Chem 465 Biochemistry II
advertisement

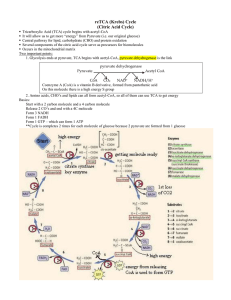
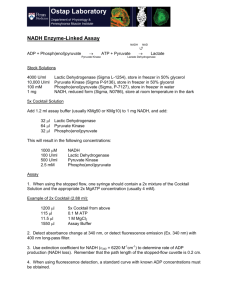
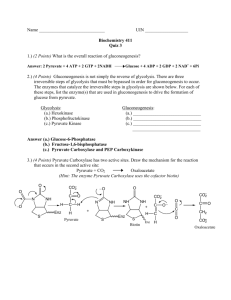
Name: 2 points Chem 465 Biochemistry II Multiple choice (4 points apiece): 1. The anaerobic conversion of 1 mol of glucose to 2 mol of lactate by fermentation is accompanied by a net gain of: A) 1 mol of ATP. B) 1 mol of NADH. C) 2 mol of ATP. D) 2 mol of NADH. E) none of the above. 2. Which of these cofactors participates directly in most of the oxidation-reduction reactions in the fermentation of glucose to lactate? A) ADP B) ATP C) FAD/FADH2 D) Glyceraldehyde 3-phosphate E) NAD+/NADH 3. An enzyme used in both glycolysis and gluconeogenesis is: A) 3-phosphoglycerate kinase. B) glucose 6-phosphatase. C) hexokinase. D) phosphofructokinase-1. E) pyruvate kinase. 4. Gluconeogenesis must use "bypass reactions" to circumvent three reactions in the glycolytic pathway that are highly exergonic and essentially irreversible. Reactions carried out by which three of the enzymes listed must be bypassed in the gluconeogenic pathway? 1) Hexokinase 2) Phosphoglycerate kinase 3) Phosphofructokinase-1 4) Pyruvate kinase 5) Triosephosphate isomerase A) B) C) D) E) 1, 2, 3 1, 2, 4 1, 4, 5 1, 3, 4 2, 3, 4 5. Cellular isozymes of pyruvate kinase are allosterically inhibited by: A) high concentrations of AMP. B) high concentrations of ATP. C) high concentrations of citrate. D) low concentrations of acetyl-CoA. E) low concentrations of ATP. 6. Which of the below is not required for the oxidative decarboxylation of pyruvate to form acetyl-CoA? A) ATP B) CoA-SH C) FAD D) Lipoic acid E) NAD+ 7. Which one of the following enzymatic activities would be decreased by thiamine deficiency? A) Fumarase B) Isocitrate dehydrogenase C) Malate dehydrogenase D) Succinate dehydrogenase E) á-Ketoglutarate dehydrogenase complex Longer questions - 15 points each - Choose any 4! 8. What is Fructose 2,6-bisphosphate? Where does it come from? And how is it important in the regulation of glycolysis and gluconeogenesis? (Include as many details as possible) Fructose 2,6 biphosphate is used primarily as an allosteric effector of the enzymes phosphofructokinase-1 and fructose 1,6 biphosphatase-1. Fructose 2,6-biphosphate is made from Fructose-6-P by the enzyme Phosphofructokinase-2 and is converted back to F-6-P by the enzyme fructose 2,6-biphosphatse-2. Both of these enzymatic activities are combined in one bifuctional protein that toggles between on activity and the other based on the phosphorylation of the enzyme. The complete control cycle looks like this. High Blood sugar insulin released phosphoprotein phosphatase stimulated protein is dephosphorylated and changes to PFK-2 active form Fructose 6-P is changed to Fructose 2,6-biphosphate [Fructose 2,6-biphosphate]8 This activates Phosphofructokinase - 1 and stimulates glycolysis to make more energy and use up glucose This also inhibits fructose 1,6 biphosphatase so gluconeogenesis is slowed so no additional glucose is made Low Blood sugar Glucagon released Adenylate cyclase makes cAMP cAMP dependent protein kinase phosphorylates enzyme and changes to FBPase-2 Active form [Fructose 2,6-biphosphate]9 Phosphofructokinase - 1 no longer activated so glycolysis is slowed and use of glucose is reduced. 1,6 biphosphatase is no longer inhibited so gluconeogenesis is can start up and additional glucose is made. 9. If I have glucose labeled with 13C in the 2 position (see diagram below) where does the 13C label end up in the following compounds? (Draw the structure of the compound and indicate the position of the label) Assume all reactions are going in the normal glycolytic direction, and no gluconeogenesis is taking place. Fructose 1,6-bisphosphate Glyceraldehyde 3-phosphate No label from initial aldolase reaction But after triose phosphate isomerase rxn ½ of the molecules will be Dihydroxyacetone phosphate Phosphoenolpyruvate ½ of the molecules Pyruvate ½ of the molecules Lactate (low oxygen) ½ of molecules (Page left blank so you can sketch out the glycolytic pathway to get your answers) 10. I am going to add glutamic acid labeled in the ä position with 13C (See diagram below) to a cell that is using the TCA cycle to generate energy. Name, and draw the structure of the first intermediate in the TCA cycle in which this label will appear. Indicate which C in this intermediate will carry the 13C label. Initially glutamate will enter the TCA as á-ketoglutarate as labeled below: Now draw the structures of these other TCA intermediates, and indicate where this label will appear in these compounds Succinyl-CoA Fumarate (Note: due to symmetry both ends equivalent) Oxaloacetate Citrate Note: due to symmetry in Succinate and Fumarate a single molecule of oxaloacetate will only have a label at one COO- or the other, but not both. Similarly a single molecule of citrate will have label at only one of the two positions indicated. Isocitrate á-Ketoglutarate Again only one of the labeled positions are labeled in any single isocitrate molecule. Only ½ of the á-ketoglutarate molecules will be carrying a label after one turn of the TCA cycle. (Page left blank so you can sketch out the TCA pathway to get your answers) 11. What is biotin? What is it used for biochemically? What enzymes have we studied that use biotin as an integral part of their mechanism. Biotin is a coenzyme or cofactor that carries activated CO2 in the active site of an enzyme. The structure of biotin is shown below, but you were not required to know this structure. We saw this cofactor in the enzyme pyruvate carboxylase which adds CO2 to pyruvate to make oxaloacetate. This oxaloacetate can either be used to replace oxaloacetate that hs been removed from the TCA cycle, or it can be used for the first step in gluconeogenesis. While Biotin is technically an vitamin because you cannot synthesize it, the bacteria in your gut can make this factor, so it is very difficult to develop a biotin deficiency. Biotin deficiencies can occur, however, in people who eat raw eggs because uncooked eggs contain the protein avidin which binds biotin so it cannot be absorbed in the gut. 12. Compare and contrast the structure, mechanism, cofactors used, and regulation of pyruvate dehydrogenase complex and á-ketoglutarate dehydrogenase complex. Pyruvate dehydrogenase catalyzes the reaction Pyruvate + CoASH 6AcetylCoA + NADH + H+ á-ketoglutarate dehydrogenase catalyzes the reaction á-ketoglutarate + CoASH 6SuccinylCoA + NADH + H+. Pyruvate dehydrogenase and á-ketoglutarate dehydrogenase are very similar in structure and mechanism. Both are large enzyme complexes that contain multiple copies of three major proteins usually called simply E1, E2, and E3. Both enzyme complexes also utilize the cofactors thiamine pyrophosphate, lippoate and FAD, ad well as the substrate CoenzymeA. In pyruvate dehydrogenase the E1 protein is actually the pyruvate dehydrogenase part of the complex. It uses TPP to cleave the terminal CO2 from pyruvate and transfers the remaining part of the pyruvate to E2 (Dihydrolipoyl transacetylase) where this acetate group is temporarily attached to lippoate. The acetate is then transferred to free CoASH to make acetyl CoA which diffuses away from the enzyme complex. When this transfer occurs the lipoate is left in a fully reduced form that must be regenerated before the enzyme can work again. This oxidation is performed by the E3 enzyme, dihydrolipoyl dehydrogenase, which transfers the protons from the reduced lipoate to NAD+ to make NADH. The E3 enzyme is virtually identical in both complexes because it does exactly the same biochemical reaction. The E1 enzyme is just a bit different in the two complexes because it must bind different substrates (pyruvate or á-ketoglutarate). The E2 complexes are close in structure because they also do the same chemistry, but in they vary slightly because different one creates acetyl CoA and the other makes sussinyl-CoA. The allosteric control of pyruvate dehydrogenase is a bit more complicated because it is the first step in the TCA pathway. Pyruvate dehydrogenase is stimulated AMP, CoA, NAD+, and Ca2+ and inhibited by ATP, acetyl-CoA, NADH. The á-ketoglutarate dehydrogenase control is simpler because it is deeper inside the TCA cycle. It is only stimulated by Ca2+ and inhibited by succinyl-CoA and NADH. 1: C 2: E 3: A 4: D 5: B 6: A 7: E