGTAC CT-GAGGCTC CT-GAGGCTC CT-GAGGTAC CT-GAGGTGC CT-GAGATGC CT-GAGGAGC CT-GAGGAGC CT-GAGGAGC CT-GAGGTGC CT-GAGGTGC CT-GAGGTGC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGAAC CT-GAGGCGC CT-GAGGCGC CT-GAGGTGC CT-GAGATAC CT-GAGATAC GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAAGGCG-T GAAG-GCG-T GAAG-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCGCT GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GCG-T GAAA-GTG-T GAAG-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GCA-T GAAA-GTA-T GAAA-GTA-T GAAA-GTA-T GAAA-GTA-T GAAA-GTA-T GGGGAGCGAA GGGGAGCGAA GGGGAGCGAA GGGGAGCGAA GGGGAGCGAA GGGGAGCGAA GGGGAGCGAA GGGGAGCGAA GGGGAGCGAA GGGGAGCGAA GGGGAGCGAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGTAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGGAGCAAA GGGTAGCAAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA GGGTAGCAAA GGGTAGCAAA GGGTAGCGAA GGGTAGCGAA GGGTAGCGAA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CGGGATTAGA CGGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CGGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CAGGA-TAGA CAGGATTAGA CAGGATTAGA CAGGATTAGA CGGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA GAGGATTAGA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCCGGTA TACCCCGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCCGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCTGGTA TACCCCGGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA TACCCTCGTA -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCACGGC GGTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-GC -GTCCAC-AC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-GC -GTCCAT-AC -GTCCAT-AC -GTCCAT-AC -GTCCAT-AC -GTCCAT-AC CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT AGTAAACGAT AGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGT-AACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT AGTAAACGAT TGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CGTAAACGAT CATAAACGAT CATAAACGAT CGTAAACGAT CATAAACGAT TGTAAACGAT 640698396_C_psychrerythraea_34H 640698404_C_psychrerythraea_34H 640698412_C_psychrerythraea_34H 640698493_C_psychrerythraea_34H 640698426_C_psychrerythraea_34H 640698400_C_psychrerythraea_34H 640698408_C_psychrerythraea_34H 640698416_C_psychrerythraea_34H 640698390_C_psychrerythraea_34H DQ027051.1_Colwellia_sp_MT41 DQ027052.1_Colwellia_sp_KT27 DQ027056.1_Psychromonas_sp_CNPT3 DQ027057.1_Psychromonas_sp_HS11 640705772_P_profundum_SS9 DQ027054.1_P_profundum_3TCK DQ027055.1_P_profundum_1230sf1 640705946_P_profundum_SS9 640705754_P_profundum_SS9 640705856_P_profundum_SS9 640705924_P_profundum_SS9 640705919_P_profundum_SS9 640705763_P_profundum_SS9 640705741_P_profundum_SS9 640705850_P_profundum_SS9 640705738_P_profundum_SS9 640705907_P_profundum_SS9 640705913_P_profundum_SS9 640705910_P_profundum_SS9 640705881_P_profundum_SS9 640705884_P_profundum_SS9 DQ027053.1_Moritella_sp_PE36 DQ027059.1_Shewanella_sp_PT48 DQ027060.1_Shewanella_sp_PT64 641466167_P_profundum_SS9 DQ027058.1_Shewanella_sp_KT99 641463346_P_profundum_SS9 DQ027061.1_Carnobacterium_sp_AT7 DQ027062.1_Carnobacterium_sp_AT12 JF303756.1_Rhodobacterales_PRT1 639101207_P_profundum_3TCK 2504111167_HIMB59b U75256.1_SAR211g2 AF353214.1_Arctic95B-1g2 2236674967_AAA288-E13 2236446051_AAA288-G21 2236452142_AAA288-N07 2236674298_AAA240-E13 2504109332_HIMB5b 2503352260_HTCC7211 2503364240_HTCC9565 640708248_HTCC1062 642513240_HTCC1002 X52172.1_SAR11g1b U75649.1_SAR193g1b Z99997.1_LD12 AB154304.1_S9D-28fresh 2503356127_HIMB114 2505687060_IMCC9063 U70686.1_OM155g3 640698396_C_psychrerythraea_34H 640698404_C_psychrerythraea_34H 640698412_C_psychrerythraea_34H 640698493_C_psychrerythraea_34H 640698426_C_psychrerythraea_34H 640698400_C_psychrerythraea_34H 640698408_C_psychrerythraea_34H 640698416_C_psychrerythraea_34H 640698390_C_psychrerythraea_34H DQ027051.1_Colwellia_sp_MT41 DQ027052.1_Colwellia_sp_KT27 DQ027056.1_Psychromonas_sp_CNPT3 DQ027057.1_Psychromonas_sp_HS11 640705772_P_profundum_SS9 DQ027054.1_P_profundum_3TCK DQ027055.1_P_profundum_1230sf1 640705946_P_profundum_SS9 640705754_P_profundum_SS9 640705856_P_profundum_SS9 640705924_P_profundum_SS9 640705919_P_profundum_SS9 640705763_P_profundum_SS9 640705741_P_profundum_SS9 640705850_P_profundum_SS9 640705738_P_profundum_SS9 640705907_P_profundum_SS9 640705913_P_profundum_SS9 640705910_P_profundum_SS9 640705881_P_profundum_SS9 640705884_P_profundum_SS9 DQ027053.1_Moritella_sp_PE36 DQ027059.1_Shewanella_sp_PT48 DQ027060.1_Shewanella_sp_PT64 641466167_P_profundum_SS9 DQ027058.1_Shewanella_sp_KT99 641463346_P_profundum_SS9 DQ027061.1_Carnobacterium_sp_AT7 DQ027062.1_Carnobacterium_sp_AT12 JF303756.1_Rhodobacterales_PRT1 639101207_P_profundum_3TCK 2504111167_HIMB59b U75256.1_SAR211g2 AF353214.1_Arctic95B-1g2 2236674967_AAA288-E13 2236446051_AAA288-G21 2236452142_AAA288-N07 2236674298_AAA240-E13 2504109332_HIMB5b 2503352260_HTCC7211 2503364240_HTCC9565 640708248_HTCC1062 642513240_HTCC1002 X52172.1_SAR11g1b U75649.1_SAR193g1b Z99997.1_LD12 AB154304.1_S9D-28fresh 2503356127_HIMB114 2505687060_IMCC9063 U70686.1_OM155g3 881 -GTCAACTAG -GTCAACTAG -GTCAACTAG -GTCAACTAG -GTCAACTAG -GTCAACTAG -GTCAACTAG -GTCAACTAG -GTCAACTAG -GTCAACTAG GGTCAACTAG -GTCTATTAG -GTCTATTAG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTTG -GTCTACTCG -GTCTACTCG -GTCTACTCG -GTCTACTCG -GTCTACTCG -GTCTACTCG -GAGTGCTAA -GAGTGCTAA -GAATGCCAG -GAATGCCAG -GGGTGCTAG -GTGTGCTAG -GTGTGCTAG -GTGTGCTAG -GTGTGCTAG -GTGTGCTAG -GTGTGCTAG -GTGTGTTAG -GTGTGTTAG -GTGTGTTAG -GTGTGTTAG -GTGTGTTAG -GTGTGTTAG -GTGTGTTAG -GATTGCTAG -GATTGCTAA -GATTGTTAG -GATTATTAA -GATTATTAG CCGTTTGTGG CCGTTTGTGG CCGTTTGTGG CCGTTTGTGG CCGTTTGTGG CCGTTTGTGG CCGTTTGTGG CCGTTTGTGG CCGTTTGTGG CCGTTCGTGG CCGTTCGTGG AAGTTTGTGG AAGTTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG AAGGTTGTGG GAGTTTGGTT GAATTTGGTG GAATTTGGTG GAATTTGGTG GAATTTGGTG GAATTTGGTG GTGTTGGAGG GTGTTGGAGG TCGTTGGATA TCGTCGGGTA TCGTC---GG ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ACGTTGGAAA ATGTTGGAAA GATGTTGGAA ATGTTGGAAA ATG--TGGGG ATG--TGGGG ACTTGA-TCC ACTTGA-TCC ACTTGA-TCC ACTTGA-TCC ACTTGA-TCC ACTTGA-TCC ACTTGA-TCC ACTTGA-TCC ACTTGA-TCC ACTTGA-TCC ACTTGA-TCC CTATAT-GCC CTATA--TCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCCTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GCC CCTTGA-GAA TCTTGA-ACA TCTTGA-ACA TCTTGA-ACA TCTTGA-ACA TCTTGA-ACA GTTTCC-GCC GTTTCCGCCC GCATG--CTA GTATA--CTA ACTT---TTG TTTA---TCT TATA---TTT TTTA---ATT TTT----ATT TTT----ATT TTT----ATT TTT----ATT TTT----ATT TTT----ATT TTT----ATT TTT----ATT TTT----ATT TTT----ATT TTT----ATT ATTTA--TTT TTT----ATT AATTA--TTT AATTA--TTT GTGAGTGGCG GTGAGTGGCG GTGAGTGGCG GTGAGTGGCG GTGAGTGGCG GTGAGTGGCG GTGAGTGGCG GTGAGTGGCG GTGAGTGGCG GTGAGTGGCG GTGAGTGGCG GTGGGTTTCA GTGGGTTTCA GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG GTGGCTTTCG CTGGGCTCTT CTGGGTTCCC CTGGGTTCCC CTGGGTTCCC CTGGGTTCCC CTGGGTTCCC TTCAGTGCTG TTCAGTGCTG TTCAGTGACA TTCGGTGACA TTCGGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTGTCG TTCAGTATCG TCAAGTATCG TTCAGTATCA CTCTGTATTA CTCTGTATCA CAGCTAACGC CAGCTAACGC CAGCTAACGC CAGCTAACGC CAGCTAACGC CAGCTAACGC CAGCTAACGC CAGCTAACGC CAGCTAACGC CAGCTAACGC CAGCTAACGC AAGCTAACGC AAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC GAGCTAACGC AAGCTAACGC AAGCTAACGC AAGCTAACGC AAGCTAACGC AAGCTAACGC AAGCTAACGC CAGCTAACGC CAGCTAACGC CACCTAACGG CACCTAACGG TAGCTAACGC CAGCAAAAGC CAGCGAAAGC CAGCAAAAGC CAGCGAAAGC CAGCAAAAGC CAGCAAAAGC CAGCGAAAGC CAGGGAAACC CAGGGAAACC CAGGGAAACC CAGGGAAACC CAGCGAA-AN CAGCGAAAGC CAGCTAACGC CAGCTAACGC CAGCTAACGC TAGCTAACGC CAGCTAACGC ACTAAGTTGA ACTAAGTTGA ACTAAGTTGA ACTAAGTTGA ACTAAGTTGA ACTAAGTTGA ACTAAGTTGA ACTAAGTTGA ACTAAGTTGA ACTAAGTTGA ACTAAGT-GA ATTAAATAGA ATTAAATAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA GTTAAGTAGA ATTAAGTAGA ATTAAGTAGA ATTAAGTAGA ATTAAGTAGA ATTAAGTAGA ATTAAGTAGA ATTAAGCACT ATTAAGCACT ATTAAGCATT ATTAAGCATT GTTAAGCACC GTTAAGCACA ATTAAGCACA GTTAAGCACA GTTAAGCACA ATTAAGCACA GTTAAGCACA GATAAACACA GATAAACACA GATAAACACA GATAAACACA GATAAACACA ATTAAACACA ATTAAACACA ATTAAGCAAT ATTAAGCAAA GTTAAACAAT GTTAAATAAT GTTAAATAAT -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG TCCGCCTGGG -CCGCCTGGG -CCGCCTGGG -CCGCCTGGG GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGGCC GAGTACGACC GAGTACGACC GAGTACGGTC GAGTACGGTC GAGTACGGTC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GAGTACGACC GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTGA GCAAGGTTGA GCAAGATTAA GCAAGATTAA GCAAGATTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAG-TTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA GCAAGGTTAA AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAAGG AACTCAAAGG AACTCAAAGG AACTCAAAGG AACTCAAATA AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AACTCAAATG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG AATTGACGGG 991 GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GG-CCCGCAC GA-CCCGCAC GA-CCCGCAC GG-CCCGCAC GG-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC GGACCCGCAC GA-CCCGCAC GA-CCCSCAC GA-CCCGCAC GA-CCCGCAC GA-CCCGCAC AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCAGTGGA AAGCAGTGGA AAGCAGTGGA AAGCAGTGGA AAGCAGTGGA AAGCAGTGGA AAGCAGTGGA AAGTAGTGGA AAGTAGTGGA AAGTAGTGGA AAGTAGTGGA AAGTAGTGGA AAGTAGTGGA AAGTAGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA AAGCGGTGGA GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGCT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT GCATGTGGTT TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGATG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGACG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG TAATTCGAAG CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-AA CAACGCG-CA CAACGCG-CA CAACGCG-AA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCG-CA ATACGCGCCA CTACGCG-CA CAACGCG-CA CAACGCG-CA GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC GAACCTT--GAACCTTACC GGACCTTACC KAACCTTACC GAACCTTACC GAACCTTACC GAACCTTACC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC ATCCCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC TACTCTTGAC AGGTCTTGAC AGGTCTTGAC AACCCTTGAC AACCCTTGAC AGCGTTTGAC AACACTTGAC AACACTTGAC AACACTTGAC AACACTTGAC AACACTTGAC AACACTTGAC AACACTTGAC AACACTTGAC AACACTTGAC AACACTTGAC AACACTTGAC --CACTTGAC AACACTTGAC AACCCTTGAC AACCCTTGAC AACACTTGAC AACCCTTGAC AACCCTTGAC A---TCCAGA A---TCCAGA A---TCCAGA A---TCCAGA A---TCCAGA A---TCCAGA A---TCCAGA A---TCCAGA A---TCCAGA A---TCCTCA A---TCCAGA A---TCCAGA A---TCCAGA A---TCCAGA A---TCCAGA A---TCCAGA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCACA A---TCCTTT A---TCCTTT ATA-CTTGTC A---TCCTGT A---TGGGAA ATG-TTCGTC ATG-TTCGTC ATGTTTCGTC ATG-TTCGTC ATG-TTCGTC ATG-TTCGTC ATG-TTCGTC ATG-TTCGTC ATG-TTCGTC ATG-TTCGTC ATG-TTCGTC ATG-TTCGTC ATG-TTCGTC ATG-TCTGTC ATG-TCTGTC ATG-TCTGTC ATG-TTTGTC ATG-TTCGTC GAAGAGACTA GAAGAGACTA GAAGAGACTA GAAGAGACTA GAAGAGACTA GAAGAGACTA GAAGAGACTA GAAGAGACTA GAAGAGACTA GAAGCCAGTA GAAGAGACTA GAATCACCTA GAATCACCTA GAAGTTACCA GAAGTTACCA GAAGTTACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAAGAGACCA GAATCTGGTA GAATTCGCTA GAATTCGCTA GAATTTGCTA GAATTCGCTA GAATTCGCTA GACAATCCTA GACAATCCTA GCGGTGACCA GCTACATTCA GTGCCGGTTA GCGAGACCAA GCGAGCCTAA GCGACTTCAA GCGACTTTAA GCGACTTTAA GCGACTTTAA GCGACTTTAA GCGACTCTAA GCGACTCTAA GCGACTCTAA GCGACTCTAA GCGACTCTAA GCGACTCTAA GCGAGTTTAG GCGAGTTTAG GCGGATTCCA GCGAAACTAA GCGAGACTAA GAGA-TAGAC GAGA-TAGAC GAGA-TAGAC GAGA-TAGAC GAGA-TAGAC GAGA-TAGAC GAGA-TAGAC GAGA-TAGAC GAGA-TAGAC GAGA-TACAS GAGA-TAGAC GAGA-TAGAT GAGA-TAGAT GAGA-TGGTT GAGA-TGGTT GAGA-TGGTT GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TGGAC GAGA-TACCT GAGA-TAGCT GAGA-TAGCT GAGA-TAGCT GAGA-TAGCT GAGA-TAGCT GAGA-TAGGA GAGA-TAGGA GAGA-TGGAC GAGA-TGAAT CAGAATGGTA GAGA-GTGGT GAGATTAGGC GAGATTGAAGAGATTAAAGAGATTGAAGAGATTAAAGAGATTAAAG GAGATTAGAG GAGATTAGAG GAGATTAGAG GAGATTAGAGAGATTAGAGAGATTAGAGAAACTAGAC GAAACTAGAC GAGA-TGGAT GAGATTAGTT GAAATTAGTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC GCGTGCCTTC TTGTGCCTTC GAGTGCCTTC GAGTGCCTTC TCGTGCCTTC TCGTGCCTTC TCGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC TTGTGCCTTC CAGTGCCTTC TAGTGCCTTC TAGTGCCTTC TAGTGCCTTC TAGTGCCTTC TAGTGCCTTC CTTTCCCTTC CTTTCCCTTC ----ACCTTC GGTTCCTTTC ----ATCTTC TTCTTCATTT TTTTCAGTTC ----GTCTTC ----GTCTTC ----GTCTTC ----GTCTTC TTTTCGGTTC TTTTCGGTTC TTTTCGGTTC TTTTCGGTTC ----GTTTTC ----GTTTTC ----GTTTTC TTTTCGGTTC TTTTCGGTTC T----CCTTC TCTTCAGTTC T-----CTTC 640698396_C_psychrerythraea_34H 640698404_C_psychrerythraea_34H 640698412_C_psychrerythraea_34H 640698493_C_psychrerythraea_34H 640698426_C_psychrerythraea_34H 640698400_C_psychrerythraea_34H 640698408_C_psychrerythraea_34H 640698416_C_psychrerythraea_34H 640698390_C_psychrerythraea_34H DQ027051.1_Colwellia_sp_MT41 DQ027052.1_Colwellia_sp_KT27 DQ027056.1_Psychromonas_sp_CNPT3 DQ027057.1_Psychromonas_sp_HS11 640705772_P_profundum_SS9 DQ027054.1_P_profundum_3TCK DQ027055.1_P_profundum_1230sf1 640705946_P_profundum_SS9 640705754_P_profundum_SS9 640705856_P_profundum_SS9 640705924_P_profundum_SS9 640705919_P_profundum_SS9 640705763_P_profundum_SS9 640705741_P_profundum_SS9 640705850_P_profundum_SS9 640705738_P_profundum_SS9 640705907_P_profundum_SS9 640705913_P_profundum_SS9 640705910_P_profundum_SS9 640705881_P_profundum_SS9 640705884_P_profundum_SS9 DQ027053.1_Moritella_sp_PE36 DQ027059.1_Shewanella_sp_PT48 DQ027060.1_Shewanella_sp_PT64 641466167_P_profundum_SS9 DQ027058.1_Shewanella_sp_KT99 641463346_P_profundum_SS9 DQ027061.1_Carnobacterium_sp_AT7 DQ027062.1_Carnobacterium_sp_AT12 JF303756.1_Rhodobacterales_PRT1 639101207_P_profundum_3TCK 2504111167_HIMB59b U75256.1_SAR211g2 AF353214.1_Arctic95B-1g2 2236674967_AAA288-E13 2236446051_AAA288-G21 2236452142_AAA288-N07 2236674298_AAA240-E13 2504109332_HIMB5b 2503352260_HTCC7211 2503364240_HTCC9565 640708248_HTCC1062 642513240_HTCC1002 X52172.1_SAR11g1b U75649.1_SAR193g1b Z99997.1_LD12 AB154304.1_S9D-28fresh 2503356127_HIMB114 2505687060_IMCC9063 U70686.1_OM155g3 640698396_C_psychrerythraea_34H 640698404_C_psychrerythraea_34H 640698412_C_psychrerythraea_34H 640698493_C_psychrerythraea_34H 640698426_C_psychrerythraea_34H 640698400_C_psychrerythraea_34H 640698408_C_psychrerythraea_34H 640698416_C_psychrerythraea_34H 640698390_C_psychrerythraea_34H DQ027051.1_Colwellia_sp_MT41 DQ027052.1_Colwellia_sp_KT27 DQ027056.1_Psychromonas_sp_CNPT3 DQ027057.1_Psychromonas_sp_HS11 640705772_P_profundum_SS9 DQ027054.1_P_profundum_3TCK DQ027055.1_P_profundum_1230sf1 640705946_P_profundum_SS9 640705754_P_profundum_SS9 640705856_P_profundum_SS9 640705924_P_profundum_SS9 640705919_P_profundum_SS9 640705763_P_profundum_SS9 640705741_P_profundum_SS9 640705850_P_profundum_SS9 640705738_P_profundum_SS9 640705907_P_profundum_SS9 640705913_P_profundum_SS9 640705910_P_profundum_SS9 640705881_P_profundum_SS9 640705884_P_profundum_SS9 DQ027053.1_Moritella_sp_PE36 DQ027059.1_Shewanella_sp_PT48 DQ027060.1_Shewanella_sp_PT64 641466167_P_profundum_SS9 DQ027058.1_Shewanella_sp_KT99 641463346_P_profundum_SS9 DQ027061.1_Carnobacterium_sp_AT7 DQ027062.1_Carnobacterium_sp_AT12 JF303756.1_Rhodobacterales_PRT1 639101207_P_profundum_3TCK 2504111167_HIMB59b U75256.1_SAR211g2 AF353214.1_Arctic95B-1g2 2236674967_AAA288-E13 2236446051_AAA288-G21 2236452142_AAA288-N07 2236674298_AAA240-E13 2504109332_HIMB5b 2503352260_HTCC7211 2503364240_HTCC9565 640708248_HTCC1062 642513240_HTCC1002 X52172.1_SAR11g1b U75649.1_SAR193g1b Z99997.1_LD12 AB154304.1_S9D-28fresh 2503356127_HIMB114 2505687060_IMCC9063 U70686.1_OM155g3 1101 640698396_C_psychrerythraea_34H GG-------G 640698404_C_psychrerythraea_34H GG-------G 640698412_C_psychrerythraea_34H GG-------G 640698493_C_psychrerythraea_34H GG-------G 640698426_C_psychrerythraea_34H GG-------G 640698400_C_psychrerythraea_34H GG-------G 640698408_C_psychrerythraea_34H GG-------G 640698416_C_psychrerythraea_34H GG-------G 640698390_C_psychrerythraea_34H GG-------G DQ027051.1_Colwellia_sp_MT41 GG-------G DQ027052.1_Colwellia_sp_KT27 GG-------G DQ027056.1_Psychromonas_sp_CNPT3 GG-------G DQ027057.1_Psychromonas_sp_HS11 GG-------G 640705772_P_profundum_SS9 GG-------G DQ027054.1_P_profundum_3TCK GG-------G DQ027055.1_P_profundum_1230sf1 GG-------G 640705946_P_profundum_SS9 GG-------G 640705754_P_profundum_SS9 GG-------G 640705856_P_profundum_SS9 GG-------G 640705924_P_profundum_SS9 GG-------G 640705919_P_profundum_SS9 GG-------G 640705763_P_profundum_SS9 GG-------G 640705741_P_profundum_SS9 GG-------G 640705850_P_profundum_SS9 GG-------G 640705738_P_profundum_SS9 GG-------G 640705907_P_profundum_SS9 GG-------G 640705913_P_profundum_SS9 GG-------G 640705910_P_profundum_SS9 GG-------G 640705881_P_profundum_SS9 GG-------G 640705884_P_profundum_SS9 GG-------G DQ027053.1_Moritella_sp_PE36 GG-------G DQ027059.1_Shewanella_sp_PT48 GG-------G DQ027060.1_Shewanella_sp_PT64 GG-------G 641466167_P_profundum_SS9 GG-------G DQ027058.1_Shewanella_sp_KT99 GG-------G 641463346_P_profundum_SS9 GG-------G DQ027061.1_Carnobacterium_sp_AT7 GG-------G DQ027062.1_Carnobacterium_sp_AT12 GG-------G JF303756.1_Rhodobacterales_PRT1 AGCTAGGCTG 639101207_P_profundum_3TCK GG-------G 2504111167_HIMB59b AGTTCGGCTG U75256.1_SAR211g2 AGTTGGGCGA AF353214.1_Arctic95B-1g2 GGCTGGACGA 2236674967_AAA288-E13 AGTTTGGCTG 2236446051_AAA288-G21 GGTTCGGCCG 2236452142_AAA288-N07 GGTTCGGCCG 2236674298_AAA240-E13 GGTTCGGCCG 2504109332_HIMB5b GGCCGGACGA 2503352260_HTCC7211 GGCCGGACGA 2503364240_HTCC9565 GGCCGGACGA 640708248_HTCC1062 GGCCGGACGA 642513240_HTCC1002 GGTTTGGCCG X52172.1_SAR11g1b GGTTCGGCCG U75649.1_SAR193g1b GGTTCGGCCG Z99997.1_LD12 GGCCGGACAG AB154304.1_S9D-28fresh GGCCGGACAG 2503356127_HIMB114 GGTTCGGCCG 2505687060_IMCC9063 GGCTGGACAA U70686.1_OM155g3 GGTTCGGCCG AACTCTGTGA AACTCTGTGA AACTCTGTGA AACTCTGTGA AACTCTGTGA AACTCTGTGA AACTCTGTGA AACTCTGTGA AACTCTGTGA AACTGAGTGA AACTCTGTGA AACTCTGAGA AACTCTGAGA AACTGTGAGA AACTCTGAGA AACTCTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA AACTGTGAGA GACAAAGTGA GACAAAGTGA GACG-AGATA GACGCAGTGA GCTTCCGC-A AAC------A AAC------A GACG-AAACA GACG-AAACA GACG-AAACA GACG-AAACA AAC------A AAC------A AAC------A AAC------A GACG-AAACA GACG-AAACA GACG-AAACA AAC------A AAC------A GACA-GAACA AAC------A GACG-AAACA CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGATGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGGTGC CAGGTGGTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC CAGGTGCTGC ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGTTGTCG ATGGTTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG ATGGCTGTCG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TCAGCTCGTG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG CTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TTGTGAAATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TCGTGAGATG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTCGGTTAAG TTCGGTTAAG TTGGGTTAAG TTGGGGTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TTGGGTTAAG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAGCG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCGGCAACG TCCGGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCACAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG TCCCGCAACG AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCA AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCACAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC AGCGCAACCC CTATCCTTAT CTATCCTTAT CTATCCTTAT CTATCCTTAT CTATCCTTAT CTATCCTTAT CTATCCTTAT CTATCCTTAT CTATCCTTAT CTATCCTTAT CTATCCTTAT TTATCCTTAC TTATCCTTAC TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTGT TTATCCTTAT TTATCCTTAT TTATCCTTAT TTATCCTTAT TTATCCTTAT TTATCCTTAT CTATTATTAG CTATTATTAG ACGTCCTTAG ACATCCCTAG TTATCGTTAG CTACGTTTAG CTACTTTTAG TTACTTTTAG TTACTTTTAG TTACTTTCAG TTACTTTTAG TCACTTTTAG TCACTTTTAG TCACTTTTAG TCACTTTTAG TCACTTTTAG TCACTTTTAG TCACTTTTAG CTACTTTTAG CTACTTTTAG CTACTTCTAG CTACTTTTAG CTACTTTTAG TTGCCAGCGC TTGCCAGCGC TTGCCAGCGC TTGCCAGCGC TTGCCAGCGC TTGCCAGCGC TTGCCAGCGC TTGCCAGCGC TTGCCAGCGC TTGCCAGCGC TTGCCAGCGC TTGCCAGCGG TTGCCAGCGG TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCAC TTGCCAGCATTGCCAGCATTGCCAGCAG TTGCCAGCATTACTAACATTACTAACATTGCCACCATTGACACCATTGCCATCATTGCCACCATTGCCATCATTGCCATCATTGCCATCATTGCCATCATTGCCATCATTGCCATCATTGCCATCATTGCCATCATTGCCATCAG TTGCCATCAG TTGCCATCAG TTGCCATCAG TTGCCATCAG 1211 640698396_C_psychrerythraea_34H GTTATG-GCG 640698404_C_psychrerythraea_34H GTTATG-GCG 640698412_C_psychrerythraea_34H GTTATG-GCG 640698493_C_psychrerythraea_34H GTTATG-GCG 640698426_C_psychrerythraea_34H GTTATG-GCG 640698400_C_psychrerythraea_34H GTTATG-GCG 640698408_C_psychrerythraea_34H GTTATG-GCG 640698416_C_psychrerythraea_34H GTTATG-GCG 640698390_C_psychrerythraea_34H GTTATG-GCG DQ027051.1_Colwellia_sp_MT41 GTTATG-GCG DQ027052.1_Colwellia_sp_KT27 GTTATG-GCG DQ027056.1_Psychromonas_sp_CNPT3 GTCATG-CCG DQ027057.1_Psychromonas_sp_HS11 GTCATG-CCG 640705772_P_profundum_SS9 GTCATG-GTG DQ027054.1_P_profundum_3TCK GTAATG-GTG DQ027055.1_P_profundum_1230sf1 GTAATG-GTG 640705946_P_profundum_SS9 GTAATG-GTG 640705754_P_profundum_SS9 GTCATG-GTG 640705856_P_profundum_SS9 GTAATG-GTG 640705924_P_profundum_SS9 GTCATGAGTG 640705919_P_profundum_SS9 GTCATG-GTG 640705763_P_profundum_SS9 GTCATG-GTG 640705741_P_profundum_SS9 GTCATG-GTG 640705850_P_profundum_SS9 GTCATG-GTG 640705738_P_profundum_SS9 GTCATG-GTG 640705907_P_profundum_SS9 GTCATG-GTG 640705913_P_profundum_SS9 GTCATG-GTG 640705910_P_profundum_SS9 GTCATG-GTG 640705881_P_profundum_SS9 GTCATG-GTG 640705884_P_profundum_SS9 GTCATG-GTG DQ027053.1_Moritella_sp_PE36 GTAATG-GTG DQ027059.1_Shewanella_sp_PT48 GTAATG-GTG DQ027060.1_Shewanella_sp_PT64 GTAATG-GTG 641466167_P_profundum_SS9 GTAATG-GTG DQ027058.1_Shewanella_sp_KT99 GTAATG-GTG 641463346_P_profundum_SS9 GTAATG-GTG DQ027061.1_Carnobacterium_sp_AT7 -TTCAG-TTG DQ027062.1_Carnobacterium_sp_AT12 -TTCAG-TTG JF303756.1_Rhodobacterales_PRT1 GTTAAG-CTG 639101207_P_profundum_3TCK GTTCGG-CTG 2504111167_HIMB59b GTTCGG-CTG U75256.1_SAR211g2 -TTTAG-TTG AF353214.1_Arctic95B-1g2 -TTTAG-TTG 2236674967_AAA288-E13 -TTTAG-TTG 2236446051_AAA288-G21 -TTTAG-TTG 2236452142_AAA288-N07 -TTTAG-TTG 2236674298_AAA240-E13 -TTTAG-TTG 2504109332_HIMB5b -TTAAG-TTG 2503352260_HTCC7211 -TTAAG-TTG 2503364240_HTCC9565 -TTAAG-TTG 640708248_HTCC1062 -TTAAG-TTG 642513240_HTCC1002 -TTAAG-TTG X52172.1_SAR11g1b -TTTAG-TTG U75649.1_SAR193g1b -TTCAG-TTG Z99997.1_LD12 GTCATG-CTG AB154304.1_S9D-28fresh GTCATG-CTG 2503356127_HIMB114 GTTAAG-CTG 2505687060_IMCC9063 GTTATG-CTG U70686.1_OM155g3 GTTAAG-CTG GGAACTCT-A GGAACTCT-A GGAACTCT-A GGAACTCT-A GGAACTCT-A GGAACTCT-A GGAACTCT-A GGAACTCT-A GGAACTCT-A GGAACTCT-A GGAACTCT-A GGAACTTT-A GGAACTTT-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCC-A GGAACTCT-A GGAACTTT-A GGAACTTT-A GGAACTTT-A GGAACTTT-A GGAACTTT-A GGCACTCT-A GGCACTCT-A GGCACTCT-A GGCACTCT-A AGGACTCT-A AGCACTCT-A GGCACTTT-A AGCACTCT-G GGCACTCT-G AGCACTCT-G GGCACTCT-G GGCACTCT-G GGCACTCT-G GGCACTCT-G GGCACTCT-G GGCACTCT-G GGCACTCT-G GGCACTCTGA GGCACTCT-G GGCACTCT-G GGCACTCT-A GGCACTCT-A GGCACTCT-A AGGAGACTGC AGGAGACTGC AGGAGACTGC AGGAGACTGC AGGAGACTGC AGGAGACTGC AGGAGACTGC AGGAGACTGC AGGAGACTGC AGGAGACTGC AGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACGGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC AGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GGGAGACTGC GTGAGACTGC GTGAGACTGC GGGAGACTGC TGGAAACTGC GCGAAACTGC AGAAGACTCC AAAGAACTGC AAAGAACTGC AAAGAACTGC AAAGAACTGC AAAGAACTGC AAAGAACTGC AAAGAACTGC AAAGAACTGC AAAGAACTGC AAAGAACTGC AAAGAACTGC AAGTAACTGC AAAGAACTGC AAAGAACTGC GTAGGACTGC ATAGAACTGC ATAGAACTGC CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA TGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAA CGGTGATAAG CCGTGATAAG CGGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CAGTGATAAG CTGTGATAAG CTGTGATAAA CTGTGATAAA CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CCGGAGGAAG CGGGAGGAAG CCGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CTGGAGGAAG CAGGAGGAAG CAGGAGGAAG CAGGAGGAAG GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGTCGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGACGA GTGGGGATGA GTGGGGATGA GTGTGGATGA GTGTGGATGA GTGGGGATGA NCGGGGATGA GTGGGGATGA GTGGGGATTA GTGGGGATGA GTGGGGATGA GTGGGGATGA GTGGGGATGA GTGGGGATGA GTGGGGATGA GTGGGGATGA GTGGGGATGA GTGTGGCTGG GCGAGGATGA GTGGGGATGA GTGGGGATGA GTTGGGATGA GTGGGGATGA GTGGGGATGA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGATA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAGTCA CGTCAAATCA CGTCAAATCA CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC ---------CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC CGTCAAGTCC TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATCGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGCCCCT TCATGCCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT ---------TCATGGCCCT TCATGGGCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TCATGGCCCT TACGGGATGG TACGGGATGG TACGGGATGG TACGGGATGG TACGGGATGG TACGGGATGG TACGGGATGG TACGGGATGG TACGGGATGG TACGGGATGG TACGGGATGG TACGGGATGG TACGGGATGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TACGAGTAGG TATGACCTGG TATGACCTGG TACGGGTTGG TACGGGTTGG TACACGCTGG TACGTGTTGG TATGTGTTGG TACGTGTTGG TACGTGTTGG TACGTGTTGG TACGTGTTGG TACGTGTTGG TACGTGTTGG TACGTGTTGG TACGTGTTGG TACGTGTTGG ---------TACGTGTTGG TACGGGTTGG TACGGGTTGG TACGTGTTGG TACGGGTTGG TACGGGTTGG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG ---------GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG GCTACACACG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG ---------TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG TGCTACAATG 1321 640698396_C_psychrerythraea_34H GCAGGTACAG 640698404_C_psychrerythraea_34H GCAGGTACAG 640698412_C_psychrerythraea_34H GCAGGTACAG 640698493_C_psychrerythraea_34H GCAGGTACAG 640698426_C_psychrerythraea_34H GCAGGTACAG 640698400_C_psychrerythraea_34H GCAGGTACAG 640698408_C_psychrerythraea_34H GCAGGTACAG 640698416_C_psychrerythraea_34H GCAGGTACAG 640698390_C_psychrerythraea_34H GCAGGTACAG DQ027051.1_Colwellia_sp_MT41 GCAGGTACAG DQ027052.1_Colwellia_sp_KT27 GCAAGTACAG DQ027056.1_Psychromonas_sp_CNPT3 GCAGATACAA DQ027057.1_Psychromonas_sp_HS11 GCAGATACAA 640705772_P_profundum_SS9 GCGTATACAG DQ027054.1_P_profundum_3TCK GCGTATACAG DQ027055.1_P_profundum_1230sf1 GCGTATACAG 640705946_P_profundum_SS9 GCGTATACAG 640705754_P_profundum_SS9 GCGTATACAG 640705856_P_profundum_SS9 GCGTATACAG 640705924_P_profundum_SS9 GCGTATACAG 640705919_P_profundum_SS9 GCGTATACAG 640705763_P_profundum_SS9 GCGTATACAG 640705741_P_profundum_SS9 GCGTATACAG 640705850_P_profundum_SS9 GCGTATACAG 640705738_P_profundum_SS9 GCGTATACAG 640705907_P_profundum_SS9 GCGTATACAG 640705913_P_profundum_SS9 GCGTATACAG 640705910_P_profundum_SS9 GCGTATACAG 640705881_P_profundum_SS9 GCGTATACAG 640705884_P_profundum_SS9 GCGTATACAG DQ027053.1_Moritella_sp_PE36 GCGCATACAA DQ027059.1_Shewanella_sp_PT48 GTCGGTACAG DQ027060.1_Shewanella_sp_PT64 GTCGGTACAG 641466167_P_profundum_SS9 GTCGGTACAG DQ027058.1_Shewanella_sp_KT99 GTCGGTACAG 641463346_P_profundum_SS9 GTCGGTACAG DQ027061.1_Carnobacterium_sp_AT7 GATGGTACAA DQ027062.1_Carnobacterium_sp_AT12 GATGGTACAA JF303756.1_Rhodobacterales_PRT1 GCAGTGACAG 639101207_P_profundum_3TCK GCATCTACAG 2504111167_HIMB59b GTAACTACAA U75256.1_SAR211g2 GCATTTACAA AF353214.1_Arctic95B-1g2 GTACTTACAA 2236674967_AAA288-E13 GCACTCACAA 2236446051_AAA288-G21 GCATTTACAA 2236452142_AAA288-N07 GCACTCACAA 2236674298_AAA240-E13 GCACTCACAA 2504109332_HIMB5b GTATCTACAA 2503352260_HTCC7211 GTATCTACAA 2503364240_HTCC9565 GCATCTACAA 640708248_HTCC1062 GCATCTACAA 642513240_HTCC1002 GCATCTACAA X52172.1_SAR11g1b ---------U75649.1_SAR193g1b GCATTTACAA Z99997.1_LD12 GTAGTGACAA AB154304.1_S9D-28fresh GTAGTGACAA 2503356127_HIMB114 GCAGTGACAA 2505687060_IMCC9063 GTAGGTACAG U70686.1_OM155g3 GTAAGTACAA AGGGCAGCAA AGGGCAGCAA AGGGCAGCAA AGGGCAGCAA AGGGCAGCAA AGGGCAGCAA AGGGCAGCAA AGGGCAGCAA AGGGCAGCAA AGGGCTGCAA AGGGCTGCAA AGGGTTGCTA AGGGTTGCTA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCCA AGGGCTGCAA AGGGTCGCAA AGGGTCGCAA AGGGTTGCAA AGGGTTGCAA AGGGTTGCAA CGAGTCGCAA CGAGTCGCAA TGGGTT---TGGGTT---CGGGACGCAA TGAGACGCAA TGGGAAGCAA TAGGAAGCAA TAGGAAGCAA TAGGAAGCGA TGGGAAGCAA CAGGAAGCAA CAGGAAGCGA CAGGAAGCAA CAGGAAGCAA CAGGAAGCAA ---------TAGGAAGCAA TGAGATGCAA TGAGATGCAA TGAGATGCTA TGAGATGCCG TGAGATGCCG TACCGCGAGG TACCGCGAGG TACCGCGAGG TACCGCGAGG TACCGCGAGG TACCGCGAGG TACCGCGAGG TACCGCGAGG TACCGCGAGG TACCGCGAGG TACCGCGAGG ACCTGCGAGG ACCTGCGAGG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCGATG ACCAGCAATG CGCCGCGAGG CGCCGCGAGG CGCCGCGAGG CGCCGCGAGG CGCCGCGAGG GGTCGCGAGG GGTCGCGAGG ------------------TACCGCGAGG AGAGGCAACT AGAGGCGACT AGATGTGAGT GATGGCGACA AGAGGCGACT AGAGGTGACT GACGGCGACG AACTGCGAAG AACGGCAACG AACGGCAACG AACGGCAACG ---------GATGGCGACA AGTGGCGACG GGTGGCGACG AACGGCGACG AGCAGCGATG AACAGCGATG TGGAGCGAAT TGGAGCGAAT TGGAGCGAAT TGGAGCGAAT TGGAGCGAAT TGGAGCGAAT TGGAGCGAAT TGGAGCGAAT TGGAGCGAAT TGGAGCGAAT TGGAGCGAAT GTATGCGAAT GTATGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTGAGCGAAT GTAAGCGAAT TCAAGCTAAT TCAAGCTAAT TCAAGCTAAT TCAAGCTAAT TCAAGCTAAT CCAAGCTAAT CCAAGCTAAT -------AAT -------AAT TGGAGCAAAT CTAAGCCAAT CCTAGCCAAT CTAAGCAAAT TCAAGCAAAT CTAAGCAAAT CTAAGCAAAT TCAAGCAAAT TTAAGCAAAT TTAAGCAAAT TTAAGCAAAT TTAAGCAAAT ---------TCAAGCAAAT CTAAGCTAAA CTAAGCTAAA TTAAGCTAAA CTTAGCTAAA TTTAGCCAAA CCCACAAAGC CCCACAAAGC CCCACAAAGC CCCACAAAGC CCCACAAAGC CCCACAAAGC CCCACAAAGC CCCACAAAGC CCCACAAAGC CCCACGAAGC CCCACAAAGC CTCATAAAGT CTCATAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCACAAAGT CCCATAAAGT CCCACAAAGC CCCACAAAGC CCCACAAAGC CCCACAAAGC CCCACAAAGC CTCTTAAAGC CTCTTAAAGC CCC-TAAAAA CCC-CAAAAG CCC-CAAAAG CTCACAAA-T CCC-TAAAAT CCTAAAAAACCTAAAAAAA CTTAAAAAACCTAAAAAACCT-TAAAAG CCC-TAAAAG CCCGAAAAGCCCGAAAAGCCCGAAAAG---------CCTAAAAAGCTCA-AAAAT CTCA-AAAAT CTCA-AAAAC CTC-CAAAAC CTCA-AAAAC TTGTCGTAGT TTGTCGTAGT TTGTCGTAGT TTGTCGTAGT TTGTCGTAGT TTGTCGTAGT TTGTCGTAGT TTGTCGTAGT TTGTCGTAGT CTGTCGTAGT TTGTCGTAGT CTGTCGTAGT CTGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT ACGTCGTAGT GCGTCGTAGT CGGTCGTAGT CGGTCGTAGT CGGTCGTAGT CGGTCGTAGT CGGTCGTAGT CATTCTCAGT CATTCTCAGT CTGTCTCAGT ATGTCTCAGT TTATCTCAGT ATGCCTCAGT GTACCTCAGT GTGCCTCAGT ATGCCTCAGT GTGCCTCAGT GTGCCTCAGT ATACCTCAGT ATACCTCAGT ATGCCTCAGT ATGCCTCAGT ATGCCTCAGT ---------ATGCCTCAGT CTACCTCAGT CTACCTCAGT CTGCCTCAGT CTACCTCAGT TTACCTCAGT CCGGATTGGA CCGGATTGGA CCGGATTGGA CCGGATTGGA CCGGATTGGA CCGGATTGGA CCGGATTGGA CCGGATTGGA CCGGATTGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATTGGG CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA CCGGATCGGA TCGGATTGCA TCGGATTGCA TCGGATTGGG TCGGATTGTC TCGGATTGCA TCGGATTGTA TCGGATTGTA TCGGATTGTA TCGGATTGTA TCGGATTGTA TCGGATTGTA TCGGATTGCA TCGGATTGCA TCGGATTGCA TCGGATTGCA TCGGATTGCA ---------TCGGATTGTA TCGGATTGCA TCGGATTGCA TCGGATTGCA TCGGATTGCA TCGGATTGCA GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GTCTGCAACT GGCTGCAACT GGCTGCAACT GTCTGCAACT GTCTGCAACT CTCTGCAACT CTCTNCAACT CTCTGTAACT CTCTGCAACT CTCTGCAACT CTCTGCAACT CTCTGCAACT CTCTGCAACT CTCTGCAACT CTCTGCAACT CTCTGCAACT CTCTGCAACT ---------CTCTGCAACT CTCTGCAACT CTCTGCAACT CTCTGCAACT CTCTGCAACT CTCTGCAACT CGACTCCATG CGACTCCATG CGACTCCATG CGACTCCATG CGACTCCATG CGACTCCATG CGACTCCATG CGACTCCATG CGACTCCATG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACCCCATG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGACTCCGTG CGCCTGCATG CGCCTGCATG CGACCCCATG CGACGGCATG CGAGTGCATG CGAGTACATG CGAGTACATG CGAGTACATG CGAGTACATG CGAGTACATG CGAGTACATG CGAGTGCATG CGAGTGCATG CGAGTGCATG CGAGTGCATG CGAGTGCATG ---------CGAGTACATG CGAGTGCATG CGAGTGCATG CGAGTGCATG CGAGTGCATG CGAGTGCATG AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTTGGAAT AAGTTGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTGGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGTCGGAAT AAGCCGGAAT AAGCCGGAAT AAGTTGGAAT AAGTCGGAAT AAGTTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT ---------AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT AAGCTGGAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT TGCTAGTAAT TGCTAGTAAT TGCTAGTAAT TGCTAGTAAT TGCTAGTAAT TGCTAGTAAT TGCTAGTAAT TACTAGTAAT TACTAGTAAT TACTAGTAAT TACTAGTAAT TACTAGTAAT ---------TACTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT CGCTAGTAAT 1431 640698396_C_psychrerythraea_34H CGTAGATCAG 640698404_C_psychrerythraea_34H CGTAGATCAG 640698412_C_psychrerythraea_34H CGTAGATCAG 640698493_C_psychrerythraea_34H CGTAGATCAG 640698426_C_psychrerythraea_34H CGTAGATCAG 640698400_C_psychrerythraea_34H CGTAGATCAG 640698408_C_psychrerythraea_34H CGTAGATCAG 640698416_C_psychrerythraea_34H CGTAGATCAG 640698390_C_psychrerythraea_34H CGTAGATCAG DQ027051.1_Colwellia_sp_MT41 CGTGGATCAG DQ027052.1_Colwellia_sp_KT27 CGTGGATCAG DQ027056.1_Psychromonas_sp_CNPT3 CGTGGATCAG DQ027057.1_Psychromonas_sp_HS11 CGTGGATCAG 640705772_P_profundum_SS9 CGTGAATCAG DQ027054.1_P_profundum_3TCK CGTGAATCAG DQ027055.1_P_profundum_1230sf1 CGTGAATCAG 640705946_P_profundum_SS9 CGTGAATCAG 640705754_P_profundum_SS9 CGTGAATCAG 640705856_P_profundum_SS9 CGTGAATCAG 640705924_P_profundum_SS9 CGTGAATCAG 640705919_P_profundum_SS9 CGTGAATCAG 640705763_P_profundum_SS9 CGTGAATCAG 640705741_P_profundum_SS9 CGTGAATCAG 640705850_P_profundum_SS9 CGTGAATCAG 640705738_P_profundum_SS9 CGTGAATCAG 640705907_P_profundum_SS9 CGTGAATCAG 640705913_P_profundum_SS9 CGTGAATCAG 640705910_P_profundum_SS9 CGTGAATCAG 640705881_P_profundum_SS9 CGTGAATCAG 640705884_P_profundum_SS9 CGTGAATCAG DQ027053.1_Moritella_sp_PE36 CGTGAATCAG DQ027059.1_Shewanella_sp_PT48 CGTGAATCAG DQ027060.1_Shewanella_sp_PT64 CGTGAATCAG 641466167_P_profundum_SS9 CGTGAATCAG DQ027058.1_Shewanella_sp_KT99 CGTGAATCAG 641463346_P_profundum_SS9 CGTGAATCAG DQ027061.1_Carnobacterium_sp_AT7 CGCGGATCAG DQ027062.1_Carnobacterium_sp_AT12 CGCGGATCAG JF303756.1_Rhodobacterales_PRT1 CGCGTAACAG 639101207_P_profundum_3TCK CGCGTAACAG 2504111167_HIMB59b CGCGGATCAG U75256.1_SAR211g2 CGCGGATCAG AF353214.1_Arctic95B-1g2 CGCGGATCAG 2236674967_AAA288-E13 CGCGGATCAG 2236446051_AAA288-G21 CGCGGATCAG 2236452142_AAA288-N07 CGCGGATCAG 2236674298_AAA240-E13 CGCGGATCAG 2504109332_HIMB5b CGTGGATCAG 2503352260_HTCC7211 CGTGGATCAG 2503364240_HTCC9565 CGTGGATCAG 640708248_HTCC1062 CGTGGATCAG 642513240_HTCC1002 CGTGGATCAG X52172.1_SAR11g1b ---------U75649.1_SAR193g1b CGCGGATCAG Z99997.1_LD12 CGTAGATCAG AB154304.1_S9D-28fresh CGTAGATCAG 2503356127_HIMB114 CGCTCATCAG 2505687060_IMCC9063 CGTGGATCAG U70686.1_OM155g3 CGTGGATCAG AATGCTACGG AATGCTACGG AATGCTACGG AATGCTACGG AATGCTACGG AATGCTACGG AATGCTACGG AATGCTACGG AATGCTACGG AACGCCACGG AATGCCACGG AATGCCACGG AATGCCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG AATGTCACGG CACGCCGCGG CACGCCGCGG CATGACGCGG CATGACGCGG CATGCCGCGG CGCGCCGCGG CGCGCCGCGG CGCGCCGCGG CGCGCCGCGG CGCGCCGCGG CGCGCCGCGG CGTGCCACGG CGTGCCACGG CGTGCCACGG CGTGCCACGG CGTGCCACGG ---------CGCGCCGCGG CGTGCTACGG CGTGCTACGG CGCGGAGCGG CGTGCCACGG CGTGC----- TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATGCGTT TGAATGCGTT TGAATGCGTT TGAATGCGTT TGAATGCGTT ---------TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT TGAATACGTT ---------- CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGCCTT CCCGGGTCTT CCCGGGTCTT CCCGGGCCTT CCCGGGCCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT ---------CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT CCCGGGTCTT ---------- GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG ---------GTACACACCG GTACACACCG GTACACACCG GTACACACCG GTACACACCG ---------- CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCACCA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA ---------CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA CCCGTCA-CA ---------- CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCACGAGAGT CCACGAGAGT CCATGGGAGT CCATGGGAGT CCATGAGAGT CCATGGAAGT CCATGGAAGT CCATGGAAGT CCATGGAAGT CCATGGAAGT CCATGGAAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT CCATGGGAGT ---------CCATGGGAGT CCATGGAAGT CCATGGAAGT CCATGGAAGT CCATGGAAAT ---------- GGGATGCAAA GGGATGCAAA GGGATGCAAA GGGATGCAAA GGGATGCAAA GGGATGCAAA GGGATGCAAA GGGATGCAAA GGGATGCAAA GGGATGCAAA GGGATGCAAA GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC GGGCTGCACC TTGTAACACC TTGTAACACC TGGGTCTACC TGGTTCTACC TGGTTTTATC TGGTTACACC TGGTTACACC TGGTTCTACC TGGTTCTACC TGGTTCTACC TGGTTCTACC TGGTTCTACC TGGTTCTACC TGGTTCTACC TGGTTCTACC TGGTTCTACC ---------TGGTTCTACC TGGTTCTACC TGGTTCTACC TGGTTTTACC TGGATTTACC ---------- -AGAAGTGGC -AGAAGTGGC -AGAAGTGGC -AGAAGTGGC -AGAAGTGGC -AGAAGTGGC -AGAAGTGGC -AGAAGTGGC -AGAAGTGGC -AGAAGTGGC -AGAAGTGGC -AGAAGTCAT -AGAAGTCAT -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTCAT -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -AGAAGTAGA -CGAAGTCGG -CGAAGTCGG -CGAAGGCAG -CGACGGCCG -TTAAGTCAG -TGAAGGCAA -TTAAGGCAA -TTAAG----TTAAG----TTAAG-------------TTAAG----TTAAGGCAA -TTAAGGCAA -TTAAGGCAA -TTAAGGCAA ----------TTAAGGCAA TTAAAGCAGA TTAAAGCAGA -TTAAGGCAT TTAAGGCAAA ---------- TAGTTTAACC TAGTTTAACC TAGTTTAACC TAGTTTAACC TAGTTTAACC TAGTTTAACC TAGTTTAACC TAGTTTAACC TAGTTTAACC TAGTTTAACC TAGTTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAACC TAGCTTAA-TAGCTTAACC TAGCTTAACC TAGCTTAACC TGAGGTAACC TGAGGTAACC TGCGCCAACC TGCGCTAACTGC------AACATAAAAC -AGCTTATAC ---------------------------------------------GGTTTTAAAC GGTTTTAAAC GGTTTTAAAC GGTTTTAAAC ---------GGTTTAAAAC TTGCT-AACC TTGCTAACCGTCACTAACC T------------------ CC-------CC-------CC-------CC-------CT-------CC-------CC-------CC-------CT-------TT-------TT-------TT-------TT-------CCGATCTTTT TT-------TT-------TT-------TT-------CCGATCTTTT TT-------TT-------CCGATCTTTT CCGATTTTTT TT-------TT-------TT-------TT-------TT-------TT-------TT-------TT-------CT----------------CT-------CT-------CT-------CT-------CT-------TC-------------------------TT-------CT----------------------------------------------------CC-------CC-------CC-------CC----------------CC-------GC----------------CT-------------------------- 1541 640698396_C_psychrerythraea_34H ------TCGG 640698404_C_psychrerythraea_34H ------TCGG 640698412_C_psychrerythraea_34H ------TCGG 640698493_C_psychrerythraea_34H ------TCGG 640698426_C_psychrerythraea_34H ------TCGG 640698400_C_psychrerythraea_34H ------TCGG 640698408_C_psychrerythraea_34H ------TCGG 640698416_C_psychrerythraea_34H ------TCGG 640698390_C_psychrerythraea_34H ------TCGG DQ027051.1_Colwellia_sp_MT41 --------CG DQ027052.1_Colwellia_sp_KT27 --------CG DQ027056.1_Psychromonas_sp_CNPT3 ------TCGG DQ027057.1_Psychromonas_sp_HS11 ------TCGG 640705772_P_profundum_SS9 AGGAGAAAGG DQ027054.1_P_profundum_3TCK --------CG DQ027055.1_P_profundum_1230sf1 --------CG 640705946_P_profundum_SS9 --------CG 640705754_P_profundum_SS9 --------CG 640705856_P_profundum_SS9 AGGAGAAAGG 640705924_P_profundum_SS9 --------CG 640705919_P_profundum_SS9 --------CG 640705763_P_profundum_SS9 AGGAGAAAGG 640705741_P_profundum_SS9 AGGAAAAAGG 640705850_P_profundum_SS9 --------CG 640705738_P_profundum_SS9 --------CG 640705907_P_profundum_SS9 --------CG 640705913_P_profundum_SS9 --------CG 640705910_P_profundum_SS9 --------CG 640705881_P_profundum_SS9 --------CG 640705884_P_profundum_SS9 --------CG DQ027053.1_Moritella_sp_PE36 --------CG DQ027059.1_Shewanella_sp_PT48 ------TCGG DQ027060.1_Shewanella_sp_PT64 ---------641466167_P_profundum_SS9 ------TCGG DQ027058.1_Shewanella_sp_KT99 ------TCGG 641463346_P_profundum_SS9 ------TCGG DQ027061.1_Carnobacterium_sp_AT7 ------TTTG DQ027062.1_Carnobacterium_sp_AT12 ---------JF303756.1_Rhodobacterales_PRT1 -----GCAAG 639101207_P_profundum_3TCK ---------2504111167_HIMB59b ---------U75256.1_SAR211g2 ---------AF353214.1_Arctic95B-1g2 ---------2236674967_AAA288-E13 ---------2236446051_AAA288-G21 ---------2236452142_AAA288-N07 ---------2236674298_AAA240-E13 ---------2504109332_HIMB5b ---------2503352260_HTCC7211 ---------2503364240_HTCC9565 ---------640708248_HTCC1062 ---------642513240_HTCC1002 ---------X52172.1_SAR11g1b ---------U75649.1_SAR193g1b ---------Z99997.1_LD12 ---------A AB154304.1_S9D-28fresh -------GCA 2503356127_HIMB114 -------CGG 2505687060_IMCC9063 ---------U70686.1_OM155g3 ---------- GGAGGACGGT GGAGGACGGT GGAGGACGGT GGAGGACGGT GGAGGACGGT GGAGGACGGT GGAGGACGGT GGAGGACGGT GGAGGACGGT GGAGGACGGT GGAGGACGGT GGAT-----GGATGGCGAT GGAGGGCGTT GGAGGGCGTT G--------GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAGGGCGAT GGAGGGCGTT ---------GGAGGGCGTT GGAGGGCGTT GGAGGGCGTT GGAG-----C ---------AGGAGGCAGC ---------------------------T ---------T ------------------------------------------------------T ---------T ---------T ---------T -----------------TT AGGAGGCGTT AGGAGGCGTT GAAGTGCGTG ------------------- C-ACCACTTT C-ACCACTTT C-ACCACTTT C-ACCACTTT C-ACCACTTT C-ACCACTTT C-ACCACTTT C-ACCACTTT C-ACCACTTT C-ACCACTTT C-ACCACTTT ---------G-ACCACGGT T-ACCACGGT T-ACCACGGT ---------T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT T-ACCACGGT G-ACCAC--------------------T-ACCACGGT T-ACCACGGT T-ACCACGGT CAGCCGC-----------TGACCACGGT ------------------TGACTACGGT TGACTACGGT ---------------------------------------------TGACCACGGT TGACCACGGT TGACCACGGT TGACCACGGT ---------GAACTACGGT TGACCAAGGT TGACCAAGGT --ACCACGGT ------------------- GTGTTTCATG GTGTTTCATG GTGTTTCATG GTGTTTCATG GTGTTTCATG GTGTTTCATG GTGTTTCATG GTGTTTCATG GTGTTTCATG GTGTTTCATG GTGTTTCATG ---------GT-------GTGGTTCATG GTGGTTCATG ---------GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG GTGGTTCATG ---------------------------GTGGTTCATG GTGGTTCATG GTGGTTCATG ------------TTT---GGGTTCAGCG ------------------ACGATCAGCG ACGATCAGCA ---------------------------------------------ATAGTCAGCG ATAGTCAGCG ATAGTCAGCG ATAGTCAGCG ---------ATAGTCAGCG ATAGTCAGCA ATAGTCAGCG ATGATCGGCG ------------------- ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ------------------ACTGGGGTGA ACTGGGGTGA ---------ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ---------------------------ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA -CTAAGGTG---------ACTGGGGTGA ------------------ACTGGGGTGA ACTGGGGTG------------------------GCAA -TTAAGGCAA ---------ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ACTGGGGTGA ---------ACTGGGGTGA ACTGGGGTGACTGGGGTGA ACTGGGGTGA ------------------- AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA ------------------AGTCGTAACA AGTCGTAACA ---------AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA ---------------------------AGTCGTAACA AGTCGTA--AGTCGTAACA ---------------------------------------------AGTCGTAACA ----------------GCA -------GCA AG-------AG--------------GCA AGTCGTAACA AGTCGTAACA AGTCGTAACA AGTCGTAACA ---------AGTCGTAACA ----------------ACA AGTCGTAACA ------------------- AGGTAACCCT AGGTAACCCT AGGTAACCCT AGGTAACCCT AGGTAACCCT AGGTAACCCT AGGTAACCCT AGGTAACCCT AGGTAACCCT AGGTAACCGT AGGTAACCGT ------------------AGGTAGCCCT AGGTAAACCT ---------AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT AGGTAGCCCT ---------------------------AGGTAGCCCT ---------AGGTAGCCCT ---------------------------------------------AGGTAGCCGT ---------AAG------AAG------------------------AGG------AGGTAGCCGT AGGTAGCCGT AGGTAGCCGT AGGTAGCCGT ---------AGGTAGCCGT ---------AGGTAACCGT AGGTAGCCGT ------------------- AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AAAGGG---AAAGGG---------------------AGGGGAACCT ------------------AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT ---------------------------AGGGGAACCT ---------AGGGGAACCT ---------------------------------------------AGGGGAA-CT ------------------------------------------------------AGGGGAACCT AGGGGAACCT AGGGGAACCT AGGGGAACCT ---------AGGGGAACCT ---------AGGGGAACCT AGGGGAACCT ------------------- GGGGTTGGAT GGGGTTGGAT GGGGTTGGAT GGGGTTGGAT GGGGTTGGAT GGGGTTGGAT GGGGTTGGAT GGGGTTGGAT GGGGTTGGAT ------------------------------------GGGGCTGGAT ------------------GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT GGGGCTGGAT ---------------------------GGGGCTGGAT ---------GGGGCTGGAT --GGACAGAT ------------------------------------GCGGTTGGAT ------------------------------------------------------GCGGCTGGAT GCGGCTGGAT GCGGCTGGAT GCGGCTGGAT ---------GCGGTTGGAT ---------GCGGTTGGAT GCGGCTGGAT ------------------- CACCTCC--CACCTCC--CACCTCC--CACCTCC--CACCTCC--CACCTCC--CACCTCC--CACCTCC--CACCTCC--------------------------------------CACCTCCTTA ------------------CACCTCCT-CACCTCCTTA CACCTCCT-CACCTCCT-CACCTCCT-CACCTCCTTA CACCTCCTTA CACCTCCT-CACCTCCTTA CACCTCCT-CACCTCCT-CACCTCCT-CACCTCCT-CACCTCCT----------------------------CACCTCCTTA ---------CACCTCCTTA ---------------------------------------------CACCTCCTTA ------------------------------------------------------TACCT----TACCT----TACCTCCTTTACCTCCTTT ---------CACCTCCTTA ---------CACCTCCTTA TACCT----------------------- ------------------------------------------------------------------------------------------------------------------------------------CCTA----CCTA------------------------------------------------------------CT--------------CCTAA -------------