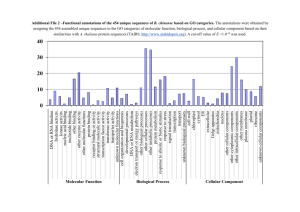
Photoperiodic effect on protein profiles of potato petiole
advertisement

Photoperiodic effect on protein profiles of potato petiole Shweta Shah1, Young Jin-Lee2, David J. Hannapel3 and A. Gururaj Rao*1 1 Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, Iowa 50011 2 Department of Chemistry, Iowa State University, Ames, Iowa 50011 3 Plant Biology Major, Iowa State University, Ames, Iowa 50011 Abstract In potato (Solanum tuberosum), StBEL5 mRNA is a mobile RNA that is involved in the signaling system that activates tuber formation. Short-day (SD) conditions induce tuberization and long-day (LD) inhibits the process. The transcriptional source of this mobile RNA is leaf veins and leaf stalks designated, petioles. Transport of StBEL5 RNA is induced by a SD photoperiod that leads to tuber formation. It is likely that the movement of StBEL5 is facilitated by the formation of RNA-protein complex(s). To further understand this proposed mechanism of downstream signaling we have undertaken a detailed proteomic analysis of proteins isolated from potato petioles (PP) grown under LD and SD photoperiod conditions using 2-dimensional gel electrophoresis (2-DE) followed by MALDI MS/MS and/or LC MS/MS. Proteins that were differentially expressed in response to changes in photoperiods were analyzed by Progenesis SameSpots software. Phosphoproteins and RNA-binding proteins were enriched using immobilized metal affinity chromatography and poly(U) Sepharose columns respectively from SD and LD PP protein extracts and similarly analyzed. In addition to establishing a catalog of PP proteins, we have so far identified nearly sixty-seven proteins that are differentially expressed in response to SD or LD photoperiods. Significantly, the RNA-binding protein, elF-5A, has been detected in multiple isoforms specifically under LD conditions. Numerous other poly(U)-binding proteins which contain RNA recognition motifs have also been isolated and identified. This is the first comprehensive proteomics study that examines and catalogs the proteins that are present in the potato petiole and are potentially differentially regulated by photoperiod. ! ! Table 3: Catalogue of Poly(U) binding proteins enriched from potato petiole extract from plants grown under LD and SD photoperiod. RNA-binding proteins are highlighted in red. Spot # Accession # 1 gi|27883932 MW % coverage MOWSE TAIR Blast (kDa) / # unique Score accession # Epeptide value 69.8 13/6 342 AT1G78900 0 Annotation 2 gi|13235340 69.1 8/5 233 AT1G78900 0 Vacuolar ATP synthase subunit A 3 gi|27883932 68.8 18/6 483 AT1G78900 0 Vacuolar ATP synthase subunit A 4 gi|28261702 55.4 17/6 446 AT2G07698 2E-163 ATP synthase alpha subunit 5 gi|28261702 55.4 11/5 320 AT2G07698 2E-163 ATP synthase alpha subunit Vacuolar ATP synthase subunit A Putative function hydrogen ion transporting ATP synthase activity, rotational mechanism, proton-transporting ATPase activity hydrogen ion transporting ATP synthase activity, rotational mechanism, proton-transporting ATPase activity hydrogen ion transporting ATP synthase activity, rotational mechanism, proton-transporting ATPase activity hydrogen ion transporting ATP synthase activity, rotational mechanism, Poly(U) binding hydrogen ion transporting ATP synthase activity, rotational mechanism, poly(U) binding 6 gi|3850926 52.7 25/6 599 AT5G08680 0 ATP synthase beta chain hydrogen ion transporting ATP synthase activity, rotational mechanism, copper ion binding hydrogen ion transporting ATP synthase activity, rotational mechanism, copper ion binding 7 gi|3850926 52.7 20/7 421 AT5G08680 0 ATP synthase beta chain 8 gi|21633353 52.9 11/5 186 AT5G08680 0 ATP synthase beta chain 9 gi|114421 59.9 32/9 461 AT5G08680 0 10 11 12 13 gi|1762130 gi|15429015 gi|15429015 gi|22094840 63 56 56 36.7 17/7 13/5 7/3 13/5 341 192 217 AT3G13470 AT2G07698 0 AT2G07698 0 AT1G13440 0 ATP synthase beta chain Chaperonin, putative ATPase subunit 1 ATPase subunit 1 Glyceraldehyde-3phosphate dehydrogenase C2 14 gi|22094840 36.7 15/4 197 AT1G13440 0 Glyceraldehyde-3phosphate dehydrogenase C2 Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity 15 No hit 16 gi|225433414 36.8 8/2 152 AT1G04690 1E-178 Oxidoreductase activity 17 gi|73808794 16.2 34/3 451 AT4G09320 2E-69 Potassium channel !subunit Nnucleoside diphosphate kinase 1 hydrogen ion transporting ATP synthase activity, rotational mechanism, copper ion binding Hydrogen ion transporting ATP synthase activity protein binding, ATP binding ATP bnding ATP binding NAD or NADH binding, glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity Nucleoside diphosphate kinase activity, ATP binding 18 gi|537313 40.9 6/2 105 AT5G11420 2E-163 19 gi|255549002 40.2 10/3 175 AT5G11420 2E-178 20 gi|118486160 40.1 7/2 106 AT5G11420 4E-171 21 gi|118486160 40.1 7/2 106 AT5G11420 4E-171 22 gi|118486160 40.1 7/2 141 AT5G11420 4E-171 23 gi|10720247 50.8 6/2 121 AT2G39730 0 Uunknown protein DUF 642 Unknown protein DUF 642 Unknown protein DUF 642 Unknown protein DUF 642 Unknown protein DUF 642 RuBisCO activase 24 gi|10720247 50.8 6/2 121 AT2G39730 0 RuBisCO activase 25 gi|10720247 50.8 6/2 121 AT2G39730 0 RuBisCO activase gi|75338883 62.1 10/5 300 AT1G60900 0 26 gi|4139264 41.8 10/5 123 AT5G09810 0 U2 snRNP auxiliary factor large subunit, putative Actin 7 27 gi|182409985 41.8 25/7 123 AT5G09810 0 Actin 7 28 gi|3328122 50.4 12/5 212 AT1G56190 0 29 gi|475736 52.4 10/5 275 0 30 gi|475736 52.4 10/5 275 0 31 gi|32332101 52.3 8/4 228 0 Phosphoglycerate kinase, putative RuBisCO large subunit RuBisCO large subunit RuBisCO large subunit Molecular function unknown Molecular function unknown Molecular function unknown Molecular function unknown Molecular function unknown ADP binding / ATP binding / enzyme regulator ADP binding / ATP binding / enzyme regulator ADP binding / ATP binding / enzyme regulator RNA binding, nucleotide binding Structural constituent of cytoskeleton Structural constituent of cytoskeleton phosphoglycerate kinase activity 32 gi|6715512 52 10/5 210 AT4G38510 0 Vacuolar ATP synthase subunit B, putative Eukaryotic release factor 1-3 embryo sacdevelopment arrest 9 Glucose -6-phosphate dehydrogenase 6 Hydrogen ion transporting ATP synthase activity 33 gi|82623383 49.1 5/2 230 AT3G26618 0 34 gi|224105607 57 36/6 420 AT4G34200 0 35 gi|585165 58 6/2 108 AT5G40760 0 36 gi|11386828 47.7 10/3 123 AT5G35630 Glutamine synthetase 2 Glutamate-ammonia ligase activity 37 gi|77416937 18 23/3 215 AT4G34412 2.00E62 AT1G06680 3.00E144 AT5G26360 0 Unknown protein molecular_function unknown 38 gi|11134035 28.1 59/5 500 Photosystem II subunit P-1 Chaperonin, putative Poly(U) binding 39 gi|159464215 61 2/2 58 40 gi|19032260 32.9 10/3 106 41 gi|37780996 16.8 18/3 186 42 43 gi|15239049 gi|76160947 39.9 22.1 5/2 57/7 78 300 44 gi|78191462 26.4 12/3 113 AT4G24770 1.00E93 AT2G37270 9.00E81 AT5G11420 0 AT1G48830 7.00E90 AT1G66680 45 gi|313129 67 17/8 215 AT3G52600 0 Translation release factor activity ATP binding Glucose-6-phosphate dehydrogenase activity Unfolded protein binding, protein binding, ATP binding 31-KDa RNA RNA binding, poly(U) binding protein binding Ribosomal protein 5B Structural constituent of ribosome Unknown protein Molecular_function unknown 40S ribosomal Structural constituent of protein S7 ribosome Sadenosylmethioninedependent methyltransferases Arabidopsis thaliana Hydrolase activity cell wall invertase 2 46 gi|24745945 49.5 30/8 192 AT5G60390 0 Elongation factor 1alpha 47 gi|255538240 71.9 13/6 188 AT4G34110 0 Poly (A) binding 2 48 gi|6752880 49 42/11 162 AT5G19990 0 Regulatory particle triple –A ATPase 6A 49 gi|197260357 51.2 27/6 397 AT3G55430 3.00E132 Glycosyl hydrolase family 17 protein Cation binding, hydrolase activity 50 gi|83283995 38.8 54/8 116 77999255 Fructose-bisphosphate aldolase activity 51 gi|77416923 34.6 40/7 453 AT4G13010 3.00E143 52 gi|6689892 63.4 22/6 514 AT3G14310 0 53 gi|24745880 54 55 gi|22326994 gi|62870979 54.5 25.8 2/1 12/3 57 147 AT5G22610 0 AT4G11010 4.00E123 Fructosebisphosphate aldolase, putative Zinc-binding dehydrogenase family protein ATPME3; pectinesterase Regulatory particle triple –A ATPase 4A F-box family protein Nucleoside diphosphate kinase 3 56 gi|38604456 41 9/3 134 AT1G66430 4.00E172 Kinase activity, ribokinase activity 57 gi|38604456 41 10/3 254 AT1G66430 4.00E172 pfkB-type carbohydrate kinase family protein pfkB-type carbohydrate kinase family protein 2.00E179 AT5G43010 0 Calmodulin binding, translation elongation factor activity RNA binding, translation initiation factor activity ATPase activity Oxidoreductase activity Pectinesterase activity ATPase activity Molecular_function unknown ATP binding Kinase activity, ribokinase activity 58 gi|38604456 41 26/7 376 AT1G66430 4.00E172 59 gi|17432522 31.3 39/6 245 60 gi|133246 30.7 14/3 188 61 gi|6015063 24.7 6/2 154 61 gi|15748154 29 6/2 126 AT2G37220 3.00E79 AT4G24770 1.00E83 AT2G18110 3.00E82 AT2G42590 5.00E120 62 gi|12229923 26.1 37/6 152 63 gi|82623423 17.5 23/4 154 64 gi|82623423 17.5 17/3 88 65 Q03878 15.7 23/3 117 66 Q03878 15.7 23/3 102 67 Q03878 15.7 23/3 100 68 Q03878 15.7 23/3 124 69 gi|82623423 17.5 34/3 247 70 gi|82623423 17.5 33/4 181 AT3G14290 2.00E135 AT4G39260 8.00E38 AT4G39260 8.00E38 AT4G39260 1.00E41 AT4G39260 1.00E41 AT4G39260 1.00E41 AT4G39260 1.00E41 AT4G39260 8.00E38 AT4G39260 8.00E38 pfkB-type carbohydrate kinase family protein 29 kDa Ribonucleoprotein 31-KDa RNA binding protein Elongation factor 1beta, putative General regulatory factor 9/14-3-3 protein Threonine-type endopeptidase AtRBP8 AtRBP8 GR-RBP8 GR-RBP8 GR-RBP8 GR-RBP8 GR-RBP8 GR-RBP8 Kinase activity, ribokinase activity RNA binding, poly(U) binding RNA binding / poly(U) binding Translation elongation factor activity Protein phosphorylated amino acid binding Peptidase activity RNA binding / nucleic acid binding / nucleotide binding RNA binding / nucleic acid binding / nucleotide binding RNA binding / nucleic acid binding / nucleotide binding RNA binding / nucleic acid binding / nucleotide binding RNA binding / nucleic acid binding / nucleotide binding RNA binding / nucleic acid binding / nucleotide binding RNA binding / nucleic acid binding / nucleotide binding RNA binding / nucleic acid binding / nucleotide binding 71 gi|224142804 47.1 38/10 411 AT3G13920 0 72 gi|25809054 47.1 34/9 363 AT3G13920 0 73 gi|50400860 47.1 44/12 751 AT3G52880 0 74 gi|6580762 70.8 9/5 219 75 gi|78191424 33.9 34/7 343 76 gi|8247363 35.9 63/13 413 77 gi|2565436 43.6 11/4 122 AT3G22640 1.00E80 AT2G40010 3.00E143 AT1G35720 2.00E122 AT3G27925 0 78 gi|1934994 15.8 18/2 150 79 gi|224102217 21 23/3 80 No hit 81 gi|132071 20.5 18/2 111 82 gi|132080 20.6 19/2 197 ! ! AT2G21660 3.00E34 AT4G14300 2.00E75 AT5G38410 4.00E79 AT5G38410 1.00E78 Eukaryotic translation initiation factor 4A1 Eukaryotic translation initiation factor 4A1 Monodehydroascorba te reductase, putative PAP85; nutrient reservoir 60S acidic ribosomal protein P0 Annexin Arabidopsis 1 DegP protease 1 ATP-dependent helicase activity Heterogeneous nuclear ribonucleoprotein, putative RNA binding, nucleotide binding RuBisCO small subunit RuBisCO small subunit ribulose-bisphosphate carboxylase activity ribulose-bisphosphate carboxylase activity ATP-dependent helicase activity Monodehydroascorbate reductase (NADH) activity Nutrient reservoir activity structural constituent of ribosome ATP binding / Protein homodimerization Serine-type endopeptidase activity Glycine rich protein 7 DNA/ RNA binding