FAMILIAL LONG QT SYNDROME (ROMANO-WARD SYNDROME)
advertisement

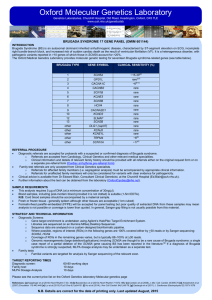
Oxford Molecular Genetics Laboratory Genetics Laboratories, Churchill Hospital, Old Road, Headington, Oxford, OX3 7LE www.ouh.nhs.uk/geneticslab FAMILIAL LONG QT SYNDROME (ROMANO-WARD SYNDROME) Introduction Autosomal dominant long QT syndrome, Romano-Ward Syndrome (RWS), is an inherited disorder of ventricular repolarisation. It has an estimated prevalence of 1 in 2,500 individuals and is characterised by a prolonged QT interval on ECG which predisposes to cardiac arrhythmia, syncope, and sudden death. RWS is genetically heterogeneous with pathogenic variants reported in > 10 genes (see table below) of which those in KCNH2, KCNQ1 and SCN5A account for 72-75% of cases [a, b, c] while the remaining genes are rare causes of LQTS [c,d]. Furthermore, large deletion/duplication variants in KCNQ1 and KCNH2 have previously been described, and an estimated 1–3% of individuals with long QT syndrome may have a variant of this type.[e,f] The Oxford Medical Genetics Laboratory provides molecular genetic testing for sixteen LQT syndrome-related genes (see table below). LQT TYPE GENE Clinical Sensitivity (%) 1 2 3 4 5 6 7 (Anderson-Tawil syndrome) 8 (Timothy Syndrome) 9 10 11 12 13 14 Other Other KCNQ1 KCNH2 SCN5A ANK2 KCNE1 KCNE2 KCNJ2 CACNA1C CAV3 SCN4B AKAP9 SNTA1 KCNJ5 CALM1 CALM2 CALM3 30-35[a, b, e, g, h] 25-30[a, b, e, g, h] 5-10[a, b, e, g, h] <1[c, d] <1[c, d] <1[c, d] <1[c, d] <1[c, d] <1[c, d] Rare[c, d] Rare[c, d] Rare[c, d] Rare[c, d] Rare[c, d] Rare[c, d] Rare[c, d] Gene of higher interest (see technical information below) Yes Yes Yes No No No No No No No No No No No No No REFERRAL PROCEDURE • Diagnostic referrals are accepted for probands with a suspected or confirmed diagnosis of Long QT. o Referrals are accepted from Cardiology, Clinical Genetics and Consultants from other relevant medical specialities. o Clinical information and details of relevant family history should be provided with all referrals either on the original request form or on a separate pre-referral form (Cardiac Arrhythmia pre-referral form). • Family test referrals are only accepted from Clinical Genetics specialists. o Referrals for affected family members (i.e. segregation analysis) must be accompanied by appropriate clinical information. o Referrals for unaffected family members will only be considered for variants with clear evidence for pathogenicity. • Clinical advice is available from Dr Edward Blair, Consultant Clinical Geneticist, at the Churchill Hospital (Ed.Blair@ouh.nhs.uk). • Further information about the test can be obtained from the laboratory (OxfordCardiac@nhs.net). Sample Requirements • This analysis requires 5 µg of DNA (at a minimum concentration of 30ng/µl). • Blood samples, including post-mortem blood (provided it is not clotted) is suitable (1-5ml EDTA). N.B. Cord blood samples should be accompanied by a maternal sample. • Fresh or frozen tissue - generally spleen although other tissues are acceptable (1cm cubed) • Formalin-fixed paraffin-embedded (FFPE) will be accepted for panel testing but poor quality of extracted DNA from these samples may mean analysis is not possible or coverage is lower than quoted. In general, Sanger gap filling is not usually possible from this material. STRATEGY AND TECHNICAL INFORMATION • Diagnostic Screens: o Gene target enrichment is undertaken using Agilent’s HaloPlex Target Enrichment System. o Libraries are sequenced on an Illumina MiSeq Desktop Sequencer. o Sequence data are analysed on a custom-designed bioinformatic pipeline. o Where possible, regions of interest (ROIs) in the following genes are 100% covered either by >30 reads or by Sanger sequencing: KCNQ1, KCNH2, SCN5A. o Coverage of ROIs in the remaining genes varies, but is typically 88–100% at >30 reads. o MLPA dosage analyses for KCNH2, KCNQ1 and KCNE2 can be undertaken additionally upon request. • Family tests o Familial variants are targeted for analysis by Sanger sequencing of the relevant exon. TARGET REPORTING TIMES Diagnostic screen: 60-80 working days Family tests: 10 days MLPA Dosage Analysis 10 days • Please see the current price list on the Oxford Genetics laboratory Molecular genetics page References: : [a] Kapplinger et al (2009) Heart Rhythm 6:1297-1303 [b] Napolitano et al (2005) JAMA 294:2975-2980 [c] Mizusawa et al (2014) Circ. J. 78(12):2827-33 [d] Griffin et al (2015) Heart Rhythm 12(2):419-422 [e] Tester et al (2010) Am J Cardiol 106:1124-1128 [f] Eddy et al. (2008). Heart Rhythm, 5: 1275-1281; [g] Splawski et al (2000); [h] Kapa et al (2009) N.B. Details are correct for the date of printing only. Last updated August 2015