Idiographic Interpolation of Hippocampal fMRI Data Introduction
advertisement

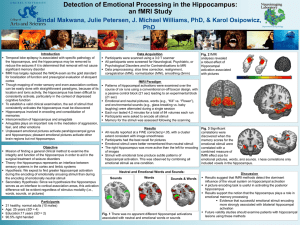
Neuroimaging Laboratory Idiographic Interpolation of Hippocampal fMRI Data Julie Petersen, J. Michael Williams, PhD, & Karol Osipowicz, PhD Department of Psychology, Drexel University Introduction Results from Standard Analysis fMRI activation of the hippocampus is elusive due to the depth of its location in the brain and tonic activity. Traditional approaches to fMRI data processing normalize the data to an MNI atlas, with a resultant resolution of 2mm3. While this approach is good for most brain areas, it does not fully utilize the potential interpolated resolution of fMRI data. Whereas standard preprocessing can produce voxel sizes of 2mm3, the native resolution of Anatomical data can reliably reach .5mm3. This increase in resolution would make finding activation of small, deep, and difficult to image structures, like the hippocampus, far less susceptible to beta-error. All results were reported at FWE corrected p<.05, with a cluster extent consistent with image smoothness. Participants had the best recall for pictures. Emotional stimuli were remembered better than neutral stimuli. The right hippocampus was more active than the left for encoding of images. See Fig. 1. Objective Department of Psychology Secondary Results from Idiographic Interpolation Analysis Fig. 3 Activation of the hippocampus on an individual-subject level Fig. 1 Standard fMRI analyses revealed an effect of posterior hippocampal activation associated with both neutral and emotional pictures In this analysis, we compare the results of two processing schemes: standard and idiographic interpolated. We hypothesize that activation of the hippocampus will be evident in the idiographic interpolated and not in the standard data. Methods 21 healthy, normal adults (10 males, Mage = 25 yrs, SDage = 4) were scanned using a 3.0 T scanner. Patterns of hippocampal activations were examined over the course of six runs. Emotional and neutral pictures, words, and environmental sounds were alternated during a single session. Each run lasted 4.2 minutes for a total of 84 volumes each run. Memory for the stimuli was assessed following the scanning. Data was analyzed using SPM (Flandin & Friston, 2008) and CONN software. Standard Analysis Data preprocessing included slice time correction, realignment, coregistration (MNI) (Schönecker et al., 2009), normalization (MNI) (Rodionov et al., 2009), and smoothing (5mm). Data underwent first and second level modeling Primary Results from Idiographic Interpolation Analysis All results were reported at FWE corrected p<.05, with a cluster extent consistent with image smoothness. Idiographic interpolation analyses revealed bilateral activation of both the anterior and posterior hippocampi for emotional v. neutral pictures (see Fig. 2 and Fig. 3), as opposed to bilateral posterior hippocampal activation present only with emotional and neutral pictures (see Fig. 1). Fig. 2 Group idiographic interpolation fMRI analyses Idiographic Interpolated Analysis revealed an effect of bilateral posterior and anterior Data preprocessing included the standard method of slice time hippocampal activation correction and realignment. Normalization utilized the standard algorithm, but each participant’s T2 data was normalized to his own associated with emotional T1 space, instead of normalizing the data to AAL Atlas. This allowed v. neutral pictures resolution to increase from the standard 2 x 2 x 2 mm per voxel to 0.5 x 0.5 x 1 mm per voxel. The AAL Atlas was then coregistered and normalized to each participant’s T1 data, giving each participant their own atlas. Data within the hippocampus, parahippocampal gyrus, and amygdala were smoothed. Each of these regions then underwent first level modeling, as data for each region was extracted from the functional data and modeled across time for each event. A sample-specific template was generated. Second level comparisons were made according to the standard method. Discussion Of our 21 participants, 6 participants were excluded from the idiographic interpolation analyses due to problematic data. Similar analyses could be run for these participants, but would require subject-specific drawing of ROIs. The idiographic interpolation analyses did not rely on any anatomical determination, but instead was purely atlas driven. The bilateral hippocampal activation of all 15 participants indicate this analytical technique as a great method for clinical hippocampal imaging. Conclusion Our results provide some support for the technique, but group level comparisons with this data are problematic, thus further research is necessary to establish the utility of this technique. References Flandin, G., & Friston, K. J. (2008). Statistical parametric mapping (SPM). Scholarpedia, 3(4), 6232. Rodionov, R., Chupin, M., Williams, E., Hammers, A., Kesavadas, C., & Lemieux, L. (2009). Evaluation of atlas-based segmentation of hippocampi in healthy humans. Magnetic resonance imaging, 27(8), 1104-1109. Schönecker, T., Kupsch, A., Kühn, A., Schneider, G.-H., & Hoffmann, K.-T. (2009). Automated Optimization of Subcortical Cerebral MR Imaging− Atlas Coregistration for Improved Postoperative Electrode Localization in Deep Brain Stimulation. American Journal of Neuroradiology, 30(10), 1914-1921.