Redacted for Privacy Lie-Fen Lin for the degree of Doctor of
advertisement

AN ABSTRACT OF THE THESIS OF
Lie-Fen Lin for the degree of Doctor of Philoso1Dh' in
Statistics presented on March 17, 1992.
Title: Uses of Bayesian Posterior Modes in Solving Complex
Estimation Problems in Statistics
Redacted for Privacy
r ,--7,1
Abstract approved:
msey
Fred L.
means are commonly used to
In Bayesian analysis,
summarize Bayesian posterior distributions.
a
number
large
of parameters
Problems with
often require numerical
In this
integrations over many dimensions to obtain means.
dissertation, posterior modes with respect to appropriate
are
measures
used
to
summarize
Bayesian
posterior
distributions, using the Newton-Raphson method to locate
modes.
Further inference of modes relies on the normal
approximation,
using
asymptotic
multivariate
normal
distributions to approximate posterior distributions. These
techniques
are
applied
to
two
statistical
estimation
problems.
First, Bayesian sequential dose selection procedures
are developed for Bioassay problems using Ramsey's prior
[28].
Two adaptive designs for Bayesian sequential dose
selection and estimation of the potency curve are given.
The relative efficiency is used to compare the adaptive
methods with other non-Bayesian methods (Spearman-Karber,
up-and-down, and Robbins-Monro) for estimating the ED50
.
Second, posterior distributions of the order of an
autoregressive (AR) model are determined following Robb's
method (1980).
Wolfer's sunspot data is used as an example
to compare the estimating results with FPE, AIC, BIC, and
CIC
Both
methods.
approximation
for
posterior results.
Robb's
estimation
method
of
the
and
order
the
have
normal
full
Uses of Bayesian Posterior Modes in Solving Complex
Estimation Problems in Statistics
by
Lie-Fen Lin
A THESIS
submitted to
Oregon State University
in partial fulfillment of
the requirements for the
degree of
Doctor of Philosophy
Completed March 17, 1992
Commencement June 1992
APPROVED:
it
z
Redacted tor Privacy
Professor of Statist'
in charg
ajor
Redacted for Privacy
Head of Department of Statisti
Redacted for Privacy
Dean of Graduate/ school
Date thesis is presented
Typed by Pao-Pao Liu for
March 17. 1992
Lie-Fen Lin
ACKNOWLEDGEMENTS
I would like to express my most sincere gratitude to
Dr. Fred L. Ramsey, my major professor and thesis advisor,
for his guidance, encouragement and patience during the
course of this work.
His direct contributions helped in the
completion of the thesis.
I also want to thank Dan Brunk,
who provided help with endless patience in the beginning of
doing this thesis.
During my Ph.D. study, many of my friends helped me in
different ways.
I especially wish to express my special
thanks to Mr. So's family, they took care of my son and
loved him so much during my research.
I would like to thank
Jane, who have helped me more than she knows.
Financial support from the department is gratefully
acknowledged.
I am also indebted to my husband, Pao-Pao, my children
Eddy and Sandra, for giving me warm and bearing up under the
strain.
Finally,
parents.
Their
I want to dedicate this work to my
love,
support
and understanding have
sustained me through all these years.
TABLE OF CONTENTS
1
2
3
4
INTRODUCTION
1.1 Introduction
1.2 The Normal Approximation to a Posterior
Distribution
1.2.1 Introduction of the Normal
Approximation
1.2.2 Bayesian Normal Approximation
1.2.3 Examples
1.3
Literature Review of the Bayesian Bioassay
1.4 Review of Autoregressive (AR) Time Series
1.5 Organization of the Dissertation
MOST PROBABLE VALUES (MPV) ESTIMATORS OF THE
POTENCIES IN BAYESIAN BIOASSAY
2.1 Model of Bayesian Bioassay
2.2 Modes of the Posterior Distribution
2.3
Point Estimator of an Effective Dose
2.4
Bayesian MPV Estimators Inference
2.4.1 Prior and Posterior Distributions of
an Effective Dose
2.4.2 Normal Approximation Method for the
Bayesian MPV Estimators Inference
ADAPTIVE DESIGNS FOR ESTIMATING THE POTENCY
CURVE
3.1 Adaptive Designs
3.2 Designs for the Adaptive Methods
3.3 One-step (Non-adaptive) Method
3.3.1 Comparisons of Non-adaptive Method
with Adaptive Methods
3.4
Sperman-nrber (Non-parametric) Method
3.4.1 Comparisons of Spearman - Karber Method
with Adaptive Methods
3.5 Up-and-down (Staircase) Method
3.5.1 Comparisons of Up-and-down Method
with Adaptive Methods
3.6 Robbins-Monro Process
3.6.1 Comparisons of Robbins-Monro Process
with Adaptive Methods
3.7
Comparisons
NORMAL APPROXIMATION METHOD TO ESTIMATE THE
ORDER OF THE AUTOREGRESSIVE (AR) MODEL UNDER
BAYESIAN POINT OF VIEW
4.1 The Bayesian Approach to Order Estimation
of AR Process
4.2 Normal Approximation Approach
4.3
Examples for Wolfer's Sunspot Data
1
1
3
3
4
8
12
15
20
21
21
24
28
30
30
31
34
34
45
47
50
50
52
52
56
56
61
61
70
70
73
78
5
CONCLUSIONS
85
BIBLIOGRAPHY
88
APPENDIX
92
LIST OF FIGURES
Fi
1.1
3.1
3.2
3.3
Page
The density functions of Beta(3, 11), N(3/14,
.03665), N(2/12, .01736), and N(3/14, .0098)
39
Determination of x2,1 for s1
step
40
1 =1
in the first
Determination of x30 for s1,1=0, s2,1=0 in the
42
Determination of x3,1 for s1,1=1, s2,1=0 in the
second step
3.6
41
Determination of x3,1 for s1,1=0, s2,1=1 in the
second step
3.5
6
Determination of x2,1 for s1,1=0 in the first
step
second step
3.4
...
42
Determination of x3,1 for s1,1=1, s2,1=1 in the
second step
43
3.7
The r.e. of Anm/A16, m=1,2,3, L=6
48
3.8
The r.e. of Bnm/B16, m=1,2,3, L=6
48
3.9
The r.e. of Anm/A112, m=1,2,3,4,6, L=12
49
3.10 The r.e. of Bnm/B112, m=1,2,3,4,6, L=12
49
3.11 The r.e. of NOnm/N016, m=1,2,3, L=6
51
3.12 The r.e. of B61/N061, A32/N016, A23/N016
51
3.13 The mse of SKNn, n=2,3,6
53
3.14 The r.e. of B61/SKN6, A32/SKN3, A23/SKN2
53
3.15 The mse of UDq, q=001, 035, 075
55
3.16 The r.e. of B61/UD035, B61/UD075
55
3.17 The r.e. of A32/UD035, A32/UD075
57
3.18 The r.e. of A23/UD035, A23/UD075
57
3.19 The mse of RM1Cc, c=.001, .2, .408
59
3.20 The mse of RM2Cc, c=.001, .25,
3.21 The mse of RM3Cc, c=.001,
.333,
3.22 The mse of RM6Cc, c=.001,
.5, 1
.5454
59
.667
60
60
3.23 The r.e. of B61/RM6Cc, c=.5, 1
62
3.24 The r.e. of A32/RM3Cc, c=.333, .667
62
3.25 The r.e. of A23/RM2Cc, c=.25, .5454
63
3.26 The r.e. of B61/RM1C2, Anm/RM6C10, Anm/RM1C4,
m=2,3, L=6
63
3.27 The r.e. of B61/N016
65
3.28 The r.e. of N061/N016
65
3.29 The r.e. of SKN6/N016
65
3.30 The r.e. of UD035/N016
65
3.31 The r.e. of RM1C2/N016
65
3.32 The r.e. of XX/N016, XX=B61, N061, SKN6, UD035,
RM1C2
66
3.33 The r.e. of Anm/N016, m=2,3, L=6
67
3.34 The r.e. of SKN2/N016
67
3.35 The r.e. of RM6C10/N016, RM1C4/N016
67
3.36 The r.e. of UD075/N016
67
3.37 The r.e. of XX/N016, XX=A32, A23, SKN2, UD075,
RM6C10, RM1C4
68
4.1
Mean corrected of the square root of yearly
averages of sunspots data 1749-1977
84
LIST OF TABLES
Table
2.1
Page
The values of (g1(r(Py ,RP2)), g2(go1)
and (H1, H2) for n1 =n2=1 and fi=1,2
2.2
god ))
29
The values of (g1 (rpo,t(P2)), g2 (rpo,r(p2)))
and (H1, H2) for n1 =n2=2 and 13=1,2
29
The estimated ED50 from means and modes for
n =n 2=2 and /3=1 and 2
31
3.1
The best designs for the adaptive methods (L=6)
47
3.2
The best designs for the adaptive methods (L=12)
47
3.3
The values of c for ni=6, 3, 2, and 1 such that
all the tested doses and ED50 are in [0,1]
58
The results of analyzing Wolfer's sunspot data
for 1749-1924 using FPE, AIC, BIC, CIC, Robb,
and normal approximation method for the order
p
81
The results of analyzing Wolfer's sunspot data
for 1749-1924 from the closed Newton-Cotes (N=3)
and Bayesian normal approximation method for
choosing the maximum order M=12
82
The results of analyzing Wolfer's sunspot data
for 1749-1977 using FPE, AIC, BIC, CIC, and
normal approximation method for the order p
assumed the maximum order M=15
83
2.3
1
4.1
4.2
4.3
Uses of Bayesian Posterior Modes in Solving Complex
Estimation Problems in Statistics
1 INTRODUCTION
1.1 Introduction
In Bayesian analysis,
means are commonly used to
summarize Bayesian posterior distributions.
For problems
with a large numbers of parameters often require numerical
integrations over many dimensions to obtain means.
In this
dissertation, posterior modes with respect to appropriate
measures
are
used
to
summarize
Bayesian
posterior
distributions, using the Newton-Raphson method to locate
Further
modes.
approximation,
inference
using
of modes
asymptotic
relies
on
multivariate
normal
normal
distributions to approximate posterior distributions. These
techniques are applied to two statistical problems.
These
are the sequential dose selection in bioassay and the
selection of the order of an autoregressive (AR) time series
process.
In
a
standard bioassay problem,
the experimenter
attempts to test the potency of a stimulus administered at
different levels to different subjects. He chooses M dosage
levels,
xl
and treats nl
levels, respectively.
subjects at these
Each subject has a response to a
given dose of drug, either positive (response) or negative
(no response).
The experimenter observes the number of
2
positive responses and records them as si,...,sm.
It is
assumed that each subject has a threshold which the given
dose must equal or exceed to produce a positive response.
However, this threshold may vary from one subject to the
next, and so is treated as a random variable with unknown
distribution
P
P.
is
often
distribution or potency curve.
called
the
tolerance
P(x) is the potency of level
x of the stimulus; that is, the tolerance distribution is
defined by the probability P(x)
of getting a positive
response to a dosage at level x for all x.
The drug dosage
levels may be the actual dosage levels or the logarithms of
these levels.
For Bayesian bioassay, Ramsey (1972) employed Ramsey
prior
and
[28]
posterior modes
developed methods
of
for
estimating
the posterior density
estimate the potencies, Pi's.
the
function to
Thus, potency curve can be
observed by linear interpolation.
Two adaptive designs for
sequentially selecting dose from the estimated potency curve
are developed using posterior modes to estimate the potency
curve in Chapter 3.
Robb
(1980)
derived the marginal posterior density
function of the order of AR model for estimating the order,
which requires numerical integrations over many dimensions
to obtain the order.
The normal approximation,
using
multivariate normal density function, will be used to get
around this problem in Chapter 4.
3
1.2 The Normal Approximation to a Posterior Distribution
1.2.1 Introduction of the Normal Approximation
Let y =(y1,
f(yi 8).
..,yn) be a random sample from a distribution
When the distribution obeys certain regularity
conditions, the likelihood function of 0 is approximately
normal and remains approximately normal under mild one-toone transformations of 0 for sufficiently large n (Johnson
1967, 1970).
In this case, the logarithm of the likelihood
is approximately quadratic, i.e.,
L(81Y) "
(emlY)
(0
a211 )
Om) 2
ae 2
le
where Om is the maximum likelihood estimator (mle) of 0.
In
general, the quantity
1.82L)L.
n a82 mm
(1.2)
is a positive function of
y.
The logarithm of a normal
density function r(x) is of the form
logr (x)
given
and,
=constant
the
1 (x- 2 ) 2
2
location
parameter
determined by its standard deviation a.
and
(1.3)
shows
that
(1.3)
a
the
standard
A,
is
completely
Comparison of (1.1)
deviation
of
the
likelihood curve is approximately equal to
n
1
a2LI-i
n (302
-m
(1.4)
The likelihood function of 0
is approximately a normal
density with mean em and variance
4
VM =
(-
)
a2L1
ae2
le
(1.5)
The approximated distribution of 0 can thus be written as
N(014,
-14
)
1 le)
(1.6)
1.2.2 Bayesian Normal Approximation
In Bayesian analysis, deriving the means and variances
(or covariances)
from the posterior distribution of the
parameters is often problematic.
In this dissertation, a
normal approximation to the posterior distribution, similar
to that just displayed for the likelihood function, is
developed and illustrated with examples.
Example 1.1
Let X be Binomial(10, p).
distribution is Beta(2, 2).
The prior
Thus the posterior distribution
is Beta(x+2, n+2-x) with density
(pix)
pl+X (1 .1)) n+1-x
w. r. t. dv
where dv is a Lebesgue measure.
(1.7)
Suppose x=1; the posterior
distribution is Beta(3, 11) with mean=3/14, variance=.01123,
and mode of this density is 2/12.
Use an asymptotic normal
distribution to approximate the posterior
distribution,
which is
P
N(P*,
aa loge
IP*)
ape
If p*=mean, p ~N(3/14, .03655).
.01736).
(1.8)
If p*=mode, p ~N(2/12,
The approximate variance evaluated at the mean is
5
much larger than the true posterior variance comparing with
that at the mode.
For a skewed distribution the second
derivative at the mode is more meaningful than the mean
because the second derivative at the mean is sometimes small
or close to zero such that the variance is overestimated.
Therefore
using
modes
meaningful
is
for
a
skewed
distribution.
In the next example, the posterior density with respect
to other measure will be displayed to see how the normal
approximation will be changed.
Example 1.2
(continuation of example 1.1)
The posterior density function of Beta(x+2, n+2-x) can
be written as
« p2+X (i_p) n+2-x
(pix)
dv
p(1-p)
w.r.t. dv-
where dv° is an improper measure.
(1.9)
The modes of above
density is 3/14, which is the same as the posterior mean,
and
the
approximate
variance
is
Posterior
.0098.
distribution of p can be approximated by N(3/14, .0098) when
x=1.
A plot of the posterior distribution Beta(3,11) and
above three normal distributions are shown in Fig. 1.1.
From Fig.
1.1 we can see which normal distribution is
appropriate
to
use
to
approximate
the
posterior
distribution.
As in example 1.1 and example 1.2,
the posterior
distribution is appropriate to be approximated by the normal
distribution with mean, which is the mode of the density
6
Fig. 1.1 The density functions of Beta(3, 11),
N(3/14, .03655), N(2/12, .01736), and
N(3/14, .0098)
N(3/14, .0098)
N(3/14, .03668)
01736)
-0.8
-0.4
0.4
0
P
8.8
1.2
7
with respect to dv°, and variance evaluated at the mode.
The posterior density function is defined with respect to
some measures specific to the problem under consideration.
The
posterior
distribution
the
of
parameter
then
is
approximated by the multivariate normal distribution.
The
approximation procedures for the one-parameter case and the
multi-parameter case are given below.
(A) One-parameter case
(1) Determine posterior density h(0) with respect to an
improper prior;
(2) Find the MODE, Om, of h(0);
(3) Evaluate
(7 11
=
[
(32
(2logh(e)
lo.
(1.10)
(4) Treat the posterior distribution of 0 as
Mem, 4)
.
(B) Multi-parameter case
(01,...,051
(1) Obtain the posterior density h(81,
,05) with respect
to an improper prior;
(2) Solve
alogh(0)
as;
(j=1,...,$)
(1.12)
for the MODE, Om, by the Newton-Raphson method;
(3) Determine
Vom
a2logh (0)
1
le.
,
(i, j=1,
.
.
.
,
s)
;
(1.13)
8
(4) Treat the posterior distribution of 0 as
0 - AARV(0m, Vilm)
This
normal
.
(1.14)
approximation
approach
will
be
used
to
approximate the posterior distribution of the potencies in
Chapter 2 and used to get around the multiple summations
problem in Chapter 4.
There are more examples in next section to illustrate
the normal approximation method using modes (with respect to
a particular measure) as means of normal distributions.
1.2.3 Examples
The means of the posterior distributions and the modes
of the posterior density functions with respect to improper
measures for some well known distributions are derived in
the following examples.
(A) One parameter case
Example 1.3 Bernoulli.
Let X1,
Xn be i.i.d.
Bernoulli B(p), and S be the sum of the Xi's.
prior distribution of p is Beta(a, /9) for 0<p<1.
Assume the
The prior
density function is written as
=
r(a) r(a)
10
+ (3)
dP
w. r . t. dvo
(1.15)
P(1
The posterior distribution is Beta(a+S, n +9 -S) with
(P)
S+a
n+a
Tfar(p)
(S+a)(n+P-S)
(n+a+P)2(n+a+P+1)
The mode and variance with respect to dv° are
(1.16)
9
S +cc
n +a +13
, Var (I)) 11«p)
Example 1.4
(S-712)+0;+
(n+pP) 3-.9)
Poisson.
(1.17)
Xn be i.i.d.
Let X1,
Poisson(0) and S be the sum of the Xi's.
Assume the prior
distribution of 0 is Gamma(a, fl) for 0>0.
The prior density
function is
r (a)
-112-0aeAni
71(0)
(a>0 ,P>0)
w.r.t. dvo = de
(1.18)
and the posterior distribution is Gamma(a+S, n+fl) with
nn+P)2
r(e) =
var(e)
+P
'94-c4
(1.19)
(
The mode of the density with respect to dv° and the variance
are the same as (1.19).
Example 1.5
Exponential.
Exp(A) and S be the sum of the Xi's.
distribution of A is Gamma(a,
Xn be i.i.d.
Let X1,
fl)
distribution is Gamma(a+n, a+0).
Assume the prior
for A>0.
The posterior
The mean equals the mode
of the density and the variance of the distribution equals
the approximated variance.
Example 1.6
i.i.d. N(0,
a2)
Normal with known a2.
Let X1,
and consider the estimation of 0.
Xn be
If we
assume the prior distribution to be N(40, a02), the posterior
distribution is
N
a
h
O22
2
n
a2 2
1
o
1
2
n
1
02
012,
(1.20)
10
Since the posterior is a symmetric distribution, the mean
equals the mode and the variance equals the approximate
variance.
Example 1.7
Normal with known A.
Let X1,
i.i.d. N(A, 0) and consider the estimation of 0.
Xn be
Assume the
prior density of u0o02/0 is x2 with uo degrees of freedom.
The posterior density of (
of freedom,
croz+sz
=E
where S2
say u1,
)/0 is x2 with uo+n degrees
2.
The posterior
distribution of 0 has
s2+1)
g'(0)
°
02
°, var(e)
2
v11 -2
(s2+vout) 2
(u1-2)2(u1-4)
(1.21)
The prior density function is
2
-uo
-U0a0
it
(0)
ec e-2-ir 0 2
w.r.t. dvo =
e
(1.22)
Therefore, the mode of the density with respect to dv° and
variance are
s2
02
M (e) =
°
,
Var (0) lice)
2
(S2+13°°)
3
(1.23)
Example 1.8
Let X V ...,
Pareto.
Pareto(0) for X1M0 and X0=1.
ln(0)
is Uniform(-02,
co).
Xn be i.i.d.
Assume the prior density of
The posterior density of 0 is
Gamma(n, n*ln(z)), where z is the geometric mean of (X1,
Xn); that is z =(11 Xe. The mean and variance of the
posterior distribution of 0 are
g()) -
1
1n(z)
Var(0)
'
1
n(ln(z) )2
(1.24)
The prior density function of 0 is proportional to one with
11
respect to d0=d0/0.
The posterior density with respect to
dv° has mode and variance
M(0)
1
1
Var (0) Im(e)
in ( z)
n(ln(z) )2
(1.25)
(B) Multi-parameter case
Example 1.9 Normal.
Let X1,
Xn be i.i.d. N(01,02),
with both parameters 01 and 82 unknown.
Assume the prior
distributions of 01 and ln(02) are independent and both are
Uniform(-00, co).
The posterior distribution of 01 is such
that
01
)
(1.26)
has a t-distribution with n-1 degrees of freedom, say v,
where
S2
=E
(Xi-Y) 2
1
(1.27)
The posterior distribution of uS2/02 is x2 with u degrees of
freedom.
The posterior means and variances of 01 and 82 are
Var (00
r(01) =
r(e,)
=
Var co o
us2
u-2
u
S2
u -2 u+1
2
(u52) 2
(u-2)2(u-4)
(1.28)
The covariance matrix of (0 11 0 2 ) is
S2
u -2 v+1
0
0
2 (uS2) 2
(u-2)2 (u-4)
(1.29)
12
The joint prior density function of (01,02) is proportional
to one with respect to dva=d01d02/02.
The joint mode of the
posterior w.r.t durl and the approximate covariance matrix
are
uS2
m(e 92) =
us21
,
cov(el, e2) =
0
(u +1) 2
0
(uS2) 2
(U4-1)3.
2
(1.30)
Modes are easier to derive than means.
When the number
of the observations n is large the means and the variances
(or covariances) of the distributions are either close or
equal to the modes and the approximate variances
(or
covariances).
1.3 Literature Review of the Bayesian Bioassay
Recently, the Bayesian non-parametric approach has been
used to estimate the tolerance distribution in the quantal
bioassay.
Ayer, et al.
(1955), proposed an estimate of P
(potency curve) based on Bernoulli data.
Brunk (1970),
Barlow, et al. (1972), and Robertson, et al. (1988) used the
estimate of the isotonic regression to estimate the potency
curve P.
The isotonic regression required the function, P,
to be monotonic where the estimators
densities
of non-decreasing
were required to satisfy order restrictions. A
Bayesian approach to estimating P was first proposed by
Kraft and Van Eeden (1964).
The prior distribution of P is
13
the Dirichlet distribution.
The main properties of the
Dirichlet distribution are discussed by Wilks (1962) and
Johnson and Kotz (1972).
Ramsey
(1972)
developed methods for computing the
posterior mode of the joint density function.
modes as estimates of the potencies.
can be
observed by
linear
He used these
Thus the potency curve
interpolation.
The
prior
distribution employed is now known as the "Ramsey's prior"
[28] and is similar to the Dirichlet process prior developed
by Ferguson (1973).
Using Ramsey's estimate, which is a
smoothed version of the isotonic regression estimator, one
may estimate the potency curve at any effective dose q (EDq)
(the effective dose which will cause q percentage of getting
a positive response).
Antoniak (1974) showed that the posterior distribution
of P is a mixture of Dirichlet process distributions and
derived the Bayes estimator of P for two dosage levels by
using the squared error loss function, where the Bayes
estimators are the means of the posterior distribution of P.
Unfortunately, analysis of this mixture of the Dirichlet
process distributions becomes increasingly intractable when
the number of stimulus levels increases.
computational
estimators,
difficulty
in
evaluating
To simplify the
these
Bayes
Kuo (1988) used linear Bayes estimators to
estimate the potency curve.
The disadvantage of this method
is that P may be non-monotonic.
14
Disch (1981) derived the marginal posterior density of
P(xk), k=1,2,..,M, and P(x) , where xk is the observational
dose and x is the non-observational dose.
He computed the
means (Bayes estimators) of the posterior distribution of P
from the marginal posterior densities for any number of
dosage levels.
Disch also derived the prior and posterior
distributions of any effective dose assuming Ramsey's prior
for the potency curve.
He pointed out that, if there are
too many experimental dose levels or too many observations
per dose level, then the computation for the posterior
cumulative distribution function (c.d.f.) of EDq becomes
numerically unmanageable.
As
noted
previously,
the
multivariate
normal
distribution will be used to approximate the posterior
distribution of
P.
Therefore,
the marginal posterior
distribution can be approximated by the normal distribution
and the posterior c.d.f. of EDq can be estimated.
The modes
of the posterior distribution of P will be used as the
estimators to estimate the potency curve (Ramsey (1972)).
The modes of the posterior will converge to the results of
mle's.
Ramsey showed in an example in that an optimal design
one experimental unit is assigned per fix dose level.
In
this situation, using Ramsey's method, we can still estimate
the potency curve.
Kuo (1983) used the Dirichlet process
prior, in which parameters are distributed uniformly over
15
[0,1], and the squared error loss function to minimize the
risk function, to derive the optimal design.
design,
In the optimal
the doses were not uniformly spaced,
but were
shifted somewhat toward the prior estimated of the ED50 for
two dosage levels.
In
this
All optimal designs are dependent on p1.
dissertation,
two
adaptive
sequentially choosing design doses
potency curves are developed.
designs
for
and estimating the
Both are sequential methods
using Ramsey's prior and use previous step information in
choosing the new dose level in each step.
1.4 Review of Autoregressive (AR) Time Series
Denote by (Xt)
order p.
an autoregressive process of finite
In general, an AR process of order p is denoted as
an AR(p) process which is given by
e(B)xt = wt
,t=1,2,...
(1.31)
where the wt are white noise with mean zero and finite
variance a2.
The stationary conditions for the Oi are very
complicated.
One way to get around the difficulty of
insuring stationarity with respect to the conditions given
above is to reparameterize the model in terms of the partial
autocorrelations.
If we let
denote the kth partial
autocorrelation, i.e. the conditional correlation between Xt
1 p is the prior sample size; it can be interpreted as
measure the strength of the belief in the prior guess. If
p is large compared to the experiment sample size, little
weight is given to observations. If (3 is small compared to
the experiment sample size, little weight is given to the
prior guess.
16
and Xt+k given the intervening X's,
Xt+i, ..
. , Xt+k_1 ,
then an
AR(p) process has Ivkl <1 for all k5p and ck=0 for all k>p
(Ramsey (1974)).
Barndorff-Nielsen and Schou (1973) showed
that there is a one-to-one mapping from
(ci,
(01,...,0p)
to
... , 9p) , and the stationarity conditions in terms of the
Bp's are simply shown as -1<91:<1 for k=1,2,...,p.
denote the kth element of 0 in an AR(p) model.
Let 0 N
k
That is, one
may solve for the O's strictly in terms of p's, i.e.,
k=1, 2, ...,p
j=1, ...,k-1
ok,k = (Pk
0k,j = ek-1,j
(Pkek-1,k-j
.
(1.32)
Suppose there are n observations Xi,X2,...,Xn from an
AR(p) process, then 0 (B) Xt=wt, t=1,2,...,n.
Since wi,...,wn
are i.i.d. N(0,02), the joint density of the n observations
is
n
1
fp (ini 6p, 02) = (27102) 2 IMpl 2 eXp{-Hp(e) /202}
(1.33)
where
01(1:0,...,0p,p) 1
14p={
TI
;4 ll a2
(i,j=1,2, ...,p)
(k>0)
Yk= r( XtXt+k )
Hp (0p) =0 11,pDp01,p
°LP
DP =
(1' ePS"
' °NI)
I
d11
-d12
-d13
-die
d22
d23
-d1,p+1 d2,p+1 d3,p+1
(1.34)
-
d2,
+1
dp+1p+1.,
17
n-i-j+1
E Xj.kXj+k
di, = di, =
k=0
the
In
above
equation,
"prime"
means
"transpose".
Expressing
1iip1
1
2
p
=H
_.(14) 2
(1.35)
in terms of T's, we can rewrite (1.33) as
_n
p
i
2
[
ji
(1-91) 2
(27,2) 9p. 02) = (27102)
4
*
expf- 1(1)(9P) 1
202
(1.36)
where
Hp(Op) = Kp(ipp)
(1.37)
Let L be the log likelihood function of (1.33) so that
L =
Hp (Op)
lna2 +
In Ihrpl
202
(1.38)
The role's for a2,0p.1,...,Opip are obtained
by solving the
following equations:
n
aL =
30
aL
a
+
Hp(9) -0
(1.39)
a3
-
p,p dp+1,j+1}
aep.i
= 0
(j=1,2,
,P)
(1.40)
where
M
2
lnIMpll
ae,j
Equation (1.39) yields the role for a 2 ,
(1.41)
18
B2 _ H(0*)
n
Unfortunately,
(1.42)
the equations of
are not easily
(1.40)
solved, since the Mj are very complex functions of the B's.
These mle's will be discussed further in Chapter 4.
One approximation to the exact role's of the B's results
from ignoring the term L,
dominates inl m I
involving
I MpI
,
because Hp(0)
for sufficiently large samples (Box and
Jenkins (1970)).
Then,
o*p = (Dp) -1 dp
where dp= (d12,d13,.,d1,p0 )
(1.43)
',Dp
first row and first column.
i
is
the same as Dp without the
These are generally called the
least squares estimates of Bp.
The primary interest here is
to estimate the order p of the autoregressive model.
Recently, increasing use has been made of goodness-of-
fit criteria, which depend on the role of the variance a2,
say,
n
E rte (P)
R
2-1
(1.44)
n
where r t (p) are the estimated residuals from a model with p
parameters
estimated.
Akaike
(1969,
1970
and
1974)
advocated a decision theoretic approach and used the future
prediction error (FPE) for the model of order p,
FPE(p)
(n+P+1)R
n -p 1 P
0,=1,2, ...,A6,
and Akaike's information criterion (AIC),
(1.45)
19
AIC(p) =
2P
(1.46)
where we may simply choose the model order p as the value
that minimizes FPE(p) or AIC(p).
Modifications to AIC(p)
have been suggested to improve the large sample performance,
since Shibata (1976) showed that we do not get a consistent
estimator for the order using AIC(p).
These suggestions
relate to replacing 2p/n in (1.46) by pn-11n(n)
1978; Schwarz 1978) or by 2pn-11n(ln(n))
(Rissenan
(Hannan and Quinn
1979), which will yield consistent estimators for order.
simulation study of Lutkepohl
A
(1985) has shown that for
multivariate autoregressions, the modification of Schwarz
(1978), sometimes called the Bayesian information criteria
(BIC),
BIC(p) = In Rp +
pinn
n
(1.47)
leads most often to correct estimates for model order and
has the smallest mean squared prediction error.
Robb (1980) also used a Bayesian approach to order
estimation of autoregressive time series.
The marginal
posterior probability distribution of the order, given the
data,
is
obtained.
The value with maximum posterior
probability is the Bayes estimate of the order with respect
to a particular loss function.
The form of the density
function of the order, given the data, is very complicated.
As noted previously, the normal approximation method is used
here to simplify complexity.
20
1.5
Organization of the Dissertation
In Chapter 2, the modes (most probable values (MPV)
estimators) of the posterior distribution, and the point
estimators
and
their
associated
normal
approximation
inferences regarding an effective dose in Bayesian bioassay
are presented.
In Chapter 3,
two adaptive methods for
sequentially choosing fixed doses and estimating the potency
curves are developed and comparisons with non-adaptive, up-
and-down, Robbins-Monro, and Spearman-Kdrber methods for
estimating the ED50 are given.
In Chapter 4, the marginal
posterior
of
density
autoregressive time
function
series
is
the
order
discussed.
approximation method applied to
The
of
the
normal
simplify the multiple
integrations required to compute their densities is given.
In Chapter
discussed.
5,
the conclusions of the dissertation are
21
MOST PROBABLE VALUES (MPV) ESTIMATORS
2
OF THE POTENCIES IN BAYESIAN BIOASSAY
2.1 Model of Bayesian Bioassay
The function P(x)
function in x.
is assumed to be an increasing
For observational doses xi
it is assumed that x1<x2<...<xm.
Pi.
M)
The random variable si has
a binomial distribution with parameters
assume that si,
(i=1,
(ni,
P(x1)).
We
sm are independent and denote P(x1) as
The joint likelihood for Pl, ..., Pm is
L (.91,
.
.
.
,
Stet
I
P1,
.
.
. ,
Pm) ac 1-1 Pis' (1-Pi)
.
(2.1)
1-1
Prior Distribution
The prior chosen is Ramsey's prior [28] (Ramsey (1972))
which is
a Dirichlet process prior with parameter alp
(i=1,2,..., M+1) (see also Ferguson (1973)).
Let a1 be non-
negative constants and the summation of a1 (1= 1,2,..., M+1)
be unity.
Let the function Q(x) be a prior guess at the
unknown potency curve P(x) and denote Q(x1) as Q1.
We can
select
al =
ai = Qi
Qi-1
am.1 = 1
QM
(i=2, 3,
, )4
(2.2)
For any observational doses xi<...<xm, the successive
differences
in
potency
have
an
ordered
Dirichlet
distribution with the density functions r and r° with
22
respect to the Lebesgue measure,
dv,
and an improper
measure, dv°, respectively, where
M +1
fl
i=1
-
Pi -1)
(Pi
ads
-1
w. r. t. dv
(2.3A)
M +1
(Pi
Pi_1) ash
i=1
w.r.t. dvQ
(2.3B)
where
dv = n (dPi)
(2.4A)
dv
dvQ
n (Pi
Pi -1)
and O<P7 <...<Pm5_1.
(2.4B)
Note that P0=Q0=0 and Pm+1=Qm+1=1.
For any non-observational dose x between xk and xk+i, we
assume an ordered Dirichlet prior over (P1,
Pk+1,
..., Pm) with parameters (91, ..., 9, 9*,
9M+1)
and 0.5_1D15_....PkP(x).5_Pk+1...Pm1 where
Pk/ P(x)
9k+1,
I
Vi = PQ1 = alP
Qi_1) =
Bpi = P (Qi
= 13 (1
(Pm+1
QM)
q)* = 13 [Q(x)
= P [Qk+3.
(p* + 0** =
E
aiP
(1=2, ...,k, k +2, ..
A)
= am+113
Qk]
Q(x) ]
ak+iP
=P
(2.5)
Then the prior is proportional to
M+1
i-1,i*k+1
(pi -Pi -1)
[P(x)
-pk],.-1 [pk+1 -p(2) ,9-1
(2.6)
23
Note that P0=0 and P14+1=1.
The marginal distribution of P(x)
is a Beta distribution,
P(x)
Q(x) *$, (l-Q(x)) *# ).
Thus, prior mode = prior mean = Q(x).
The constant
is
non-negative and can be used to specify the degree of
smoothing in the posterior estimate.
For #=0 and #-00, the
posterior estimators are the isotonic regressor and Q,
respectively.
Also, fl can be interpreted as a measure of
the strength of belief in the prior guess Q.
Posterior Distribution
The joint density of the posterior distribution for the
observational doses is proportional to
M+1
[ H Pisi
w.r. t .
[
i=1
dv
(2.7A)
or
M+1
(1-Pi)
[
i=1
[
H
aiP]
i=1
w.
r. t. dvo
(2.7B)
If there is a dose x between xk and xk+i, the joint density
of the posterior distribution is proportional to
[P(x)
-4] 9-1
M+1
[pk+i-P(x)
9---1 H
(P1 -P2_1) 9i
-1}
i=1,i*k+1
M
1=1
or
PiSi
w.
r . t . dP(x)ildPi
(2.8A)
24
M+1
{[P(X)Pk]*.[Pic+1-1)(A7) ] il
II
(Pi P1-1)14)1}
1=1,1*k+1
* TIPP
i=1
II dPi dP (x)
w. r. t.
M +1
[P(x) -Pk] [Pk+i-P(x)]
i=1,i*k+1
(2.8B)
2.2 Modes of the Posterior Distribution
For the potency curve, P(x), both the joint modes and
means
of
the posterior density may be considered
estimators
.
as
For calculating the means, we need to derive
the marginal posterior density functions of Pi from (2.7A)
(Disch, 1981).
In what follows,
we will concentrate on the
joint posterior modes as estimators, and we will refer to
them as
"most probable value"
g(P1,...,
Pm)
be expression
density function.
(MPV)
estimators.
Let
which is the joint
(2.7B),
Setting the partial derivatives of the
logarithm of g(P1,..., Pm) with respect to the potencies to
zero, we have the expressions
api
log g(P1,
(P1,
. . .
Pm)
Si
1
Pi
n.-si +
3. -Pi
(1 =1, 2,
where
P=(P1,
aiP
...,
Po=0, and Pm+1=1.
...,
Pm),
i,j=1,...,M, where
21=(H1,
ai#141
Pi
(2.9)
Let f = (f1,
and
df=[dfu]
fm)
,
for
25
=
logg (P)
a
si
ni-si
Pi
1-Pi
aip
ai+iP
Pi -pi -1
Pi+1-Pi
(i = 1,
=0
a2
dfL i =
logg(P)
...,
(1=1,
(3 .P1
ni-si
-si
(1....pi)
aPi aPi-1
32
logg(P) =
32
apiapi
(2.11)
aiP
(2.12)
(Pi-Pi-1) 2
a = +11
(i= 1,...,M)
(2.13)
(Pi+l-Pi) 2
aPiaP14.1
df
AO
( pi.cpi) 2
(Pi-pi -1) 2
logg(P)
=
(2.10)
ai+10
2
32
df.,
,
logg(P) = 0
for li
jI
z
2
(2.14)
We can solve the M equations in (2.10) for modes H by using
the Newton-Raphson iteration process with
Pr
Pr+i
f (Pr) dri (Pr)
(2.15)
for the (r+l)th iteration P"1 until the sequence converges
to H.
These modes are unique since (2.7B)
function.
is a concave
In the Newton-Raphson method in the bioassay
problem, the solutions of modes are very sensitive to the
initial values
Po.
introduced by Ayer
The
et
isotonic regression estimator
al.
(1955),
Brunk
(1970),
and
Robertson (1988), will be used as the initial values of Po.
The simple form of this estimator is
s
s
P(x) = min max (Esj/En)
i
ilsOf 1li
For
the
j =r
Dirichlet
(2.16)
.1'-r
distribution,
the
means
of
the
26
distribution equal the modes of the density e with respect
to measure dv °, which is a property of the Dirichlet prior
distribution.
It will not be so in the posterior.
In the
following example, we will see how different the modes are
from the means.
Example 2.1 Consider the dose levels M=2 and the prior
guess Q(x)=x for 0 .x1.
Assume the observational doses are
x1=1/3 and x2=2/3 and get ai=1/3 for i=1, 2, and 3.
Then the
prior density function is proportional to
1
1
P13 (P2-P1)
1
3 (1-P2) 3
w. r. t. otO
(2.17)
and the posterior density function is
g(Pli P2)
4122-32
S
P13
(1-P1)n1-51(P2-P1) 3 P22 (1-132) 3
w. r. t. dvo
(2.18)
where
dvo
dP1 dP2
P1(P2 -P1) (1-P2)
(2.19)
Let B(a,b) be a Beta distribution with parameters a and b.
The posterior means of the posterior distribution, of P1 and
P2,
with respect to dvadPi for i=1,2 should be
s2
8T1,1) = c
E{(-1) I CgAB(i+n2-s2+1,
* B(s +1+1,
i+ni-si+n2-s2+
3
}
3
th-s,
X(P2) = C
E {(-1)-itnisiBu+s1+4,
j =0
k
1
* B(j+si+s,+
+1, n2-s2+1)}
(2.20)
27
in which the normalizing constant C can be obtained from the
following formula
n -s 1
= E {(_1)ichis1B(J+s1+4, 4)*
(2.21)
B(j+s1 +s2+4, n2 -s2 +2)}
Define gl and g2 as
)
(
=
alogg
s +.3
n1 -s1
8133.
P1
1 -P1
(pi, P2) = f_o_gs
1
I.3
P2-P1
aP2
2
P2 -pi.
s2 - 13 +n2 -s2
(2.22)
1 -P2
P2
The modes of P1 and P2 will be such that g1 and g2 equal
zero.
of
The forms of the modes are not explicit but the forms
the means are known (2.20).
In order to evaluate the
differences between the modes and the means we replace the
modes of P1 and P2 by the means in (2.20) and evaluate
gi(r(Pi) r(P2)) a
g2
g'(P2)
(r(Pi.)
)
" 0
(2.23)
In the following two examples, the difference between the
modes and the means are examined.
The values of g1 and g2 are evaluated at r(P0 and r(P2)
from (2.19)
n1 =n2=2 ,
for the cases both of
)9=1,2
n1 =n2=1,
(see Table 2.1 and 2.2)
.
)3=1,2 and of
In Table 2.1,
n1 =n2=1, /3=1,2 and the absolute values of gi (r(P1), g'(P2)) and
g2(g1P1),
(1,0)
,
are less than .00005 if
g'(P2))
and (1,1)
.
For (s1,s2) = (0,1)
,
(si,s2)
= (0,0),
the absolute values
28
of gl and g2 are a greater than 0.2.
In Table 2.2, n1 =n2=2,
and the absolute values of
0=1,2
g2(r(P1),
r(P2))
g1(r(P1),
are less than .0002 if
(1,0), (2,0), (2,1), and (2,2).
(s1,s2)
r(P2))
and
= (0,0),
For (si,s2) = (0,1), (0,2),
(1,1), and (1,2), the absolute values of functions g1 and g2
are greater than 0.15.
From Table 2.1 and Table 2.2 we see that the means are
equal to the modes (g1.0) when s1 >s2 and the means are not
equal to the modes (g1 #0) when s1<s2.
We can also observe
that H1 5r(P1) and H2r(P2) for s1 5s2, where Hi is the mode of
Pi.
In general, for any M dosage levels, if there exists
any si5si.o,
then H1 5..r(P1) and Holr(Pm).
The joint modes
at the peak of the posterior distribution will converge to
the mle's.
The joint modes of the posterior density will be
used to estimate the actual potency curve.
2.3 Point Estimator of an Effective Dose
Ramsey (1972) developed a method for computing the mode
which is used to estimate the true potency curve.
estimator can estimate any effective dose.
mode of ith observational dose.
His
Let Hi be the
The estimator H(x), of
P(x), at dose level x is
If (A7)
= 11.
2
+
Q(x)-Qi
H1 -Hi
{(21+1
Qi}
(2.24)
where Q0=H0=0 and Qm+1=Hm+1=1.
When estimating the effective
dose EDq, the experimenter observes which dose xh yields
H(xh)=q.
In other words, if the observational dose xi is
29
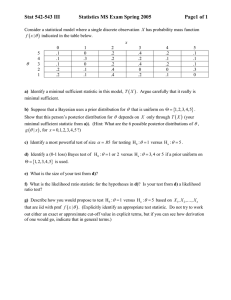
Table 2.1 The values of (gi (r(Pl) , '(P2)) , g2
and (H1, H2) for n1 =n2=1 and /3=1,2
ray ,
P (si, so ('(P1) , r(Po
(H1, HO
(0,0)
(1,0)
(1,1)
(.11111,.28889)
(.20635,.79365)
(.44445,.55556)
(.71111,.88889)
(-.00003,.00001)
(-.21220,.21219)
(-.00001,.00001)
(.00000,-.00005)
(.11111,.28889)
(.18765,.81235)
(.44445,.55556)
(.71111,.88889)
(0,0)
(0,1)
(1,0)
(1,1)
(.16667,.40476)
(.25758,.74242)
(.41667,.58333)
(.59524,.83333)
(-.00002,.00002)
(-.13372,.13371)
(-.00001,.00001)
(.00000,-.00003)
(.16667,.40476)
(.24994,.75005)
(.41667,.58333)
(.59524,.83333)
1 (0,1)
2
I A
(g1, g,)
r(p2) ) )
:
evaluated at (r(P1)
,
r(P2) )
Table 2.2 The values of (g1
.
('(P1)
Rp2) )
g2(rp1) , Rp2) ) )
and (H1, HO for n1 =n2=2 and /3=1,2
(si,s,)
('(P1)
,
ray )
(g1, gO
(H1,
(0,0)
(0,1)
(0,2)
(1,0)
1 (1,1)
(1,2)
(2,0)
(2,1)
(2,2)
(.06667,.18333)
(.12917,.49834)
(.13446,.86554)
(.26667,.35833)
(.40952,.59048)
(.50417,.87083)
(.46667,.53333)
(.64167,.73333)
(.81667,.93334)
(-.00005, .00001)
(-.62512, .28126)
(-.28769, .28767)
(-.00002, .00002)
(-.27985, .27985)
(-.28129, .62514)
(-.00001, .00001)
(.00001, -.00002)
(.00003, .00016)
(.06667,.18333)
(.10613,.51484)
(.12159,.87841)
(.26667,.35833)
(.40055,.59945)
(.48515,.89387)
(.46667,.53333)
(.64167,.73333)
(.81667,.93334)
(0,0)
(0,1)
(0,2)
(1,0)
2 (1,1)
(1,2)
(2,0)
(2,1)
(2,2)
(.11111,.28889) (-.00004, .00002)
(.16945,.53056) (-.31998, .18069)
(.19834,.80166) (-.23857, .23857)
(.27778,.42222) (-.00002, .00002)
(.38384,.61616) (-.15042, .15042)
(.46945,.83056) (-.18069, .31975)
(.44444,.55556)
(- .00001,-.00000)
(.57778,.72222) (.00000, - .00000)
(.71111,.88889) (.00002, - .00009)
(.11111,.28889)
(.16057,.53889)
(.18828,.81172)
(.27778,.42222)
(.37994,.62006)
(.46110,.83943)
(.44444,.55556)
(.57778,.72222)
(.71111,.88889)
'A': evaluated at (r(P1), r(P2)).
30
such
that
H1 =q,
then
the
estimated
EDq
is
dose
x1.
Otherwise, we can determine a pair of (xi,xi+i) which will
satisfy Hi<q<Hi.o.
Q(xh)
in which H(xh)=q.
Q(xh) = 0- +
1
We have the equation
Q(xh)
H(xh)
Qi
(2.25)
Therefore, we have
(1-111
1/141
Qi)
111(Q1+1
(2.26)
Once we obtain Q(xh), we can find the estimated EDq which is
xh from Q-1.
Example 2.2 (continuation of example 2.1)
For n =n2 =1 and 19=1,
1
2,
we know that the estimated
potency curve by using means and modes are different for
(s1,s2)=(0,1) but the estimated ED50 (=0.5) are the same in
both cases.
In this case, both means and modes give the
same estimated ED50.
Similarly, for n1 =n2=2 and fl=1, 2, the
estimated means and modes are different for (s1,s2)
{(0,1),(0,2),(1,1),(1,2)).
e
Both means and modes have the
same estimated ED50=0.5 for (s1,s2)=(0,2) and (1,1).
For
(s1 ,s2)=(0,1) and (1,2), the estimated ED50 for means and
modes are displayed in Table 2.3.
2.4 Bayesian MPV Estimators Inference
2.4.1 Prior and Posterior Distributions of an Effective
Dose
In section 2.3, we determined the point estimate of an
effective dose.
Disch (1981) derived a prior c.d.f. of the
31
Table 2.3 The estimated ED50 from means and modes for n1 =n2=2
and /3 =1 and 2
g
(s1, s2)
1
(0,
ED50 from
means
1)
(1, 2)
2
(0,
1)
(1, 2)
Ed50 from
modes
.66777
.33223
.65456
.34544
.63846
.36153
.63240
.36760
effective dose q (EDq) which is an incomplete Beta(1-q;b,a),
a=pQ(x) and b=P(1-Q(x)). The posterior c.d.f. (derived from
the marginal posterior distribution of P(x)) of the EDq is
a linear combination of the incomplete Beta distribution
functions.
Computation
distribution of P(x)
of
the
marginal
posterior
involves multiple summations which
cause complexity of computation.
The normal approximation
method can be used here, so the posterior distribution of
the potencies can be approximated by the multivariate normal
distribution and the marginal posterior distribution of P(x)
can be approximated by an univariate normal distribution.
2.4.2 Normal Approximation Method for the Bayesian MPV
Estimators Inference
The posterior density function is proportional to
M
{ II P:71
i=1
M+1
1-1
w.r. . t. dvo
.z=1
= g(p)
(2.27)
The modes H=(H/,...,EW
can be calculated by using the
Newton-Raphson iteration process described in section 2.2.
32
The approximate covariance matrix, VH, is defined as
a2 logg(P)
( vm)
1
(2.28)
the approximate joint posterior distribution of
So,
Pi
(i=1,...,M) is
P
MVN( H,
)
VH
(2.29)
and the approximate marginal posterior distribution of Pi
(i=1,
M) for the observational dose xi is N(H1, V(Hi)),
where V(Hi)= [VH]fl.
For any non-observational dose x between xi and xi+i, we
have (Ramsey (1972))
Q(x) -Qi
P(x)
Pi
Q1+1- Q(x)
P(x)
(xi <x <
xi.1)
.
(2.30)
From (2.30), we can find
P (x) = P (xi) +
Q(x) -Qi
(Pi+i
Qi+1
Pi)
P1)
= Pi + t
(2.31)
where
Q(x)
wi+3.
(2.32)
Qi
Therefore, the marginal posterior distribution of the nonobservational dose x for xi<x<xi+i can be approximated by
P(x) ....N(r(P(x)),
Var(P(x)))
(2.33)
where
'(P(x)) = Hi + t (Hi+i
Var(P(x)) = (1
t) 2
Hi)
Val- (Pi) +
+ 2 t (1
t2 Va.
r(Pi+i)
t) Cov(Pi, P1 +1)
(2.34)
33
and, the approximated posterior c.d.f. of EDq is
1
-E. f(y) dy = 1
0(q- r(P(x)))
(2.35)
slVar (P (x)) )
where 0 is a c.d.f. of standard normal distribution.
If the
estimated EDq is the observational dose xi, f(y) will be the
density function of N(Hi, V(Hi))
density of (2.33)
.
However, f(y) will be the
if the estimated EDq is between xi and
xi+1'
Example 2.3
We want to estimate the first quartile,
of the distribution of the EDq.
T1,
If there is an
observational dose such that FEDq(xi)=Pr(EDq5.xi)=.25, we have
Ti=xi.
(xi,
If not, we determine the pair of observational doses
xi+1) such that FEDg (Xi) ) < . 25<FEDg (Xj+1 ) , i.e. xi<Ti<xi+i and
FEDq (TO = 1
(D(qVVar (P(71))
(2.36)
We rewrite (2.36), which should be a quadratic function of
,
as
q-ir(P(ri))
- 0-1(.75)
IlVar (P(Ti))
(2.37)
where '(P (T1)) and Var(P(TO)
then solve
for T1.
are described in (2.34) and
34
3 ADAPTIVE DESIGNS FOR ESTIMATING THE POTENCY CURVE
3.1 Adaptive Designs
The adaptive designs developed here are for sequential
selection of the test doses for estimating the potency
curve.
Then use the estimated potency curve to obtain the
final ED50 estimate in the last step.
The prior is assumed to be the Ramsey's prior [28]
(Ramsey (1972))
method
is
in this Bayesian analysis.
sequential,
the
prior
modal
The adaptive
function
incorporating into a non-informative function.
Q(x)
is uniform over
analysis.
available
[0,1]
testing
at
We assume
in the first step of the
If there are L (=m*n)
for
Q(x)
fixed
experimental subjects
doses
chosen
by
the
experimenter, Ramsey (1972) showed that the best design (in
the sense of estimating the ED50 by comparing bias, standard
deviation, and the mean square error) used one subject per
dose.
In this adaptive method, we will perform n steps and
assign one subject per dose to m doses.
In the first step,
we choose doses uniformly for dose i/(m+1), i=1,..,m.
Then,
the estimated potency curve is used as the prior modal
function to find the new dosage levels for the next step.
Algorithms for evaluating two adaptive designs using (A)
full information and (B) reduced information follow.
an example for L=6, m=1 and n=6,
these two adaptive designs.
Also
is given to illustrate
35
(A) Full Information Method
The prior is proportional to
(P(xj)-P(xj_1))4i13
(3.1)
We choose 0=2 for all n steps and use m doses in each step.
In the last step we use L (=m*n)
doses and full prior
distribution, incorporating all L parameters, to estimate
the potency curve and the ED50.
The algorithm is shown
below.
Step 1
:
Q(x)=x, sZox.51.
Selected dosage levels:
xLi= i /(m +l), i=1,...,m.
m+1
Prior ..: II (P(xl,i) -P(xi,i_l) )441,
p1=2, al= n74-1
Likelihood ac
where
(3.2)
,
1
P (xi)
8
(1-P (2ci,i)
Posterior c< Likelihood * Prior.
)i-s,.i
(3.3)
Evaluate the new m doses
from the posterior modal function, 111(x), such
that x2,i=1.11-1(i/(m+1)), i=1,...,m.
Step k (k=2,...,n-1)
:
Selected dosages (X1,...,X0, where
.xiadi xm
"1= (x1.1,
x
J,I, ,m
=(X
,
Sort
Sil
i
)l
'
SI kin x 2
ILT3.
=41'
J-1
k/ ..
1
m+1
Lr-1
III'
/
M \\/
m+1
j=2
,
,
k
36
by the first column, xj,i, to (xt,st),
where X= (xi ,
,xkm)
Ey=(sko,
,
i sk,m)
131=-"(s1
, " I skm)
km
Likelihood
cc f P(xi) si (1-P (xi) ) 1 -si
(3.4)
i-3.
Jart+1
cc H
Prior
( P (xi ) -P
(xi
where
) ) 41115k ,
i=3.
Pk=2, ai=xi-x1_1, i=1,...,m+1.
(3.5)
Posterior c< Likelihood * Prior.
Evaluate the new m doses
from the posterior modal function Hk(x) such that
xk+i,i=Hk-1(i/(m+1)), i=1,...,m.
Step n :
Selected dosages (Xi,
21=(x1,1,.",xim)
,Xn)
, where
i xm
(11;11 (
j
j=2,...,n
zf
Sort
,
'
II
(
rij-1 k
m+1
M
))
.
sit
SI Lx2
by the first column, xbi, to (xt,st), where
X--(x1,...,x), 8=(si,...,s1.).
Likelihood
oci=iI P (xi) si (1-P (xi) ) 1-si
(3.6)
L+1
Prior
cc
(P(xi)
-P(xj_1)) ai/Sn,
where
(3.7)
Pn=2,
Posterior oc Likelihood * Prior.
The estimated potency
curve Hn(x) is the estimated posterior modal function.
37
(B) Reduced Information Method
The prior is proportional to (3.1), where ai=1/(m+1),
i=1,...,m+1.
We begin by choosing /3=2 and then, for each
step, increase Q by the increment m.
The posterior modes
are computed in each step and the posterior modal function
Hk(x) is adjusted from the previous step modal function
Hk_1(x).
The new m doses are chosen from the adjusted modal
function Hk(x).
Step 1
The algorithm is shown below.
:
Q(x)=x,
Selected dosage levels:
xL= i /(m +l), i=1,...,m.
m+1
Prior oc II (p(x) -p(x,) )441-,
01=2, a1=
m1
+1
1
where
4
-=-,..,m+1.
(3.8)
Likelihood cc 11 P(x1,i)s"(1P(x1,i))1 -s,,i
(3.9)
iii
Posterior c< Likelihood * Prior.
Evaluate the new m doses
from the estimated posterior modal function H1(x) such
that x2, i=1-11-1( i/ (m+1) )
Step k (k=2,...,n-1)
,
i=1 ,
. . . ,m.
:
Selected dosages Xk=(xko,...,xk,m)
m
Likelihood cc fl P(x.k,i) S" (1--P(xk,i)) 1-sk1
(3.10)
m+1
Prior ac fl (p(xk,i)-p(xk,i_1))
i-1
where
(3.11)
Pk=Pk_l+m, ai=
m1
1
,
i=1, ...,m+1.
38
Posterior
ec
Likelihood * Prior.
The estimated posterior
modal function is
0 sxsxk,i
14(Xk,l) Hk-1(X)
[ (i +1) Hk(Xk, i) 1Hic(XJ, i+1))
(m+1)
1
+ (111+1) (lik(XL i+3.) lik(XL i) ) Hki. (X)
14(x) =
r
for Xk,15 X SXk.ifi, i=1,2,...,m-1
[(m+1)Hk(X.kad m] + (Ri+1) (1Hk (Xk,m) ) Hk_i (X) ,
for xk,m
where
Hic_1
sx s 1
(xk,i)=i/(m+1), i=1,...,m.
chosen as xk+iii=Hk-1(i/ (m+1) )
Step n
(3.12)
,
The new m doses are
1=1, ... ,m.
:
Xn=(xn,i,...,xn,m)
Selected dosages
m
Likelihood oc II P
i-i
(x, i)84" (1-P (x, i)
)1-sn'i
(3.13)
m..1.
Prior oc if (P(x,i)-P(x,i..3.))ailln, where
i=i
Pm=i3m-i+m, ai=m+1
Posterior
ec
i,
1=1, ...,m+1.
Likelihood * Prior.
(3.14)
The estimated posterior
modal function is
(m+1)Hn(Xn,i)Hn-1
(x)
,
0 Sx5xn,i
[(i+1)H(x,i)-iHn(xn,i+1))
Hm(x)=
+(m+1) (Hm(xn,i+1) -11,2(xn,i))Hn_1(x) ,
for xn,is x sxn,i+1,
i=1,2,...,m-1
[(m+1) H (xn,m) -m)+(m+1) (1-Hn(xn,,))Hn_1(x)
for xn,m
sx s 1
where H 11-1(x0=i/(m+1), i=1,.
,
(3.15)
.. ,m.
The estimated potency
39
curve function Hn(x) is the estimated posterior modal
function.
Example 3.1
Let L=6, m=1 and n=6.
The procedures for
design A and design B are the same in the first step.
Step 1:
Prioroc13(x1,1)111u1 (1-P (x1,1) ) I31a2
(3.16)
where fl1 =2, ai=1/2, i=1,2.
Fixed dose x141=1/2.
so, Prior .2c P(1/2)[1-P(1/2)]
Likelihood 04
Posterior
cc
P(1/2) 81.1(1- P(1 /2) )1 -81.1
(3.17)
P(1/2) 81.1+1(1-P(1/2)) 2 -S'.1
(3.18)
-> The estimated mode is
H1(1/2)
S1.+1
3
=1
2/3
6
(3.19)
H1(1/2)=1/3, so (see Fig. 3.1)
1-x2,1
1-1/2
1-x1,1
1 -111.(xl_i)
-
x2,1=5/8
Fig. 3.1 Determination of x21
for s1,1=0 in the first step
0.9
0.3
0.7
0.6
03
0.4
0.3
0.333
Di
0.1
V
s1
1
=1
H (1/2)=2/3, so (see Fig. 3.2)
1
(3.20)
40
Ic2,1 _
1/2
x2,1 =3
x1,1
/8
(3.21)
After the first step the procedures diverge.
illustrated separately.
They are
For ease of explanation, design B
is discussed first.
Fig. 3.2 Determination of x2,1
for si,f=1 in the first step
1
110
0
6
5
3
2
.1
n
n.
n ,
n i
A A
fl
ft76
f i ...7
11:11
09
1
X2,1
Design B
Step k: (k=2,...,6)
Prior°c P(xk,1)13kul (1-P (xk,l) ) giku2
(3.22)
fixed dose x1(,1 is selected from the (k-1) th step, pk=1+k,
a.=1/2, i=1,2.
So,
1+k
Prior e< P (xk,i) 2 (1_p(xk,1) )
Likelihoodoc P(xk,i)
1+k
2
(3.23)
(1-P(xk,1) )1-8k>_
(3.24)
1+k+sk.i
Poster ior
<P(xk,i)
1+k
(1P(Xic,i) )
The estimated mode is
(3.25)
41
k+1
Hk(Xk,
sk,14- 2
k+2
1
1
2
2 (A*2)
' fssk,1=1
(3.26)
For example k=2,
H2 (x2,1)
(1)
3/8 , 52,1=0
5/8
S2,1=1
s1,1=0 s21=0 : the fixed doses are x11=1/2, x2,1=5/8, and
H1 (x1.1) =1/3, H2(x20)=3/8.
so
(3.27)
H2(x1,1)
112(x) can be expressed as (3.12),
= 2*112(x2,1) *Hi (x1,1)
=
2 (3/8) (1/3)=1/4.
H2(x3,1)=1/2, H2(x2,1)<H2(x3,1)<1, (see Fig.
1-x3,1
1-x2,1
1-1/2
1 -H22- (X2,1)
Since
3.3)
x3,1=0.7
(3.28)
Fig. 3.3 Determination of x31
for s10=0 s2,1=0 in the second
step
a9
it2(4
06
9.7
9
05
0
:-..9
3
ol
2
0. 1
0
f
uo20.30.40.5umuu
i
X3,1
(2) s11=0 s21=1 : the fixed doses are x11=1/2, x21=5/8, and
H1 (x1,1) =1/3
so H (
)
112 (X2,1)
5/8.
2*H2 (x2,1)
*Hi
H2 (x)
(x1,1)
can be expressed as (3.12),
= 2 (5/8) (1/3)=5/12.
H2(x3,1)=1/2, H2(x1,1)<H2(x3,1)<H2(x2,1), (see Fig. 3.4)
Since
42
2 X1,1
* ( .5-H
())
X2.1 -X1.1
x3.1
x11+ H2 (X2.1) H2 (Xi,i)
=
x3,1=0.55
(3.29)
x31
Fig. 3.4 Determination of
for s 1°=0 s2,1=1 in the second
step
.
1
09
10(1)
a
07
.923
04
0S
.
04
i
Ufa
0. 3
0. 2
t
1
0
0
0.1
0.2
03
OA
OS a
0.7
0:8
09
i
(3) s10=1 s2,1=0 : the fixed doses are x1,1=1/2, x2,1=3/8, and
H2(x) can be expressed as (3.12),
ili(xi,i)=2/3/ H2 (X2,1)=3/8*
so
H (
[2*H2(x2,1) -1
=
]-1-2*
(1-H2(x2,1 ) *Hi (x1,1)=0.58333.
Since H2(x3,1)=1/2, H2(x2,1)<H (x3,1)<H2(xi,i),
L1X2,1
X3,1 = X2'1+
H2 (X1X,1)
H2 (X2,1)
*
(see. Fig. 3.5)
5-112 (X2,1) )
X3,1=0 45
(3.30)
Fig. 3.5 Determination of x3,1
for s 1,1 =1 s 2,1 =0 in the second
step
i
09
09
0. 7
6
6 -SU 4..
05
O
03/
0 3
02
0.
00A0.20.3040.50.60/0209
x3 , 1'
1
43
(4) s1,1=1 s2,1=1 : the fixed doses are x1,1=1/2, x2,1=3/8, and
H1 (x1 1)=2/3, H2(x2,1)=5/8.
H2 (x) can be expressed as (3.12),
so H2 ( XL1 ) = [2*H2(x2,1)-1] +2*(1-H2(x2,1))*H/(x0=0.75.
H2(x3,1)=1/2, O<H2(x3,1)<H2(x2,1)
x3,1
X2,1
1/2
H2 (X2,1)
,
Since
(see Fig. 3.6).
x 3,1 =0 3
(3.31)
Fig. 3.6 Determination of x3,1
for s1 1 =1 s21=1 in the second
step
1
mod
09
0
0 77
0.
1
5
04
03
02
0.
1
o
0
0.1
0.2
0.3
OA
0.5
Ob
X3,1
This continues from k=2 to k=6 and the final selected dose
in the last step is the final ED50 estimate.
The six
selected doses and the final ED50 estimate are shown in the
Appendix.
Design A
Step k: (k=2,...,6)
Let the fixed doses X=(x1,1,fxko), where x 1(0 is selected
from the (k-1)th step, and the corresponding responses are
.
Sort X in ascending order, such that
xi<...<xk, with corresponding S=(si,...,sk).
k +1
Prior 41 (P(xi)-P(xi_0)ailik
11=1.
(3,32)
44
ai=xi-xi_1,
where 13k=2,
i=1,
,k+1, (xo=0, xio0=1, P(xo)=0,
P(xk+.1)=1).
k
Likelihood oil
P(xi)si (1-P(xj))1-si
(3.33)
11-1.
k
k+1
Posterior .11 P (xi) si (1-P (xi) )1-si -r-r
n (p(x,) -p(xi_o ) 2ai
i1
1=1
There is no explicit form of H(x1).
(3.34)
The Newton-Raphson
method is used to calculate H(x).
(1)
So,
s11 =0 s2,1=0
7L=(x1,
ai=1/2,
x2)=(1/2, 5/8) and El= (si
a2=1/8,
H2(x2)=.34375.
X3=1, H2 (X0)=0
X
the fixed doses are x1,1=1/2, x2,1=5/8.
:
and
a3=3/8.
If H2(x1_i)
,
H2 (X3)=1) ,
We
H2(x30)=0.5
, s2) = (0,0) ,
have
p2=2,
/31=2,
H2 (X1) =0 . 25
and
H2(x1), i=1,2,3 (x0=0,
then
Xi-Xi_1
* ( . 5-H2 (Xi_i) )
3'1=X2-1+ H2 (Xi) -H2 (Xi_i)
(3.35)
From (3.35), x3,1 can be calculated as .71429.
(2) sl=0 s2,1=1
:
the fixed doses are x11=1/2, x2,1=5/8.
So, X =(x1, x2)=(1/2,
5/8) and S=(si,s2)=(0,1),
ai=1/2, a2=1/8,
a3=3/8.
H2(x2)=.68038.
and
From
(3) s1=1 s2,1=0
:
(3.35),
We have H2(x1)=0.39522
a2=1/8,
and
x3,1 can be calculated as .54593.
the fixed doses are x1,1=1/2, x2,1=3/8.
So, X =(x1, x2)=(3/8, 1/2) and 8=(si,s2)=(0,1),
a1=3/8,
132=2,
13 =2,
and a3=1/2.
pi=2,
We have H2(x1)=0.31962
132=2,
and
H2(x2)=.60478. From (3.35), x3,1 can be calculated as .45407.
(4)
sio=1 s21=1
:
So, X =(x1, x2)=(3/8,
the fixed doses are x11=1/2, x2,1=3/8.
1/2)
and 121=(s1,s2)=(1,1),
/31=2,
132=2,
45
a1=3/8,
a2=1/8,
H2(x2)=.75000
and a3=1/2.
We have H2(x1) =O. 65625 and
From (3.35), x3,1 can be calculated as .28571.
This continues from k=2 to k=6 and the final selected dose
is the final ED50 estimate.
The six selected doses and the
final ED50 estimate are shown in the Appendix.
An
example
of
computing
the
exactly
sampling
distribution of the final ED50 estimate will be given in
next
section to evaluate the efficiency
of different
combinations of m and n in estimating the true ED50.
3.2 Designs for the Adaptive Methods
Assume that the actual potency curve is P(x)=xd, d>0,
The goal of this section is to construct the best
design for estimating the true ED50 of a given L subjects.
For L=6, four experimental designs are examined.
With
the possible {n;m} arrangements, we consider {6;1), {3;2),
{2;3), and {1;6) experiments where n is the number of steps
and m is the number of doses.
subject in each step.
Each dose is assigned to one
There are 64 possible outcomes for
all experiments.
For L=12, six experimental designs are examined.
With
the possible {n;m) arrangements, we consider {12;1), {6;2 },
{4;3),
{3;4),
{2;6), and {1;12) experiments.
There are
212=4096 possible outcomes for all experiments.
The values of d are chosen from [ .001, 20].
For any d,
calculate exactly sampling distribution for the final ED50
estimate. The relative efficiency (r.e.) is used to compare
46
(n;m) designs with (1;L) design in adaptive method A (full
information)
and method B
(reduced information).
For
convenience, we will adopt the following to describe the
experiments:
Anm : adaptive method A for n steps and m doses per
step. (fl=2 for all steps);
Bnm : adaptive method B for n steps and m doses per
step. (31=2, Pi=pi.i+m for ith step, i=2,...,n).
AIL and B1L are equivalent designs.
The selected doses
x,,...,x6 and estimated ED50, L=6, for Anm and Bnm designs
are shown in the Appendix.
The r.e. of design Y relative to
design X is defined as
r.e. (Y/A)
mse(X)
100%
mse(Y)
(3.36)
For convenience the notation "Y/X" is used for r.e.(Y/X).
Note that the scales of all figures for d in this chapter
are logarithmic.
The notation d-1 will be used for
de(.5,4); otherwise, d#1 will be used.
The best designs of adaptive methods for L=6 and 12
are shown in Table 3.1, 3.2, respectively (see Fig. 3.7
3.10).
From above computation, we see that design B, with
one dose (close to the prior ED50) per step, is the best
(highest r.e.)
for estimation of the true ED50 if the
initial uniform prior modal function Q(x)
true potency curves (d-1).
is close to the
If the prior guess is bad (prior
ED50 is not close to the true ED50 for d4.1), design A for
man appears the best.
Assuming P(x)=xd, d>0, Ox5.1 and L=6 (L=m*n) , there are
47
Table 3.1
The best designs for the adaptive methods
(L=6)
Method A
A61
B61
A23, A32
B16
d.-1
d+1
Table 3.2
Method B
Both A and B
B61
A23, A32
The best designs for the adaptive methods
(L=12)
Method A
Method B
Both A and B
B121
d-1
A121
B121
d+1
A34, A43
B112
A34, A43
four methods which will be described in the following
sections for estimating the ED50: (A) one-step method (nonadaptive), (B) Spearman-Karber method (non-parametric) (27]
(Finney (1964)), (C) up-and-down (staircase) method (Dixon
(1948, 1965), Little (1974)), and (D) Robbins-Monro process
(Cochran and Davis (1965)).
3.3 One-step (Non-adaptive) Method
This
is
a
one-step method
of
Bayesian bioassay.
Bayesian posterior modes for /3 =2, ai=1/(m+1), i=1,...,m, are
used to estimate the potency curve.
We assign n subjects
per dose to m equally spaced doses (x1=i/(m+1), i=1,...,m)
and find the estimated ED50.
For convenience
,
we will
adopt the following to describe the experiments:
NOnm
:
non-adaptive method assigning n subjects to
each m doses.
(Q =2, one-step).
The fixed doses and estimated ED50 of all possible outcomes
48
Fig. 3.7 The r.e. of Anm/A16, m=1,2,3, L=6
-4-
A61/A16
A32/A16
220
see
140
lee
60
0.01
0.1
10
100
d
Fig. 3.8 The r.e. of Bnm/B16, m=1,2,3, L=6
Bsi/Bis
-4-
1332/1316
170
150
130
I;
118
C.
90
70
50
0.01
0.1
s
d
10
see
49
Fig. 3.9 The r.e. of Anm/A112, m=1,2,3,4,6, L=12
-+
800
A121/A112
A62/A112
.94#1.41-2
-a' 04042
i=i8e/ii112
600
I
400
200
0
0.01
0.1
100
10
d
Fig. 3.10 The r.e. of Bnm/B112, m=1,2,3,4,6, L=12
-+
180
A
-a
150
834/"12
026/6112
... ...
....
8121/8112
B82/8112
B43/BiL2
..........
**if- -"/:!'
120
'449
oct -
:
-0 -04:
44Kx*--4(-4-01
98
L
.. -
89
* ....
30
0
0.01
8.1
1
d
10
100
50
for NO16, NO23, NO32, and NO61 are shown in the Appendix.
The best design of the non-adaptive method is NO61 for
d-+1; otherwise, NO16 is the best (see Fig. 3.11).
The NO61
design will be compared with other methods for estimating
ED50 for d-a in section 3.7; otherwise, NO16 will be used.
3.3.1 Comparisons of Non-adaptive Method with Adaptive
Methods
NO61 will be compared with B61 for d-l.
will be compared with NO16 for d+1.
NO61
A32 and A23
Comparing the r.e.,
is more efficient than the B61 design for
d-41;
otherwise, A23 and A32 dominate the NO16 design (see Fig.
3.12).
3.4 Spearman-Kirber (Non-parametric) Method
The Spearman-Kdrber estimator
estimator of the ED50.
is
a
non-parametric
If the levels are ordered such that
xi<...<xm, this estimator is defined by
mx-.1
(Pi +1-P1) (xi+x1 +1)
2
(3.37)
provided that P1=0 and Pm=1, where Pi=si/ni, ni is the number
of the observations and si is the number of the positive
responses at dose xi,
i=1,...,m.
If P1>0, then an extra
level is added below x1, where no responses are assumed to
occur.
Similarly, if Pm<1, an extra level is added above xm,
where responses are assumed to occur.
The levels are
assumed to be equally spaced and nin, i=1,...,m.
So, we
will have n1 =6, 3, and 2 possible experiments and the test
51
Fig. 3.11 The r.e. of NOnm/N016, m=1,2,3, L=6
--- NO61/N016
-4NO32/N018
6
200
160
120
L
80
40
0
0.01
0.1
1
10
100
d
Fig. 3.12 The r.e. of B61/N061, A32/N016, A23/N016
-+-
B81/N081
A32/N016
400
300
U
200
C.
100
0
9.91
0.1
3.
d
10
100
52
dosage levels x1=i/(m+1), i=1,..,m.
For convenience, we
will adopt the following to describe the experiments:
SKNn
Spearman-Kdrber methods for ni=ne(2, 3, 6).
:
The fixed doses and the estimated ED50 of all possible
outcomes for SKN6, SKN3, and SKN2 are shown in the Appendix.
SKN6 has smaller mse when compared with SKN2 and SKN3
when the prior is close to the true potency curve (d-,1) (see
Fig. 3.13).
SKN6 is more efficient for d-,1; otherwise, SKN2
is more efficient.
The SKN6 design will be compared with
the other methods for estimating ED50 when d-+1 in section
3.7; otherwise, SKN2 will be used.
3.4.1 Comparisons of Spearman-Kerber Method with Adaptive
Methods
SKN6 will be compared with B61,
compared with Anm for n=2, 3.
and SKNn will be
Comparing the r.e., SKN6 is
more efficient than the adaptive B61 design
for
d-+1;
otherwise, A23 and A32 are more efficient than the SKN2 and
SKN3 designs respectively (see Fig. 3.14).
3.5 Up-and-down (Staircase) Method
The up-and-down method is another non-parametric method
for estimating ED50.
sequentially.
The dose levels
are determined
A series of test dose levels is chosen with
equal spacing between doses.
The first level should be
chosen as near as possible to the ED50.
Then, a series of
53
Fig. 3.13 The mse of SKNn, n=2,3,6
-4-
SKN6
SKN3
0.1
0.08
0.06
8
p
E
0.04
0.02
a
0.01
0.1
1
180
10
d
Fig. 3.14 The r.e. of B61/SKN6, A32/SKN3, A23/SKN2
-4-
861/SKN6
A32/SKN3
600
400
300
U
L.
200
100
e
0.01
0.1
i
d
10
lee
54
trials
increasing the
performed,
is
dose
following
a
negative response and decreasing the dose following
a
positive response.
The estimated ED50 is Xf+kD, where Xf is
the last dose administered, k is a value from the provided
table (Dixon, 1965), and D is the interval between doses.
In this example, an experiment is conducted on six
subjects.
We will choose x1=0.5, which is close to the true
ED50 for d close to 1.
Since the true potency curve is
assumed to be P(x)=xd, d>0 and (:))cl, the D value (dose
interval) should be chosen in (0, .075) such that all doses
and ED50 lie in (0, 1).
Three values of De(.001,
.035,
.075) are chosen in computing the exact distribution of the
ED50.
For convenience
,
we will adopt the following to
describe the experiments:
UDq
:
up-and-down method using cle(001, 035, 075)
as dose interval.
The fixed doses and the estimated ED50 of all possible
outcomes for UD001,
and UD075 are shown in the
UD035,
Appendix.
UD001 has smaller mse when compared with UD035 and
UD075 for d-41 (see Fig. 3.15).
UD001 is more efficient for
d-41; otherwise, UD075 is more efficient.
Since D is .001,
the fixed doses and estimated ED50 are all close to 0.5 for
all 64 outcomes.
Also, UD001 is more efficient for d-)1, but
extremely less efficient for d#1.
however, is not realistic.
This extreme case,
So the UD035 will be used to
55
Fig. 3.15 The mse of UDq, q=001, 035, 075
-+-
U0991
UD035
9.25
9.2
0.15
a
N
E
0.1
0.05
9
0.01
0.1
1
lee
10
d
Fig. 3.16 The r.e. of B61/UD035,
B61/UD075
261/UD035
-I-
E161/UD075
409
300
I;
200
C.
100
e
0.01
0.1
1
d
10
lee
56
compare with other methods for estimating ED50 for d-41 in
section 3.7; otherwise, UD075 will be used.
3.5.1 Comparisons of Up-and-down Method with Adaptive
Methods
Comparing the r.e., UD035 is more efficient (UD075 is
less efficient) than the B61 design for d -+1, and UD035 and
UD075 are less efficient than the A23 and A32 designs for
d#1, but UD075 is more efficient than A23 and A32 designs
when d is extremely small (d<.003) or large (d>10) (see Fig.
3.16 ,...3.18).
3.6 Robbins-Monro Process
The Robbins-Monro stochastic approximation process is
used in sequential experiments to estimate the ED50.
To
start the experiment, an initial guess xi is made at the
ED50, and n1 subjects are given the dose x1.
have positive responses and
pi=si/ni
is
If si subjects
the proportion
response, a second group of n2 subjects is tested at the
dose level x2=x1-c(p1 -.5). More generally, the dose level at
which the (r+l)th group of subjects is tested is found from
xr by
=
(P1-4)
(3.38)
where c is a suitable chosen constant.
We will let x1=0.5 (the prior ED50) and choose c such
that all the tested doses and ED50 are in [0,1].
The
experimenter conducted on six subjects resulting in n1 =6, 3,
57
Fig. 3.17 The r.e. of A32/UD035, A32/UD075
A32/U13836
-4-
A32/UD076
1699
1600
1200
908
L
600
300
0
0.01
0.1
1
100
10
d
Fig. 3.18 The r.e. of A23/UD035, A23/UD075
-4-
A23/UD035
A23/U0876
1599
1200
900
U
600
300
0
0.01
8.1
1
d
19
108
58
2, and 1 different experiments to be examined by using the
Robbins-Monro process.
The appropriate c values for the
different n1 are listed in Table 3.3.
For convenience, we
will adopt the following to describe the experiments:
RMnCc : Robbins-Monro process for ni=n and constant c
(see Table 3.3).
Table 3.3
The values of c for n1=6, 3, 2, and 1 such that
all the tested doses and ED50 are in [0,1]
ni=6
ni=3
ce(0,1)
cc(0,2/3)
n1=2
c(0,.5454)
ni=l
c(0,.4082)
The following examples of Robbins-Monro methods are used in
this section.
RM6C0
RM6C5
RM6C10
RM3C0
RM3C3
RM3C6
RM2C0
RM2C2
RM2C5
RM1C0
RM1C2
RM1C4
n1 =6
n1 =6
n.=6
n.1 =3
n.1 =3
n.1 =3
n1 =2
n1 =2
n-=2
n.1 =1
n1 =1
n1 =1
c=.001
c=.5
c=1
c=.001
c=.333
c=.667
c=.001
c=.25
c=.5454
c=.001
c=.2
c=.408
The fixed doses and estimated ED50 of all possible outcomes
for the above examples are shown in the Appendix.
Fig. 3.19 ~3.22 indicate that RMnC0 for fixed n is
more efficient for d-,1 but extremely less efficient for d#1,
and so is excluded from the comparisons.
RM6C10 has the
smallest mse when compared with RM1C6, RM2C5, and RM3C6 for
d#1.
RM2C2 has the smallest mse when compared with RM1C2,
RM3C3, and RM6C5 for d -+1 (when .7d51.5), but rose's of all
59
Fig. 3.19 The mse of RM1Cc, c=.001, .2, .408
-4-
RM1C0
RM1C2
0.25
0.2
0.15
U
E
0.1
9.95
e
0.01
0.1
10
199
d
Fig. 3.20 The mse of RM2Cc, c=.001, .25, .5454
-4-
RM2C0
RM2C2
0.25
0.2
0.15
U
U
0.1
0.05
0
0.01
0.1
1
d
10
100
60
Fig. 3.21 The mse of RM3Cc, c=.001, .333, .667
-4-
RM3C0
RM3C3
0.25
0.2
0.15
II
U
E
E. 1
0.95
e
0.01
8.1
1
10
109
d
Fig. 3.22 The mse of RM6Cc, c=.001, .5,
--
1
RM6C0
RM6C5
0.25
8.2
0.16
S
U
E
8.1
0.06
0
0.01
0.1
1
d
10
199
61
four are similar.
For RM1Cc designs, there are 6 fixed
doses sequentially chosen, making them more comparable with
adaptive designs.
methods
for
So, the RM1C2 will be compared with other
estimating
ED50
for
d -+l
in
section
3.7;
otherwise, RM6C10 and RM1C4 will be used for dill..
3.6.1 Comparison of Robbins-Monro Process with Adaptive
Methods
Comparing the RM6Cc method with the B61 design for d-41,
RM6Cc is more efficient for small c, but less efficient for
large c (see Fig. 3.23).
Comparing RMnCc (n=2, 3) with the
Anm design for d4,1, RMnCc is less efficient than Anm design
for small c, but more efficient for large c (see Fig. 3.24,
3.25) .
Comparing the higher efficiency of the Robbins-Monro
method with adaptive designs, RM1C2 is more efficient than
B61 for d-,1, and RM6C10 and RM1C4 both are more efficient
than the A23 and A32 designs for d+1 (see Fig. 3.26).
Robbins-Monro process
methods
The
is more efficient than adaptive
in comparing the uniformly optimal
cases with
respect to each method.
In the Robbins-Monro process and the previous up-and-
down method, estimation the ED50 depends on the initial
chosen dose, x1, and the constants, c and D.
This results
in making them more difficult to use than the other methods.
3.7 Comparisons
62
Fig. 3.23 The r.e. of B61/RM6Cc, c=.5,
-+.
1
561/RM6C5
861/RM6C10
300
250
280
S
150
100
50
e
6.01
0.1
1
lee
Le
d
Fig. 3.24 The r.e. of A32/RM3Cc,
c=.333, .667
-4-
A32/RM3C3
A32/RM3C6
1280
1000
800
S
I:
600
498
280
e
0.01
0.1
i
d
10
190
63
Fig. 3.25 The r.e. of A23/RM2Cc, c=.25, .5454
A23/RM2C2
A23/RM2C6
1280
1000
600
a
600
488
200
0
0.01
0.1
1
109
10
d
Fig. 3.26 The r.e. of B61/RM1C2, Anm/RM6C10,
Anm/RM1C4, m=2,3, L=6
-._4-
B61/RM1C2
A32/RM6C10
10
390
000041b4
A2#/R01$4
260
200
160
L
100
50
0
0.01
0.1
1
d
10
100
64
The best designs for the adaptive methods have been
compared with non-adaptive, Spearman-Kdrber, up-and-down,
and Robbins-Monro methods in previous sections.
Since the
complexities of comparing other methods with up-and-down and
Robbins-Monro methods are many, these comparisons will be
discussed in two parts.
For d-,1, we would like to compare
{B61, NO61, SKN6, UD035, RM1C2).
For d#1, we would like to
compare {A23, A32, NO16, SKN2, UD075, RM6C10, RM1C4).
All
the comparisons are based on the r.e. of the chosen designs
relative to the NO16 design (see Fig. 3.32 and Fig. 3.37).
The individual r.e. plots of the chosen compared cases
for d-fl are shown in Fig. 3.27
UD035 and RM1C2 have
the highest efficiencies when d=1, but the efficiencies
decrease quickly (become lower efficiency) when d is shifted
from 1 (see Fig. 3.32).
UD035 and RM1C2 have the steepest
curves, demonstrating more variability in their relative
efficiencies than the others.
SKN6 and NO61 have higher
efficiencies than B61 for d-41 (see Fig. 3.32).
The individual r.e. plots of the chosen compared cases
for d#1 are shown in Fig. 3.33
Both RM6C10 and
RM1C4 dominate the other cases (see Fig. 3.37).
Fig. 3.34
indicates that SKN2 is the least efficient when the prior
modal function is far away from the true model (d+1).
r.e. of UD075 is less than 100% when dc(.08, .6)
3.36).
The
(see Fig.
UD075 is more efficient than A23 and A32 when d is
extremely small (d<.05) or large (d>9); otherwise, A23 and
65
Fig. 3.27 The r.e. of
B61/N016
Fig. 3.28 The r.e. of
N061/N016
1.7e
298
150
160
139
120
U
L
89
90
C.
49
"re
60
.....
...
.....
.
1.11:1
0.01
0.1
1
10
.. :
.11
0.01
100
........
0.1
d
.....
,11
1
10
100
d
Fig. 3.29 The r.e. of
SKN6/N016
Fig. 3.30 The r.e. of
UD035/N016
699
608
480
1300
L
200
100
t-N14;i^.
8
0.01
0.1
1
d
Fig. 3.31 The r.e. of
RM1C2/N016
250
200
169
100
50
e
0.01
0.1
1
d
10
100
10
100
99
qqd ZVE
ST.UL
91
'9NNS
Jo '9TON/XX 'T9G=XX 'T90N
'gum ZDIW
9TON/T99
+-
9TON/T9ON
9
009
9T
9T
SOS
00V
00C
5
see
ear
0
*0 TO
0 T
OT
ØØT
67
Fig. 3.33 The r.e. of Anm/N016,
m=2,3, L=6 A32/N016
-4
Fig. 3.34 The r.e. of
SKN2/N016
A23/N016
240
171
218
151
180
/1.1.131
111
120
98
0.01
91
9.1
lee
1
0.01
0.1
d
1
10
100
d
Fig. 3.35 The r.e. of
RM6C10/N016,
RM1C4/N016
Fig. 3.36 The r.e. of
UD075/N016
RM6C10/N016
-+ RM1C4/N016
1500
500
1200
408
900
300
I.
600
280
300
lee
0
0
0.01
8.1
1
d
10
108
0.01
0.1
1
10
100
68
Fig. 3.37 The r.e. of XX/N016, XX=A32, A23,
SKN2, UD075, RM6C10, RM1C4
A32/N016
A23/N016
-41
I
i
1
I I
1
1
1
1
1
ink
360
...
AkNge4,4Q16
240
180
U
*.1
120
60
0
8.81
0.1
1
d
10
100
69
A32 are more efficient (see Fig. 3.37).
Since the Spearman-
Kdrber and non-adaptive methods are less efficient for d41,
and the Robbins-Monro and up-and-down methods demonstrate
more variability in estimating the ED50,
the adaptive
designs A23 and A32 are more consistently efficient.
The Spearman-Kdrber estimator, Robbins-Monro process,
and up-and-down method are designed for estimating the ED50.
They cannot be used to estimate the potency curves.
The
adaptive designs cannot only be used to estimate the ED50,
but also can be used to estimate the potency curves.
70
4 NORMAL APPROXIMATION METHOD TO ESTIMATE THE ORDER OF THE
AUTOREGRESSIVE (AR) MODEL UNDER BAYESIAN POINT OF VIEW
4.1 The Bayesian Approach to Order Estimation of AR Process
Let v. be the prior probability that the order of a
stationarity autoregressive time series is j (j=0,1,...,M)
such that E vi=1, where M is the maximum order of the AR
model.
Let Xn:=(Xl,
Xn) '
be a vector of n consecutive
observations and
90e=(91,...,9m)
autocorrelations,
from a stationary AR(p)
unknown.
be
a vector of partial
model with p
The joint density of X, p, and 0 (Robb (1980)) is
given as
41,1:40(Zni j
41)//)
= Gn
II
m
2-3 [
k=j+1
8 (9k) ]
[
II
-7:=0
(1(PD 2]
[2-1Km(pm)
n
(4.1)
where
__n
2 r (yn)
Gn = (2v)
(n+3)
Yn
2
(p0 s 0
M
II 8 (tpk)
E 1
k=14+1
(4.2)
6() is the Dirac delta function and Km(%) is defined as
(1.37).
From (4.1), Robb (1980) expressed the marginal posterior
probability density of the order given the data as
71
ff1
1
4, (in,
chpi
'Pm) cl(Pm
(2En, k, (pm) dcpm
k=0
civ1
1
n+3
r i (1-4)1)
Ki ( gi)
chpi
2
chpi
n+3
1:
k=o
-
4 (9 k)
(1-91)1]
-1 nic2-jc[II i=0
1
2 dVk
dV1
I
E Ik
k=0
(4.3)
where
1
=
n+3
1
-I ICk2-ic
(1-91)-511c(1Pk)
2
thp
chpi
.
(4.4)
The Bayes estimator is used to minimize the Bayes risk
with
respect
to
the
loss
Robb chose
function.
the
particular loss function as:
L(0, a) = 0
= 1
if e=a (decision is correct)
otherwise (decision is wrong)
.
(4.5)
The Bayes risk is defined by
R(0, 8) = E( L(0,8(2))]
=
f F(8,1) dlix
(4.6)
where
F (a, I )
f(X,O)
=
fe
L(e,
a) f(.7f, 0) do (0)
.
is the density of X given 0,
(4.7)
v(0)
is the prior
probability distribution for 0, and x and 6 are the domain
72
of X and 0,
respectively.
minimizing F(6(8), X).
F(L.in) = E
So, R(0,
6)
is minimized by
In the present case, we have
L(k,j) f(Jrn,k)Irk
k=0
L(k,j) [f(27,,;k)/nk]uk
k=0
= E L(k,j)
k=0
= krj
E 1k
(4.8)
for some je(0, 1, ..., M).
If j is chosen to be the integer
between 0 and M such that I.
is a maximum, the function
F(j,8n) is minimized and j should be a Bayes estimator of
the order p.
Robb derived the approximated posterior
probability of the order given data to simplify the multiple
integration in (4.4).
And Robb wrote (4.3) as
Itpir(i lira)
7C.12-i (21c/ (2yn+1)
i+1
1
32[
2
1=1
kk
Enk2-k[2n/(2yn+1] 2 H
k=0
(1-91)
II
(4.9)
2
.1=0
where (ei is the approximated mle of
p1.
One way to deal with the multiple integrations in (4.3)
is to use the closed Newton-Cotes integration formulas
(Burden (1981)) to approximate Ij in (4.4).
the closed Newton-Cotes formulas
is
in
The boundary of
(-1,
quadrature formula for Newton-Cotes is given as
1].
The
73
Ef (x) dx z E
ci f (xi)
(4.10)
where Ci and xi (1=1,
N) are the weight coefficients and
the roots of the function, respectively.
In (Burden, 1981),
we can find the corresponding Ci and x. for N up to 5.
With
Newton-Cotes closed formula, we have
/k = 7C k 2 -k
E
N
. . .
E {C1i
ii=0
2
cik (3.-(pid
21
k
(1-(pL)7KkOpii, ..., (pi)
for k=1, ..., M.
.
ik.°
-(
n4.3)
2
(4.11)
}
In general, the answer is accurate enough
if N=3 because the degree of precision is up to (2N-1) for
Newton-Cotes closed formula.
To deal with the multiple
summations is still a big problem because we have to run Nk
iterations to get the final answer and Kk(tpk) is difficult
to calculate.
It takes a very long time to get the answer
if k is large even with modern computers.
In the following
section the normal approximation method will be applied
again to solve the computation problem mentioned above.
4.2
Normal Approximation Approach
The marginal probability density of order p given the
data is shown as (4.3).
Ik= ji. il f
ri
k (fp k )
The Ik in (4.4) can be expressed as
k41
(4.12)
where
74
-(n+3)
fk(+11
f(pk)
= 7tk2-k [11 (1 -(p j) 2] Kk(9k)
1=0
2
(4.13)
To apply the normal approximation we will approximate fk(9k)
by a constant
Ck
multiplied the multivariate normal density
function of ifk with mean c
and covariance E* and express
as
f k (9 k )
fk ((Pi'
(Pk)
Ck MVN(111k; op*, E*)
k
-1
Ck(±)
71E* I2- exp{--21
271
(,,*) Es"
(9-9*)1
(4.14)
where 9* has to be satisfied with
=
affik
(4.15)
and
-1
-a2logfic(9k)
E*
1(
a9 &pi
)idtp.
(4.16)
Substitute 9* into (4.14) and get
_1
k
fk(11)*)
E* I
C k (+; )
2
(4.17)
Substituting (4.16) into (4.17), we can find
-1
Ck
fk(C) (2 a)
-a2logfk(4pk)
Ck
as
1
2
2
[(
a(Pkavi
(4.18)
From (4.12) and (4.14) we have
75
Ik
p
=
i
pi
Li fk (4x) dq)k
chPi
mvN(Ipk; "% Es) dpk
Ck
d(pi.
= Ck
(4.19)
The multiple integrations of Ik has been simplified and
approximated only by a constant Ck in (4.19).
So we can
approximate (4.3) as
C
'Piz('
f.
(i=o,
M
E Ck
. . .
,
(4.20)
k=0
where C k is given in (4.18).
However,
before approximating Ik by Ck.
used here to find 9* and Z.
Ui =
alogfk(pk)
api
dUij
The Newton-Raphson method is
Define
(1=1,
.
.
. ,
k)
a Icopkd
n+3
(4.21)
'
2(9k)
1-1
,
and E* are needed
1Cic
a2 log fk (tpk)
acpiacpi
i (1+(p1)
(1_91) 2
+
n+3
2 Kk(41$k)
(n+3)
[
[ a2
vlopk,,
a(pi.,...k
a 4.k
re-%.ric,
(, \
i=j
2Kk (4k) ' °, i
24 (9k)
,
a2
(n+3)
a(PiU(Pi
n+3
21(k(f p
1
(4.22)
gki
a 41..k
w.f..,
%Irk,
v tin
--k
kil
ij
.
oc)
In order to obtain the first and second derivative of Kk(tpk)
76
with respect to
we we need to express all of the functions
of 9k in terms of O's.
From (1.34) and (1.37), we have
Kk (41k) = Hk (0k)
(4.23)
1, kDke1, k
Applying chain rule we obtain
a
k.
---=( 2 D I 8 1 .
K (9 k )
(4.24)
where
aelk
a9,
Dk*
(aek,i
_
is same as
Dk
(4.25)
without the first row and 01,k = (1, 0k,1
The expressions Oksi (j=1,
ek,k)
k) in terms of 9
is shown in (1.32), i.e.,
ek,k = Ok
0
0 k-1, j
fk°k-1,k-j
(j=1,
k-1)
Therefore, each element in (4.25) can be found as
aek,,
_
aek-1,m _A
aok-i,k_m
s'k.k
k-1)
(4.26)
aek,,
uk-1,k-m
avk
(m=1,
..., k-1)
(4.27)
aok,k _ 0
ay,
a ek,k
(i,m=1,
(i=1,
k-1)
(4.28)
1
a (Pk
Differentiate (4.24) and (4.25) and get
(4.29)
77
a2
)
414;
alEk
*
r
aoia9i
L2/21,01, ki +
[2 D*k*
c#3°
(4.30)
aqvk
aPe
( a2eLl
.
a(piapj
Lk
(i,J=1,...,k)
a(Pia9.1'...- &Piaci
(4.31)
where Dk** are same as Dk without the first row and the first
column of Dk.
Similarly, we can express each element in
(4.31) as
aNkm
a2ek-1,m
4i4j
a2ek,m
_
a2ok,rn
aq) jay
a2ek,n,
439 jag)
89,
(i,j=1,2,
- 0
41
a20k-1,k-m _
(4.32)
aok-i,
4payi
.3914k
a213k,
k, k
aq, jag,
(4.33)
k-1)
(4.34)
for m=1,
..., k-1 and
""
424,
for m=k.
0
(i,j =1,
...,
(4.35)
Let
9 = (91,
cd
u
(4.36)
dU = [dUu]
(i,j=1,
k) .
The mode 9* can be found by using Newton-Raphson method
shown as
(pro = yr - U(4r) de (yr)
for (r+1)
to
iteration.
(4.37)
From (1.43) we can evaluate the least
78
square estimates 13*=(0 p1 *
.
. . ,0
*)
which will be used to
choose the initial value of cp, i.e.,
0* = [D "Yid
(p=1,
1.4 = ((Qi,
In
(4) I r =0 =
(4.38)
k)
u1,1
*
(4.39)
ekA)
If the sequence converges, say, to 9*, then (1)*=((p1 *,
is the mode.
,ck*)
Substituting e* into (4.22), we can solve for
E*, i.e.,
E* =
d0) -11
(4.40)
according to (4.16) and (4.25).
Once we obtain 9* and E*,
C k can be calculated from (4.13) and (4.18), i.e.,
Ck = IC k2-Ic{k
IT (1-
(C)2) i Kk(r)
2
-(n+3)
2
k
1
(270 21E4.12
i=0
4.3
(4.41)
Examples for Wolfer's Sunspot data
Wolfer's sunspot data consists monthly means of daily
relative sunspot number which are based upon counts of spots
and groups of spots beginning in 1749.
The yearly means of
sunspot data observed in 176 consecutive years which can be
found in most of time series analysis books.
current
data
is
collected
by
observatory from 1749 to 1977.
the
Tokyo
The most
Astronomical
We transform the data by
taking square root of this sunspots data from 1749 to 1924
corrected for the mean.
to comparing each other.
The following methods will be used
Akaike's future prediction error
(FPE) and information criterion (AIC), Schwarz's Bayesian
information
criterion
(BIC)
and
Hannan
and
Quinn's
79
information criterion (CIC), Robb's Bayes estimation of the
order and the Bayesian normal approach all choose a maximum
order M=15 to estimate the order of AR time series.
For
Robb's Bayes estimator of order and the Bayesian normal
approach, let /7)=1/16, j=0,1,
function as (4.5).
..., 15 and define the loss
The estimation criteria for AR order are
list as following.
FPE(p) =
min/FPE(j) =
AIC(p) = minIAIC(j) =
BIC(p) = minIBIC(j)
CIC(p) =
minICIC(j)
InRobb (p) = max
n +j+1R
nj1
j = 0, 1,
1nRi + 2j
n
I j = 0, 1, ..., M}
= 1nR. + i inn
.7
=
+ j lnlnn
1nR
n
.7
iiii}
-1
j = 0, 1, ..., MI
1
I
j
.7
= 0 , 1,
M
1
1
Ail,
1
Ii
pix(jlin)
. . .
..; = 0, 3., ...
,m
E 1k
k=0
Normal (p)
= max
pix( j lin) =
mCi
E ck
I j = 0, 1,
. . .
,M
kO
where R. (1.44) is the role of the variance a2 from a model
J
with j parameters estimated and rpfx(ji Xn) are estimated by
(4.9) and Cj can be calculated from (4.41).
The estimation results of analyzing the transformed
sunspot data from these methods is given in Table 4.1.
Akaike's FPE and AIC both have the absolute minimum value at
80
the 9th order but Schwarz's BIC, Hannan and Quinn's CIC and
Robb's and the Bayesian normal approach all have the same
estimated order at the
2nd
order,
which means all the
consistent estimators for order yield the same results.
The
asymptotic results of Schwarz Bayesian criterion do not
depend on the prior distribution and our Bayesian normal
approach has the same results as Schwarz's BIC results and
both put very large weights on the 2nd and 3"1 orders.
The closed Newton-Cotes method mentioned in section 4.1
is very accurate, but the running time is very much.
It is
good to compare this method with the normal approximation
method for M=12 and n=3.
The results is shown in Table 4.2
Both methods have the
same
results
that the maximum
posterior probability of the order is at j=2.
However, the
running time of the normal approximation approach is much
less than the Newton-Cotes method.
Consider the most current sunspot data collected from
1749 to 1977.
By using FPE, AIC, BIC, CIC, and Bayesian
normal approach to analyze the mean corrected square root of
the yearly averages (the maximum order M=15), the results is
shown in Table 4.3 and plot of the data is given in Fig.
4.1.
The best fitted AR model is the 9th order for the most
current transformed sunspots data for all criteria.
FPE,
AIC,
BIC,
and CIC do not give full posterior
results, but the Robb's and normal approximation approach
do.
81
Table 4.1
The results of analyzing Wolfer's sunspot data
for 1749-1924 using FPE, AIC, BIC, CIC, Robb, and
normal approximation metnoa ror rne oraer p
701(01 Xn)
j
FPEJ .
AICJ
.
BIC.
CIC.
J
J
Robb
Normal
0
5.3565
1.6669
1.6669
1.6669
.0000
.0000
1
1.7905
0.5711
0.5891
0.5784
.0000
.0000
2
1.0395
0.0273
0.0633*
0.0419*
.7677^
.6678^
3
1.0383
0.0262
0.0802
0.0481
.2070
.2793
4
1.0496
0.0370
0.1091
0.0663
.0204
.0272
5
1.0584
0.0454
0.1354
0.0819
.0024
.0042
6
1.0535
0.0407
0.1487
0.0845
.0008
.0018
7
1.0371
0.0249
0.1511
0.0761
.0008
.0053
8
1.0295
0.0177
0.1618
0.0761
.0000
.0023
9
1.0115*
0.0000*
0.1621
0.0658
.0000
.0107
10
1.0201
0.0834
0.1885
0.0814
.0000
.0011
11
1.0276
0.0157
0.2138
0.0960
.0000
.0002
12
1.0394
0.0270
0.2432
0.1147
.0000
.0000
13
1.0511
0.0381
0.2723
0.1331
.0000
.0000
14
1.0508
0.0378
0.2899
0.1401
.0000
.0000
1.0627
0.0489
0.3192
0.1586
.0000
'A': indicates that the value is maximum.
'*': indicates that the value is minimum.
.0000
15
82
Table 4.2
j
The results of analyzing Wolfer's sunspot data
for 1749-1924 from the closed Newton-Cotes (N=3)
and Bayesian normal approximation method for
choosing the maximum order M=12
closed Newton-Cotes
Normal
0
0.0000
0.0000
1
0.0000
0.0000
2
0.5556*
0.6678*
3
0.2469
0.2793
4
0.1098
0.0272
5
0.0488
0.0042
6
0.0217
0.0018
7
0.0096
0.0053
8
0.0043
0.0023
9
0.0019
0.0107
10
0.0008
0.0011
11
0.0004
0.0002
12
0.0002
'*': indicates that the value is maximum.
0.0000
83
Table 4.3
j
The results of analyzing Wolfer's sunspot data
for 1749-1977 using FPE, AIC, BIC, CIC, and
normal approximation method for the order p
assumed the maximum order M=15
FPE.
AICJ
.
BICJ .
J
CIC.J
701E01 Xn)
0
6.3096
1.8333
1.8333
1.8333
0.0000
1
2.0921
0.7294
0.7444
0.7355
0.0000
2
1.1353
0.1182
0.1482
0.1303
0.0022
3
1.1308
0.1142
0.1592
0.1324
0.0009
4
1.1408
0.1229
0.1829
0.1471
0.0000
5
1.1440
0.1258
0.2008
0.1561
0.0000
6
1.1109
0.0964
0.1864
0.1327
0.0000
7
1.0879
0.0755
0.1804
0.1178
0.0006
8
1.0634
0.0526
0.1726
0.1011
0.0011
9
1.0088*
0.0000*
0.1349*
0.0544*
0.9109
10
1.0173
0.0084
0.1583
0.0689
0.0769
11
1.0261
0.0169
0.1819
0.0835
0.0066
12
1.0349
0.0255
0.2054
0.0980
0.0006
13
1.0439
0.0341
0.2290
0.1127
0.0000
14
1.0392
0.0295
0.2394
0.1142
0.0000
15
1.0479
0.0378
0.2627
0.1285
'A': indicates that the value is maximum.
'*': indicates that the value is minimum.
0.0000
.,
Fig. 4.1 Mean corrected of the square root of yearly
averages of sunspots data 1749-1977
0
50
100
150
200
85
CONCLUSIONS
5
In Bayesian analysis, means are commonly used to
summarize Bayesian posterior distributions.
a large number of parameters,
Problems with
often require numerical
integrations over many dimensions to obtain means.
In this
dissertation, posterior modes with respect to appropriate
measures
were
distributions.
used
to
summarize
Bayesian
posterior
Two statistical estimation problems were
investigated here. These were the sequential dose selection
in
bioassay,
and
the
selection
of
the
order
of
an
autoregressive model.
First, for Bayesian bioassay, two adaptive designs were
developed for sequential dose selection to estimate the
potency curve by using posterior modes.
The first was a
full information method employing the full likelihood for
all collected doses and using a full prior distribution to
obtain modes and estimate the potency curve.
the
reduced
information
method
for
The second was
simplifying
complexity of the full information method.
the
In this second
procedure, the Dirichlet prior was modified by updating the
prior parameters in each step.
Modes were obtained at
experimental doses in each step, so the full modal potency
curve was determined according to these modes and the
previously estimated potency curve.
The relative efficiencies of the adaptive designs for
estimating the
ED50
were compared.
For prior guess
86
close
functions
to
the
true
function,
the
reduced
information method involving assignment of subjects to doses
close to the ED50,
is more efficient.
For prior guess
functions which are not close to the true function, the full
information method of choosing nearly equal numbers of doses
and steps is more efficient.
A comparison of the relative efficiencies of the
adaptive designs with other non-Bayesian methods (SpearmanKarber, up-and-down, and Robbins-Monro) shows that the full
information is appropriate for estimating the ED50 when the
prior guess function is not close to the true function.
The
reduced information method is less efficient than other
methods for estimating the ED50 when the prior guess is
close to the true function. These non-Bayesian methods were
designed for estimating the ED50 only, while the adaptive
designs were designed for estimating both the ED50 and the
potency curve.
Second, determination of the order of an autoregressive
model following Robb's method
marginal
posterior
considered.
(1980)
probabilities
of
by evaluating the
the
order
was
The normal approximation method was used to
approximate a function in the posterior density such that
the
integrations
simplified.
over
many
dimensions
problem
was
This method was compared with other methods
(FPE, AIC, BIC, and CIC) by choosing Wolfer's sunspot data
an example.
FPE,
AIC,
BIC,
and CIC were developed to
87
estimate the order of an autoregressive model.
In contrast
to Robb's method and the normal approximation approach,
these methods do not give full posterior results.
Bayesian methods
(Robb's,
BIC,
All the
and normal approximation
approach) have the same estimated order.
For
further
inference,
approximate
posterior
distributions can be based on the multivariate normal
distribution.
88
BIBLIOGRAPHY
1. Akaike, H. (1969), "Fitting Autoregressive Models for
Prediction," Annals of the Institute of Statistical
Mathematics, 21, 243-7.
2. Akaike, H. (1970), "Statistical Predictor
Identification," Institute of Statistical Mathematics,
21, 203-17.
3. Akaike, H. (1974), "A New Look at the Statistical
Model Identification," IEEE Transactions on Automatic
Control, v.19, 716-23.
4. Antoniak, C.E. (1974), "Mixtures of Dirichlet Process
with Applications to Bayesian Nonparametric Problems,"
Ann. Statist. 2, 1152-74.
5. Ayer, M., Brunk, H.D., Ewing, G.M., Reid, W.T., and
Silverman, E. (1955), "An Empirical Distribution
Function for Sampling with Incomplete Information,"
Ann. Math. Statist., 26, 641-7.
6. Barlow, R.E., Bartholomew, D,J., Bremner, J.M., Brunk,
H.D. (1972), Statistical Inference under Order
Restrictions, Wiley.
7. Barndorff-Nielsen, O., Schou, G. (1973), "On the
Parametrization of Autoregressive Models by Partial
Autocorrelations," Journal of Multivariate Analysis,
3,
408-19.
8. Box, G.E.P. and Jenkins, G.M. (1970), Time Series
Analysis: Forecasting and Control, Holden-Day, San
Francisco.
9. Box, G.E.P. and Tiao, G.C. (1972), Bayesian Inference
in Statistical Analysis, Addison Wesley, California.
10. Broemeling, L.D. (1985), Bayesian Analysis of Linear
Models, Dekker.
11. Brunk, H.D. (1970), Estimation of Isotonic Regression,
Nonparametric Techniques in Statistical Inference,
M.L. Puri (ed), Cambridge University Press, 177-94.
12. Brunk, H.D. (1975), An Introduction to Mathematical
Statistics, 3rd ed., Xerox.
13. Burden, R.L., Faires, J.D., Reynolds, A.C. (1981),
Numerical Analysis, 2nd ed., Prindle, Weber & Schmidt.
89
14. Cochran, W.G. and Davis, M. (1965), "The RobbinsMonro Method for Estimating the Median Lethal Dose,"
Journal of the Royal Statistical Society, B, 27,
28-44.
15. Diaconis, P. and Ylvisaker, D. (1983),
"Quantifying
Prior Opinion," Tech. Report NO. 207, Department of
Statistics, Stanford University, Stanford, California.
16. Disch, D. (1981), "Bayesian Nonparametric Inference for
Effective Doses in a Quantal-Response Experiment,"
Biometrics, 37, 713-22.
17. Dixon, W.J. (1965), "The Up-and-down Method for Small
Samples," Journal of the American Statistical
Association, 60, 967-978.
18. Dixon, W.J. and Mood, A.M. (1948), "A Method for
Obtaining and Analyzing Sensitive Data," Journal of the
American Statistical Association, 43, 109-126.
19. Ferguson, T.S. (1973), "A Bayesian Analysis of Some
Nonparametric Problems," Ann. Statist., 1, 209-30.
20. Ferguson, T.S. and Phadia, E.G. (1979), "Bayesian
Nonparametric Estimation Based on Censored data," Ann.
Statist., 7, 163-86.
21. Finney, D.J. (1964), Statistical Method in Biological
Assay, 2nd ed., New York, Hafner Publishing Company.
22. Hannan, E.J., and Quinn, B.G. (1979), "The Determination
of the Order of an Autoregression," Journal of Royal
Statistics, Soc. B, 41, 190-95.
23. Jeffreys, H. (1966), Theory of Probability, 3rd ed.,
Oxford University Press.
24. Johnson, N.L. and Kotz, S. (1972), Distributions in
Statistics, Continuous Multivariate Distributions,
Wiley, New York.
25. Johnson, R.A.(1967), "An Asymptotic Expansion for
Posterior Distributions," Ann. Math. Statist, 38, 18991907.
26. Johnson, R.A. (1970), "Asymptotic Expansions
Associated with Posterior Distributions," Ann. Math.
Statist, 41, 851-64.
27. Kotz, S., Johnson, N.L., Read, C.B. (1981),
Encyclopedia of Statistical Sciences, 4, 354-7, Wiley
90
Interscience.
28. Kotz, S., Johnson, N.L., Read, C.B. (1981),
Encyclopedia of Statistical Sciences, 7, 503-5, Wiley
Interscience.
29. Kraft, C.H. and Van Eeden, C. (1964), "Bayesian
Bioassay," Ann. Math. Statist, 35, 886-90.
30. Kuo, L. (1983), "Bayesian Bioassay Design," Ann.
Statist., 11, 886-95.
31. Kuo, L. (1988), "Linear Bayes Estimators of the Potency
Curve in Bioassay," Biometrics, 75, 91-6.
32. Lindley, D.V. (1965), Introduction to Portability and
Statistics from a Bayesian Viewpoint, Part 2,
Inference,
Cambridge University Press.
33. Little, R.E. (1974), "A Mean Square Error Comparison
of Certain Median Response Estimates for the Up-andDown Method with Small Samples," Journal of the American
Statistical Association, 69, 202-206.
34. Lutkepohl, H. (1985), "Comparison of Criteria for
Estimating the Order of a Vector Autoregressive
Process," Journal of Time Series Anal., 6, 35-52.
35. Press, J.S. (1989), Bayesian Statistics, Wiley
Interscience.
36. Ramsey, F.L.(1972), "A Bayesian Approach to Bioassay,"
Biometrics, 28, 841-58.
37. Ramsey, F.L.(1974), "Characterization of the Partial
Autocorrelation Function," Annals of Statistics, 2,
1296-1301.
38. Rissanen, J. (1978), "Modeling by Shortest Data
Description," Automatica, 14, 465-71.
39. Robb, L.J. (1980), "Estimation of the Order of an
Autoregressive Time Series -- A Bayesian Approach," Ph.
D. Dissertation, Oregon State University, Corvallis.
40. Robertson, T., Wright, F.T., Dykstra, R.L. (1988),
Order Restricted Statistical Inference, Wiley.
41. Schwarz, G. (1978), "Estimating the Dimension of a
Model," Ann. Stat, 6, 461-64.
42. Shibata, R. (1976), "Selection of the Order of an
91
Autoregressive Model by Akaike's Information Criteria,"
Biometrika, 63, 117-26.
43. Wetherill, G.B. (1963), "Sequential Estimation of
Quantal Response Curves," Journal of the Royal
Statistical Society, B, 25, 1-48.
44. Wilks, S.S. (1962), Mathematical Statistics, Wiley,
New York.
APPENDIX
92
The fixed doses and the estimated ED50 for 64 possible
outcomes when L=6 (L=m*n) for Anm and Bnm designs.
Al6=B16
X1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1 1
1
0 0
0 0
0
0
0
1
1
1
1
0
0
0
1
1
0
1
1
1
0
0
0
0
1
1
1
1
0
1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
1
0 0 0
0 0 1
0 1 0
0
1
1
1
1 1
0 0
0 1
1 0
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
1
1
1
0
0
0
0
1
1
1 1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
1
0
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
X2
X3
X4
X5
X6
ED50
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.88088
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.75466
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.70695
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.62746
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.80740
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.59947
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.54918
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.50000
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.84868
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.70227
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.61619
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.50000
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.58150
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.42056
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.40795
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.37254
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.86009
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.72231
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.64861
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.57944
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.67704
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.50000
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.46987
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.40053
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.72269
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.50000
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.41513
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.29773
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.38546
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.27769
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.27154
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.24534
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.86314
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.72846
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.66341
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.59205
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.71972
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.53013
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.50000
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.45082
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.75553
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.58487
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.50000
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.38381
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.45005
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.35139
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.33659
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.29305
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.77069
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.61454
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.54995
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.41850
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.50000
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.32296
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.28028
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.19260
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.50000
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.27731
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.24447
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.15132
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.22931
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.13991
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.13686
0.14286 0.28571 0.42857 0.57143 0.71429 0.85714 0.11912
93
A23
S
X1
0
0
0
0
0 0
0 1
1 0
1 1
1 0 0
1 0 1
1
0
1
1
1
0
0
0
0
0 0
0 1
1 0
1 1
1
1 0 0
1 0 1
1 1 0
1
1
0
0
0
0
1
0 0
0 1
1 0
1 1
1 0 0
1 0 1
1
0
1
1
1
0
0
0
0
1
0 0
0 1
1 0
1 1
0 0
1
1 0 1
1 1 0
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
1 0 0
1 0 1
1 1 0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
1
0
0
0
1
1
1 0
1 0
1 1
0
1
1
1
0
0
0
0
0
0
0
0
1
1
1
0
1
0
1
0
0 0
0 1
1 0
1 1
1 0 0
1 0 1
1
1
0
1
1
1
X2
X3
X4
X5
X6
ED50
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.91389
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.81711
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.80461
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.75612
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.89677
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.79034
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.72100
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.59958
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.73701
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.64491
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.59235
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.54288
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.72158
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.60265
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.52031
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.41999
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.84712
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.69663
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.59456
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.46488
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.74798
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.52913
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.43092
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.31239
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.58001
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.45712
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.39735
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.35509
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.47969
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.40765
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.27842
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.26299
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.87042
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.75936
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.70830
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.56017
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.80182
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.65353
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.53717
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.34171
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.68761
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.53512
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.47087
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.30337
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.56908
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.40544
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.25202
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.15288
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.65829
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.43983
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.34647
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.24064
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.46283
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.29170
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.19818
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.12958
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.40042
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.24388
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.20966
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.18289
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.27900
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.19539
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.10323
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.08611
94
A32
S
X1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
1
1
1
0
0
0
0
0 0
0 1
1 0
1 1
1
1 0 0
1 0 1
1 1 0
1
0
0
0
0
1
1
1
1
1
0 0
0 1
1 0
1 1
0 0
0 1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
1
0
1
0
0
0
0
1
1
1
0
0
1
1
0
0
1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
1
0
1
0
1
X2
X3
X4
X5
X6
ED50
0.33333 0.66667 0.56667 0.81333 0.74704 0.89766 0.92088
0.33333 0.66667 0.56667 0.81333 0.74704 0.89766 0.83865
0.33333 0.66667 0.56667 0.81333 0.74704 0.89766 0.89815
0.33333 0.66667 0.56667 0.81333 0.74704 0.89766 0.73875
0.33333 0.66667 0.56667 0.81333 0.63702 0.76797 0.78463
0.33333 0.66667 0.56667 0.81333 0.63702 0.76797 0.70641
0.33333 0.66667 0.56667 0.81333 0.63702 0.76797 0.76484
0.33333 0.66667 0.56667 0.81333 0.63702 0.76797 0.62898
0.33333 0.66667 0.56667 0.81333 0.49356 0.86510 0.87643
0.33333 0.66667 0.56667 0.81333 0.49356 0.86510 0.76993
0.33333 0.66667 0.56667 0.81333 0.49356 0.86510 0.77721
0.33333 0.66667 0.56667 0.81333 0.49356 0.86510 0.54524
0.33333 0.66667 0.56667 0.81333 0.41558 0.71583 0.72316
0.33333 0.66667 0.56667 0.81333 0.41558 0.71583 0.55282
0.33333 0.66667 0.56667 0.81333 0.41558 0.71583 0.54787
0.33333 0.66667 0.56667 0.81333 0.41558 0.71583 0.40480
0.33333 0.66667 0.38889 0.61111 0.54585 0.67564 0.70564
0.33333 0.66667 0.38889 0.61111 0.54585 0.67564 0.63324
0.33333 0.66667 0.38889 0.61111 0.54585 0.67564 0.63265
0.33333 0.66667 0.38889 0.61111 0.54585 0.67564 0.52829
0.33333 0.66667 0.38889 0.61111 0.42746 0.57254 0.58914
0.33333 0.66667 0.38889 0.61111 0.42746 0.57254 0.50000
0.33333 0.66667 0.38889 0.61111 0.42746 0.57254 0.50000
0.33333 0.66667 0.38889 0.61111 0.42746 0.57254 0.41086
0.33333 0.66667 0.38889 0.61111 0.34884 0.65116 0.65375
0.33333 0.66667 0.38889 0.61111 0.34884 0.65116 0.50000
0.33333 0.66667 0.38889 0.61111 0.34884 0.65116 0.50000
0.33333 0.66667 0.38889 0.61111 0.34884 0.65116 0.34625
0.33333 0.66667 0.38889 0.61111 0.32436 0.45415 0.47171
0.33333 0.66667 0.38889 0.61111 0.32436 0.45415 0.36676
0.33333 0.66667 0.38889 0.61111 0.32436 0.45415 0.36735
0.33333 0.66667 0.38889 0.61111 0.32436 0.45415 0.29436
0.33333 0.66667 0.26667 0.73333 0.41763 0.82800 0.84524
0.33333 0.66667 0.26667 0.73333 0.41763 0.82800 0.74260
0.33333 0.66667 0.26667 0.73333 0.41763 0.82800 0.76086
0.33333 0.66667 0.26667 0.73333 0.41763 0.82800 0.53023
0.33333 0.66667 0.26667 0.73333 0.30458 0.69542 0.69885
0.33333 0.66667 0.26667 0.73333 0.30458 0.69542 0.50000
0.33333 0.66667 0.26667 0.73333 0.30458 0.69542 0.50000
0.33333 0.66667 0.26667 0.73333 0.30458 0.69542 0.30115
0.33333 0.66667 0.26667 0.73333 0.21739 0.78261 0.75831
0.33333 0.66667 0.26667 0.73333 0.21739 0.78261 0.50000
0.33333 0.66667 0.26667 0.73333 0.21739 0.78261 0.50000
0.33333 0.66667 0.26667 0.73333 0.21739 0.78261 0.24169
0.33333 0.66667 0.26667 0.73333 0.17200 0.58237 0.46977
0.33333 0.66667 0.26667 0.73333 0.17200 0.58237 0.25740
0.33333 0.66667 0.26667 0.73333 0.17200 0.58237 0.23914
0.33333 0.66667 0.26667 0.73333 0.17200 0.58237 0.15476
0.33333 0.66667 0.18667 0.43333 0.28417 0.58442 0.59520
0.33333 0.66667 0.18667 0.43333 0.28417 0.58442 0.44718
0.33333 0.66667 0.18667 0.43333 0.28417 0.58442 0.45213
0.33333 0.66667 0.18667 0.43333 0.28417 0.58442 0.27684
0.33333 0.66667 0.18667 0.43333 0.23203 0.36298 0.37102
0.33333 0.66667 0.18667 0.43333 0.23203 0.36298 0.29359
0.33333 0.66667 0.18667 0.43333 0.23203 0.36298 0.23516
0.33333 0.66667 0.18667 0.43333 0.23203 0.36298 0.21537
0.33333 0.66667 0.18667 0.43333 0.13490 0.50644 0.45476
0.33333 0.66667 0.18667 0.43333 0.13490 0.50644 0.23007
0.33333 0.66667 0.18667 0.43333 0.13490 0.50644 0.22279
0.33333 0.66667 0.18667 0.43333 0.13490 0.50644 0.12357
0.33333 0.66667 0.18667 0.43333 0.10234 0.25296 0.26125
0.33333 0.66667 0.18667 0.43333 0.10234 0.25296 0.16135
0.33333 0.66667 0.18667 0.43333 0.10234 0.25296 0.10185
0.33333 0.66667 0.18667 0.43333 0.10234 0.25296 0.07912
95
A61
S
>C1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
1
1
0
0
0
0
1
1
1 1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
1
0
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0 0 0
0 0 1
0
0
1
1
1
1 0
1 1
0 0
0 1
1 0
1
1
1
0
0
0
0
1
1
1
0
0
1
1
0
0
1
0
1
0
1
0
1
0
1
1
1
0
0
0
0
1
1
1
0
0
1
1
0
0
1
0
1
1
1
0
1
0
1
0
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1 1
1
>C2
>C3
)C4
C 5
x6
E r) 5 0
0.50000 0.62500 0.71429 0.78306 0.83727 0.88009 0.91354
0.50000 0.62500 0.71429 0.78306 0.83727 0.88009 0.85127
0.50000 0.62500 0.71429 0.78306 0.83727 0.80076 0.81595
0.50000 0.62500 0.71429 0.78306 0.83727 0.80076 0.78813
0.50000 0.62500 0.71429 0.78306 0.73699 0.75737 0.77029
0.50000 0.62500 0.71429 0.78306 0.73699 0.75737 0.74457
0.50000 0.62500 0.71429 0.78306 0.73699 0.72044 0.72736
0.50000 0.62500 0.71429 0.78306 0.73699 0.72044 0.71568
0.50000 0.62500 0.71429 0.65537 0.68373 0.70079 0.70914
0.50000 0.62500 0.71429 0.65537 0.68373 0.70079 0.69100
0.50000 0.62500 0.71429 0.65537 0.68373 0.66627 0.67450
0.50000 0.62500 0.71429 0.65537 0.68373 0.66627 0.65903
0.50000 0.62500 0.71429 0.65537 0.63257 0.64287 0.64942
0.50000 0.62500 0.71429 0.65537 0.63257 0.64287 0.63649
0.50000 0.62500 0.71429 0.65537 0.63257 0.62635 0.62885
0.50000 0.62500 0.71429 0.65537 0.63257 0.62635 0.62520
0.50000 0.62500 0.54593 0.58856 0.61200 0.62150 0.62424
0.50000 0.62500 0.54593 0.58856 0.61200 0.62150 0.61672
0.50000 0.62500 0.54593 0.58856 0.61200 0.59910 0.60589
0.50000 0.62500 0.54593 0.58856 0.61200 0.59910 0.59266
0.50000 0.62500 0.54593 0.58856 0.56329 0.57633 0.58332
0.50000 0.62500 0.54593 0.58856 0.56329 0.57633 0.56900
0.50000 0.62500 0.54593 0.58856 0.56329 0.55185 0.55723
0.50000 0.62500 0.54593 0.58856 0.56329 0.55185 0.54768
0.50000 0.62500 0.54593 0.50993 0.52846 0.53894 0.54349
0.50000 0.62500 0.54593 0.50993 0.52846 0.53894 0.53348
0.50000 0.62500 0.54593 0.50993 0.52846 0.51758 0.52310
0.50000 0.62500 0.54593 0.50993 0.52846 0.51758 0.51271
0.50000 0.62500 0.54593 0.50993 0.50050 0.50475 0.50757
0.50000 0.62500 0.54593 0.50993 0.50050 0.50475 0.50212
0.50000 0.62500 0.54593 0.50993 0.50050 0.50000 0.50019
0.50000 0.62500 0.54593 0.50993 0.50050 0.50000 0.50000
0.50000 0.37500 0.45407 0.49007 0.49950 0.50000 0.50000
0.50000 0.37500 0.45407 0.49007 0.49950 0.50000 0.49981
0.50000 0.37500 0.45407 0.49007 0.49950 0.49525 0.49788
0.50000 0.37500 0.45407 0.49007 0.49950 0.49525 0.49243
0.50000 0.37500 0.45407 0.49007 0.47154 0.48242 0.48729
0.50000 0.37500 0.45407 0.49007 0.47154 0.48242 0.47690
0.50000 0.37500 0.45407 0.49007 0.47154 0.46106 0.46652
0.50000 0.37500 0.45407 0.49007 0.47154 0.46106 0.45651
0.50000 0.37500 0.45407 0.41144 0.43671 0.44815 0.45232
0.50000 0.37500 0.45407 0.41144 0.43671 0.44815 0.44277
0.50000 0.37500 0.45407 0.41144 0.43671 0.42367 0.43100
0.50000 0.37500 0.45407 0.41144 0.43671 0.42367 0.41668
0.50000 0.37500 0.45407 0.41144 0.38800 0.40090 0.40734
0.50000 0.37500 0.45407 0.41144 0.38800 0.40090 0.39411
0.50000 0.37500 0.45407 0.41144 0.38800 0.37850 0.38328
0.50000 0.37500 0.45407 0.41144 0.38800 0.37850 0.37576
0.50000 0.37500 0.28571 0.34463 0.36743 0.37365 0.37480
0.50000 0.37500 0.28571 0.34463 0.36743 0.37365 0.37115
0.50000 0.37500 0.28571 0.34463 0.36743 0.35713 0.36351
0.50000 0.37500 0.28571 0.34463 0.36743 0.35713 0.35058
0.50000 0.37500 0.28571 0.34463 0.31627 0.33373 0.34097
0.50000 0.37500 0.28571 0.34463 0.31627 0.33373 0.32550
0.50000 0.37500 0.28571 0.34463 0.31627 0.29921 0.30900
0.50000 0.37500 0.28571 0.34463 0.31627 0.29921 0.29086
0.50000 0.37500 0.28571 0.21694 0.26301 0.27956 0.28432
0.50000 0.37500 0.28571 0.21694 0.26301 0.27956 0.27264
0.50000 0.37500 0.28571 0.21694 0.26301 0.24263 0.25543
0.50000 0.37500 0.28571 0.21694 0.26301 0.24263 0.22971
0.50000 0.37500 0.28571 0.21694 0.16273 0.19924 0.21187
0.50000 0.37500 0.28571 0.21694 0.16273 0.19924 0.18405
0.50000 0.37500 0.28571 0.21694 0.16273 0.11991 0.14873
0.50000 0.37500 0.28571 0.21694 0.16273 0.11991 0.08646
96
B23
S
X1
0 0 0
0 0 1
0
0
1
0
1
1
1
1 1
0 0
0 1
1 0
1
1
0
0
0
0
0 0
0 1
1 0
1 1
1
1
0
0
1 0
1
1
1
0
1
1
1
0 0 0
0 0 1
0 1 0
0
1
1
1 0 0
1 0 1
1
1
0
0
0
0
1
1
1
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
0 0
0 1
1 0
1 1
0 0
1
0
1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
0
0
0
1
1
1
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
X2
X3
X4
X5
x6
ED50
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.85739
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.81063
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.79552
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.75727
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.81756
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.77238
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.75034
0.25000 0.50000 0.75000 0.52778 0.78395 0.89198 0.63153
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.68589
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.62705
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.60803
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.55988
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.63576
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.57890
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.55117
0.25000 0.50000 0.75000 0.39938 0.59347 0.72942 0.50104
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.79152
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.66949
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.60322
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.47150
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.69985
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.50170
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.45810
0.25000 0.50000 0.75000 0.31404 0.55246 0.84208 0.38099
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.49896
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.44012
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.42110
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.37295
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.44883
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.39197
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.36424
0.25000 0.50000 0.75000 0.27058 0.40653 0.60062 0.31411
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.81664
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.75653
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.71131
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.56374
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.76543
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.62203
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.53704
0.25000 0.50000 0.75000 0.20833 0.66667 0.86111 0.32510
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.61901
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.52850
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.49830
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.33051
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.54190
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.39678
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.30015
0.25000 0.50000 0.75000 0.15792 0.44754 0.68596 0.20848
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.67490
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.43626
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.37797
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.24347
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.46296
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.28869
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.23457
0.25000 0.50000 0.75000 0.13889 0.33333 0.79167 0.18336
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.36847
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.24273
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.22762
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.18937
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.24966
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.20448
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.18244
0.25000 0.50000 0.75000 0.10802 0.21605 0.47222 0.14261
97
B32
S
0
0
0
0
1
1
X1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
1
0
0
0
0
1
0 0
0 1
1 0
1 1
0 0
1
0
1
1
1
0
1
1
1
0 0 0
0 0 1
0
1
0
0 1 1
1 0 0
1 0 1
0
1
1
1
1
1
0 0
0 0
0
0
0
1
1
0 1 1
1 0 0
1 0 1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
1
0 0 0
0 0 1
0
1
0
0 1 1
1 0 0
1 0 1
1
1
0
1
1
1
0 0 0
0 0 1
0
1
0
0 1 1
1 0 0
1 0 1
1
1
0
1
1
1
>C2
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
0.33333 0.66667
x3
X4
X5
X6
0.56667 0.81333 0.70000 0.87429
0.56667 0.81333 0.70000 0.87429
0.56667 0.81333 0.70000 0.87429
0.56667 0.81333 0.70000 0.87429
0.56667 0.81333 0.61442 0.79423
0.56667 0.81333 0.61442 0.79423
0.56667 0.81333 0.61442 0.79423
0.56667 0.81333 0.61442 0.79423
0.56667 0.81333 0.50000 0.84000
0.56667 0.81333 0.50000 0.84000
0.56667 0.81333 0.50000 0.84000
0.56667 0.81333 0.50000 0.84000
0.56667 0.81333 0.41429 0.74000
0.56667 0.81333 0.41429 0.74000
0.56667 0.81333 0.41429 0.74000
0.56667 0.81333 0.41429 0.74000
0.38889 0.61111 0.47619 0.70068
0.38889 0.61111 0.47619 0.70068
0.38889 0.61111 0.47619 0.70068
0.38889 0.61111 0.47619 0.70068
0.38889 0.61111 0.41163 0.58837
0.38889 0.61111 0.41163 0.58837
0.38889 0.61111 0.41163 0.58837
0.38889 0.61111 0.41163 0.58837
0.38889 0.61111 0.35714 0.64286
0.38889 0.61111 0.35714 0.64286
0.38889 0.61111 0.35714 0.64286
0.38889 0.61111 0.35714 0.64286
0.38889 0.61111 0.29932 0.52381
0.38889 0.61111 0.29932 0.52381
0.38889 0.61111 0.29932 0.52381
0.38889 0.61111 0.29932 0.52381
0.26667 0.73333 0.42857 0.82041
0.26667 0.73333 0.42857 0.82041
0.26667 0.73333 0.42857 0.82041
0.26667 0.73333 0.42857 0.82041
0.26667 0.73333 0.29396 0.70604
0.26667 0.73333 0.29396 0.70604
0.26667 0.73333 0.29396 0.70604
0.26667 0.73333 0.29396 0.70604
0.26667 0.73333 0.22857 0.77143
0.26667 0.73333 0.22857 0.77143
0.26667 0.73333 0.22857 0.77143
0.26667 0.73333 0.22857 0.77143
0.26667 0.73333 0.17959 0.57143
0.26667 0.73333 0.17959 0.57143
0.26667 0.73333 0.17959 0.57143
0.26667 0.73333 0.17959 0.57143
0.18667 0.43333 0.26000 0.58571
0.18667 0.43333 0.26000 0.58571
0.18667 0.43333 0.26000 0.58571
0.18667 0.43333 0.26000 0.58571
0.18667 0.43333 0.20577 0.38558
0.18667 0.43333 0.20577 0.38558
0.18667 0.43333 0.20577 0.38558
0.18667 0.43333 0.20577 0.38558
0.18667 0.43333 0.16000 0.50000
0.18667 0.43333 0.16000 0.50000
0.18667 0.43333 0.16000 0.50000
0.18667 0.43333 0.16000 0.50000
0.18667 0.43333 0.12571 0.30000
0.18667 0.43333 0.12571 0.30000
0.18667 0.43333 0.12571 0.30000
0.18667 0.43333 0.12571 0.30000
ED50
0.85333
0.81000
0.81000
0.73667
0.76949
0.72000
0.72000
0.67051
0.81333
0.72000
0.72000
0.56667
0.70333
0.57500
0.57500
0.46667
0.65873
0.60714
0.60714
0.51984
0.55891
0.50000
0.50000
0.44109
0.61111
0.50000
0.50000
0.38889
0.48016
0.39286
0.39286
0.34127
0.79048
0.72857
0.72857
0.55952
0.67070
0.50000
0.50000
0.32930
0.73333
0.50000
0.50000
0.26667
0.44048
0.27143
0.27143
0.20952
0.53333
0.42500
0.42500
0.29667
0.32949
0.28000
0.28000
0.23051
0.43333
0.28000
0.28000
0.18667
0.26333
0.19000
0.19000
0.14667
86
"I9E
s
0 0 0
0 0 I
0 I 0
0 I I
I 0 0
T 0 I
I I 0
I
T
I
0 0 0
0 0 1
0 T 0
0 I I
I 0 0
T 0 I
I
I
0
I
I
I
0 0 0
0 0 T
0 1 0
0
T
T
0 0
0 I
T I 0
I
T
I
I
I
0 0 0
0 0 1
0 T 0
0 T 1
i 0 0
I 0 I
I T 0
T
I
I
0 0 0
0 0 T
0 I 0
0 I I
I 0 0
I 0 I
I I 0
I
T
I
0 0 0
0 0 T
0 T 0
0 T I
1 0 0
I 0 T
I I 0
I
I
T
0 0
0 0 I
0 T 0
0 I 1
I 0 0
I
0 I
0
I
I
I
0
I
I
0 0 0
0 0 T
0 I 0
0
T
T
I
1
0 0
0 I
1 0
I
T
T
T
Ix
ZX
Ex
4X
1x
9x
OSCIE
00005'O 00SZ9'0 0000L'0 0005C0 ILS81:0 OSZI8'0 EEEE8'0
00005'0 00S19'0 0000L'0 000SC0 ILS8C0 OSZT8'0 L9T6L'0
00005'0 00SZ9'0 0000L'0 0005CO ILS8C0 E68SL'0 9L6LCO
00005'0 00SZ9'0 0000L'0 0005C0 IL58C0 E685C0 ETEEL*0
00005*0 00SZ9'0 0000L'0 00054:0 6ZPIL'0 LOTWO L999L'0
00005*0 00529'0 000000 0005C0 6Z4IC0 LOTWO PZOZCO
00005*0 00SZ9'0 0000L'0 0005CO 6Z4IL-0 SZT89'0 U80L:0
0000S-0 00529'0 000000 0005L'0 6n/1t:0 SZT89'0 000;9'0
00005*0 00529'0 0000L'0 00059'0 ILS89'0 SL81L*0 0005C0
0000S-0 00529'0 000000 00059'0 1L589'0 SL8TCO L9169'0
00005'0 00519'0 0000L'0 00059'0 ILS89-0 E68S9'0 9L6L9'0
00005'0 00519'0 0000L'0 00059'0 IL589'0 £6859'0 EEEE9'0
00005'0 00SZ9'0 0000L'0 00059'0 6IL09'0 L0T49'0 L9999'0
00005'0 00529-0 0000L'0 00059'0 4TL09'0 LOT69'0 90119'0
00005'0 00SZ9'0 0000L'0 00059'0 4IL09'0 05295'0 ZZL65'0
00005'0 00SZ9'0 0000L'0 00059'0 4TL09'0 05295'0 8LLZ5'0
00005-0 00SZ9'0 00055-0 00009'0 98249'0 OSL89-0 ZZZZL-0
0000S.0 00529'0 00055-0 00009'0 98Z49'0 OSL89'0 8L1S9'0
00005'0 00SZ9'0 00055-0 00009'0 98249'0 £6809'0 46Z£9-0
00005*0 00SZ9'0 00055-0 00009'0 98ZP9'0 £6809'0 EEE8S'0
00005-0 00SZ9'0 00055-0 00009'0 61495'0 L0165'0 L9919'0
0000S.0 00SZ9'0 00055'0 00009'0 6095'0 LOT6S'0 4ZOLS'0
00005'0 00529'0 00055'0 00009'0 61495'0 SZTES'0 E£855"0
0000S'0 00SZ9'0 00055-0 00009'0 61795'0 SZTES*0 00005'0
00005'0 00SZ9'0 00055*0 00005'0 ILSE5'0 SL895'0 00009'0
00005*0 00519'0 00055'0 00005-0 ILSES*0 1L89S*0 L9145'0
00005'0 00SZ9-0 00055'0 0000S-0 ILSES*0 £6805-0 9L6ZS-0
00005'0 00SZ9'0 00055-0 0000S-0 ILSE5'0 E6805'0 L990"0
00005*0 005Z9'0 00055'0 00005-0 LS8W0 41Z84'0 L99[5'0
00005'0 00SZ9'0 00055'0 00005'0 LS8W0 PIZWO 84044'0
00005*0 00SZ9'0 00055*0 00005'0 LS8Z4'0 005LE-0 L9916'0
00005*0 00529'0 00055*0 00005'0 LS8Z4'0 005LE'0 EEEEE'0
00005'0 00SLE'0 0000'0 00005*0 E41L5*0 00SZ9'0 L9999'0
00005*0 00SLE'0 0000'0 00005-0 E4TL5-0 00SZ9'0 EEE8S-0
0000S.0 4E-0 00 0000'0 00005*0 EPILS-0 98LIG*0 ZS6S5'0
00005-0 00SLU0 0000'0 0000S.0 £PIL5'0 98LIS*0 EEEWO
00005'0 00SLE'0 0000'0 00005'0 61494'0 LOI64'0 EEEES*0
00005*0 004E'0 0000'0 00005*0 614917'0 LOTWO 4ZOL6*0
0000S-0 00SLE'0 0000-0 00005'0 61494'0 SZTE4'0 EE8S6'0
00005*0 00SLUO 0000'0 00005*0 61494'0 SZIWO 0000'0
00005*0 00SLE'0 0000'0 000017-0 ILSWO SL894'0 00005'0
00005"0 005LE*0 0000'0 00001e0 IL50'0 SL80'0 L9I64'0
00005'0 004E*0 0000'0 000017'0 USW() E6804-0 9L6W0
00005*0 00SLE'0 0000'0 00004'0 USW() £6800-0 ££E8C0
00005'0 00SLE'0 0000'0 00004'0 4ILSE-0 LOIWO L99T4*0
0000S.0 005LCO 0000'0 00004'0 4ILSE-0 LOI6E-0 90L91"0
00005'0 00SLUO 0000'0 00004'0 4ILSE*0 OSZTUO ZZLVE*0
00005*0 00SLE'0 0000-0 000017'0 4ILSE-0 OSZIE'0 8LLI.r0
00005*0 00SLE'0 0000E"0 000SE-0 98Z6E*0 05LE4*0 ZZZL4*0
00005'0 00SLE'0 00001'0 0005E*0 9816£'0 OSLE4°0 8LZ06*0
0000S.0 00SLE'0 0000£'0 0005£'0 98Z6C0 £685C0 46Z8E'0
00005-0 00SLUO 0000£'0 000SE-0 9816E'0 £685E-0 EEEEE'0
00005*0 00SLE'0 00001'0 0005E-0 60TE-0 LOTPUO L9991'0
0000S.0 00SLE'0 0000£'0 000SE*0 6ZPIE*0 LOI4£'0 4ZOZE'0
0000S.0 00SLE'0 0000E-0 000SE'0 6Z4I£'0 SZT8Z-0 £E80£'0
00005-0 00SLE'0 0000E-0 0005E'0 6Z4TE'0 SZI8Z°0 00051-0
00005'0 00SLE'0 0000E'0 0005Z-0 ILS8Z'O SL8TE'0 0005E-0
00005*0 00SLE'0 0000E'0 00051'0 TLS8Z'0 5L8T1'0 L916Z'0
00005*0 00SLE'0 0000E-0 00051*0 ILS8Z*0 E68SZ*0 9L6LZ'0
00005'0 00SLE'0 0000E*0 000SZ-0 ILS8r0 £68SZ'0 EEEEZ'O
00005*0 00SLE'0 0000£'0 00051'0 6Z6IZ'0 LOIVZ'O L999r0
00005'0 00SLE'0 0000£'0 000S1*0 6ZPIZ*0 LOI4Z*0 PZOZZ'O
0000S.0 00SLE'0 0000£'0 00051'0 6Z4IZ*0 OSL8T*0 E£80Z'O
0000S.0 005LE-0 0000£'0 00051'0 60TZ'O OSL8I*0 L9991-0
99
The fixed doses (xj=3./(m+1), i=1,...,n) and the estimated
ED50 for NOnm designs when L=6 (L=m*n).
(Note
:
N016=A16=B16)
NO23
S
0 0
0
0
2
1
0
1
1
1
2
2
2
2
0
0
0
0
1
1
2
0
1
2
1
0
1
1
1 2
2 0
2 1
2
2
0
0
0 1
0 2
1 0
1
1
1
2
2
0
2
1
2
2
X1
X2
X3
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
N032
s
0 0
0
0
0
1
1
2
3
0
1
1
1 2
1 3
2 0
2
1
2
2
3
3
3
3
2
3
0
1
2
3
xi.
x2
ED50
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.78512
0.71275
0.57797
0.50000
0.75120
0.67356
0.50000
0.42203
0.70455
0.50000
0.32644
0.28725
0.50000
0.29545
0.24880
0.21488
NO61
s
0
1
2
3
4
5
6
xl
ED50
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.71429
0.66667
0.60000
0.50000
0.40000
0.33333
0.28571
ED50
0.82400
0.71617
0.60902
0.78386
0.60000
0.50000
0.57638
0.43724
0.39098
0.79692
0.66013
0.56276
0.75165
0.50000
0.40000
0.45179
0.33987
0.28383
0.76000
0.54821
0.42362
0.50000
0.24835
0.21614
0.24000
0.20308
0.17600
100
The fixed doses
(x1 =i/(m+1), i=1,...,n) and the estimated
ED50 for SKNn (n=2,3,6) designs when L=6 (L=m*n)
SKN2
S
X1
x2
x3
0
0
0
1
0
1
2
0
1
1
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.25000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
0.75000
1 2
2 0
2
1
2
0
0
0
1
1
1
2
2
2
0
0
0
1
2
0
1
2
0
1
2
0
1
2
0
1
2
0
1
1
1
2
2
2
2
0
1
2
SKN3
s
0
0
0
0
1
1
0
1
2
3
0
1
1 2
1 3
2 0
2
1
2
2
3
3
3
3
2
3
0
1
2
3
xl
x2
ED50
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.33333
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.66667
0.83333
0.72222
0.61111
0.50000
0.72222
0.61111
0.50000
0.38889
0.61111
0.50000
0.38889
0.27778
0.50000
0.38889
0.27778
0.16667
SKN6
s
0
1
2
3
4
5
6
xl
ED50
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.75000
0.66667
0.58333
0.50000
0.41667
0.33333
0.25000
ED50
0.87500
0.75000
0.62500
0.75000
0.62500
0.50000
0.62500
0.50000
0.37500
0.75000
0.62500
0.50000
0.62500
0.50000
0.37500
0.50000
0.37500
0.25000
0.62500
0.50000
0.37500
0.50000
0.37500
0.25000
0.37500
0.25000
0.12500
.
101
The fixed doses and estimated ED50 for 64 possible outcomes
when L=6 for UDq (q=001) design.
UD001
S
X1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
1
0
0
0
0
1
1
1
1
0
0
1
1
0
0
1
0
1
0
1
0
1
0
1
0
1
1
1
0
0
0
0
1
1
1
0
0
1
1
0
0
1
0
1
1
0
1
0
1
0
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
1
0
0
1
1
0
0
0
1
1
0
1
0
0
0
0
1
1
1
1
0
0
1
1
0
0
1
1
1
1
1
0
1
0
1
0
1
0
1
0
1
0
1
>C2
X3
>c4
X5
X6
EI) 5 0
0.50000 0.50100 0.50200 0.50300 0.50400 0.50500 0.50589
0.50000 0.50100 0.50200 0.50300 0.50400 0.50500 0.50462
0.50000 0.50100 0.50200 0.50300 0.50400 0.50300 0.50389
0.50000 0.50100 0.50200 0.50300 0.50400 0.50300 0.50303
0.50000 0.50100 0.50200 0.50300 0.50200 0.50300 0.50332
0.50000 0.50100 0.50200 0.50300 0.50200 0.50300 0.50257
0.50000 0.50100 0.50200 0.50300 0.50200 0.50100 0.50214
0.50000 0.50100 0.50200 0.50300 0.50200 0.50100 0.50150
0.50000 0.50100 0.50200 0.50100 0.50200 0.50300 0.50285
0.50000 0.50100 0.50200 0.50100 0.50200 0.50300 0.50214
0.50000 0.50100 0.50200 0.50100 0.50200 0.50100 0.50174
0.50000 0.50100 0.50200 0.50100 0.50200 0.50100 0.50117
0.50000 0.50100 0.50200 0.50100 0.50000 0.50100 0.50137
0.50000 0.50100 0.50200 0.50100 0.50000 0.50100 0.50083
0.50000 0.50100 0.50200 0.50100 0.50000 0.49900 0.50050
0.50000 0.50100 0.50200 0.50100 0.50000 0.49900 0.49990
0.50000 0.50100 0.50000 0.50100 0.50200 0.50300 0.50245
0.50000 0.50100 0.50000 0.50100 0.50200 0.50300 0.50175
0.50000 0.50100 0.50000 0.50100 0.50200 0.50100 0.50137
0.50000 0.50100 0.50000 0.50100 0.50200 0.50100 0.50083
0.50000 0.50100 0.50000 0.50100 0.50000 0.50100 0.50102
0.50000 0.50100 0.50000 0.50100 0.50000 0.50100 0.50050
0.50000 0.50100 0.50000 0.50100 0.50000 0.49900 0.50017
0.50000 0.50100 0.50000 0.50100 0.50000 0.49900 0.49961
0.50000 0.50100 0.50000 0.49900 0.50000 0.50100 0.50070
0.50000 0.50100 0.50000 0.49900 0.50000 0.50100 0.50017
0.50000 0.50100 0.50000 0.49900 0.50000 0.49900 0.49983
0.50000 0.50100 0.50000 0.49900 0.50000 0.49900 0.49930
0.50000 0.50100 0.50000 0.49900 0.49800 0.49900 0.49950
0.50000 0.50100 0.50000 0.49900 0.49800 0.49900 0.49896
0.50000 0.50100 0.50000 0.49900 0.49800 0.49700 0.49860
0.50000 0.50100 0.50000 0.49900 0.49800 0.49700 0.49789
0.50000 0.49900 0.50000 0.50100 0.50200 0.50300 0.50211
0.50000 0.49900 0.50000 0.50100 0.50200 0.50300 0.50140
0.50000 0.49900 0.50000 0.50100 0.50200 0.50100 0.50104
0.50000 0.49900 0.50000 0.50100 0.50200 0.50100 0.50050
0.50000 0.49900 0.50000 0.50100 0.50000 0.50100 0.50070
0.50000 0.49900 0.50000 0.50100 0.50000 0.50100 0.50017
0.50000 0.49900 0.50000 0.50100 0.50000 0.49900 0.49983
0.50000 0.49900 0.50000 0.50100 0.50000 0.49900 0.49930
0.50000 0.49900 0.50000 0.49900 0.50000 0.50100 0.50039
0.50000 0.49900 0.50000 0.49900 0.50000 0.50100 0.49983
0.50000 0.49900 0.50000 0.49900 0.50000 0.49900 0.49950
0.50000 0.49900 0.50000 0.49900 0.50000 0.49900 0.49898
0.50000 0.49900 0.50000 0.49900 0.49800 0.49900 0.49917
0.50000 0.49900 0.50000 0.49900 0.49800 0.49900 0.49863
0.50000 0.49900 0.50000 0.49900 0.49800 0.49700 0.49825
0.50000 0.49900 0.50000 0.49900 0.49800 0.49700 0.49755
0.50000 0.49900 0.49800 0.49900 0.50000 0.50100 0.50155
0.50000 0.49900 0.49800 0.49900 0.50000 0.50100 0.50225
0.50000 0.49900 0.49800 0.49900 0.50000 0.49900 0.49863
0.50000 0.49900 0.49800 0.49900 0.50000 0.49900 0.49917
0.50000 0.49900 0.49800 0.49900 0.49800 0.49900 0.49898
0.50000 0.49900 0.49800 0.49900 0.49800 0.49900 0.49950
0.50000 0.49900 0.49800 0.49900 0.49800 0.49700 0.49583
0.50000 0.49900 0.49800 0.49900 0.49800 0.49700 0.49639
0.50000 0.49900 0.49800 0.49700 0.49800 0.49900 0.49930
0.50000 0.49900 0.49800 0.49700 0.49800 0.49900 0.49983
0.50000 0.49900 0.49800 0.49700 0.49800 0.49700 0.49783
0.50000 0.49900 0.49800 0.49700 0.49800 0.49700 0.49670
0.50000 0.49900 0.49800 0.49700 0.49600 0.49700 0.49650
0.50000 0.49900 0.49800 0.49700 0.49600 0.49700 0.49704
0.50000 0.49900 0.49800 0.49700 0.49600 0.49500 0.49340
0.50000 0.49900 0.49800 0.49700 0.49600 0.49500 0.49411
102
The fixed doses and estimated ED50 for 64 possible outcomes
when L=6 for UDq (q=035) design.
UD035
S
X1
0
0
0
0
0 0
0 1
1 0
1 1
1 0 0
1 0 1
1 1 0
1
1
0
0
1
1
1 0
1 0
1 1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
0
0
0
1
1
1
0
1
0
1
0
1
0
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
1
0
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
0 0
0 1
1 0
1 1
0 0
1
0
1
1
1
0
1
1
1
0
0
0
0
0 0
0 1
1 0
1 1
1 0 0
1 0 1
1 1 0
1
1
1
X2
X3
x4
x5
x6
ED50
0.50000 0.53500 0.57000 0.60500 0.64000 0.67500 0.70626
0.50000 0.53500 0.57000 0.60500 0.64000 0.67500 0.66184
0.50000 0.53500 0.57000 0.60500 0.64000 0.60500 0.63629
0.50000 0.53500 0.57000 0.60500 0.64000 0.60500 0.60598
0.50000 0.53500 0.57000 0.60500 0.57000 0.60500 0.61603
0.50000 0.53500 0.57000 0.60500 0.57000 0.60500 0.58988
0.50000 0.53500 0.57000 0.60500 0.57000 0.53500 0.57487
0.50000 0.53500 0.57000 0.60500 0.57000 0.53500 0.55250
0.50000 0.53500 0.57000 0.53500 0.57000 0.60500 0.59961
0.50000 0.53500 0.57000 0.53500 0.57000 0.60500 0.57487
0.50000 0.53500 0.57000 0.53500 0.57000 0.53500 0.56080
0.50000 0.53500 0.57000 0.53500 0.57000 0.53500 0.54092
0.50000 0.53500 0.57000 0.53500 0.50000 0.53500 0.54802
0.50000 0.53500 0.57000 0.53500 0.50000 0.53500 0.52909
0.50000 0.53500 0.57000 0.53500 0.50000 0.46500 0.51750
0.50000 0.53500 0.57000 0.53500 0.50000 0.46500 0.49640
0.50000 0.53500 0.50000 0.53500 0.57000 0.60500 0.58586
0.50000 0.53500 0.50000 0.53500 0.57000 0.60500 0.56125
0.50000 0.53500 0.50000 0.53500 0.57000 0.53500 0.54802
0.50000 0.53500 0.50000 0.53500 0.57000 0.53500 0.52909
0.50000 0.53500 0.50000 0.53500 0.50000 0.53500 0.53577
0.50000 0.53500 0.50000 0.53500 0.50000 0.53500 0.51750
0.50000 0.53500 0.50000 0.53500 0.50000 0.46500 0.50592
0.50000 0.53500 0.50000 0.53500 0.50000 0.46500 0.48639
0.50000 0.53500 0.50000 0.46500 0.50000 0.53500 0.52464
0.50000 0.53500 0.50000 0.46500 0.50000 0.53500 0.50592
0.50000 0.53500 0.50000 0.46500 0.50000 0.46500 0.49409
0.50000 0.53500 0.50000 0.46500 0.50000 0.46500 0.47536
0.50000 0.53500 0.50000 0.46500 0.43000 0.46500 0.48250
0.50000 0.53500 0.50000 0.46500 0.43000 0.46500 0.46350
0.50000 0.53500 0.50000 0.46500 0.43000 0.39500 0.45110
0.50000 0.53500 0.50000 0.46500 0.43000 0.39500 0.42625
0.50000 0.46500 0.50000 0.53500 0.57000 0.60500 0.57375
0.50000 0.46500 0.50000 0.53500 0.57000 0.60500 0.54890
0.50000 0.46500 0.50000 0.53500 0.57000 0.53500 0.53651
0.50000 0.46500 0.50000 0.53500 0.57000 0.53500 0.51750
0.50000 0.46500 0.50000 0.53500 0.50000 0.53500 0.52464
0.50000 0.46500 0.50000 0.53500 0.50000 0.53500 0.50592
0.50000 0.46500 0.50000 0.53500 0.50000 0.46500 0.49409
0.50000 0.46500 0.50000 0.53500 0.50000 0.46500 0.47536
0.50000 0.46500 0.50000 0.46500 0.50000 0.53500 0.51362
0.50000 0.46500 0.50000 0.46500 0.50000 0.53500 0.49409
0.50000 0.46500 0.50000 0.46500 0.50000 0.46500 0.48250
0.50000 0.46500 0.50000 0.46500 0.50000 0.46500 0.46423
0.50000 0.46500 0.50000 0.46500 0.43000 0.46500 0.47092
0.50000 0.46500 0.50000 0.46500 0.43000 0.46500 0.45198
0.50000 0.46500 0.50000 0.46500 0.43000 0.39500 0.43875
0.50000 0.46500 0.50000 0.46500 0.43000 0.39500 0.41414
0.50000 0.46500 0.43000 0.46500 0.50000 0.53500 0.55415
0.50000 0.46500 0.43000 0.46500 0.50000 0.53500 0.57875
0.50000 0.46500 0.43000 0.46500 0.50000 0.46500 0.45198
0.50000 0.46500 0.43000 0.46500 0.50000 0.46500 0.47092
0.50000 0.46500 0.43000 0.46500 0.43000 0.46500 0.46423
0.50000 0.46500 0.43000 0.46500 0.43000 0.46500 0.48250
0.50000 0.46500 0.43000 0.46500 0.43000 0.39500 0.35408
0.50000 0.46500 0.43000 0.46500 0.43000 0.39500 0.37361
0.50000 0.46500 0.43000 0.39500 0.43000 0.46500 0.47536
0.50000 0.46500 0.43000 0.39500 0.43000 0.46500 0.49409
0.50000 0.46500 0.43000 0.39500 0.43000 0.39500 0.42408
0.50000 0.46500 0.43000 0.39500 0.43000 0.39500 0.38464
0.50000 0.46500 0.43000 0.39500 0.36000 0.39500 0.37750
0.50000 0.46500 0.43000 0.39500 0.36000 0.39500 0.39650
0.50000 0.46500 0.43000 0.39500 0.36000 0.32500 0.26889
0.50000 0.46500 0.43000 0.39500 0.36000 0.32500 0.29374
103
The fixed doses and estimated ED50 for 64 possible outcomes
when L=6 for UDq (q=075) design.
UD075
X1
S
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
1
1
1
0
0
0
0
1
1
1
0
0
1
1
0
0
1
0
1
1
0
1
0
1
0
1
1
1
0
0
0
0
1
1
1
1
0
0
0
0
1
1
1
0
0
1
1
0
0
1
1
0
0
1
1
0
0
1
0
1
0
1
0
1
0
1
0
1
0
1
0
1
0
1
1
1
0
0
0
0
1
1
1
0
0
1
1
0
0
1
0
1
1
1
1
0
1
0
1
0
0 0 0
0 0 1
0 1 0
0 1 1
1 0 0
1 0 1
1
1
0
1
1
1
0
0
0
0
1
1
1
1
0
0
0
0
1
1
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1 0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1 0
1 1
X2
X3
X4
X5
X6
0.50000 0.57500 0.65000 0.72500 0.80000 0.87500
0.50000 0.57500 0.65000 0.72500 0.80000 0.87500
0.50000 0.57500 0.65000 0.72500 0.80000 0.72500
0.50000 0.57500 0.65000 0.72500 0.80000 0.72500
0.50000 0.57500 0.65000 0.72500 0.65000 0.72500
0.50000 0.57500 0.65000 0.72500 0.65000 0.72500
0.50000 0.57500 0.65000 0.72500 0.65000 0.57500
0.50000 0.57500 0.65000 0.72500 0.65000 0.57500
0.50000 0.57500 0.65000 0.57500 0.65000 0.72500
0.50000 0.57500 0.65000 0.57500 0.65000 0.72500
0.50000 0.57500 0.65000 0.57500 0.65000 0.57500
0.50000 0.57500 0.65000 0.57500 0.65000 0.57500
0.50000 0.57500 0.65000 0.57500 0.50000 0.57500
0.50000 0.57500 0.65000 0.57500 0.50000 0.57500
0.50000 0.57500 0.65000 0.57500 0.50000 0.42500
0.50000 0.57500 0.65000 0.57500 0.50000 0.42500
0.50000 0.57500 0.50000 0.57500 0.65000 0.72500
0.50000 0.57500 0.50000 0.57500 0.65000 0.72500
0.50000 0.57500 0.50000 0.57500 0.65000 0.57500
0.50000 0.57500 0.50000 0.57500 0.65000 0.57500
0.50000 0.57500 0.50000 0.57500 0.50000 0.57500
0.50000 0.57500 0.50000 0.57500 0.50000 0.57500
0.50000 0.57500 0.50000 0.57500 0.50000 0.42500
0.50000 0.57500 0.50000 0.57500 0.50000 0.42500
0.50000 0.57500 0.50000 0.42500 0.50000 0.57500
0.50000 0.57500 0.50000 0.42500 0.50000 0.57500
0.50000 0.57500 0.50000 0.42500 0.50000 0.42500
0.50000 0.57500 0.50000 0.42500 0.50000 0.42500
0.50000 0.57500 0.50000 0.42500 0.35000 0.42500
0.50000 0.57500 0.50000 0.42500 0.35000 0.42500
0.50000 0.57500 0.50000 0.42500 0.35000 0.27500
0.50000 0.57500 0.50000 0.42500 0.35000 0.27500
0.50000 0.42500 0.50000 0.57500 0.65000 0.72500
0.50000 0.42500 0.50000 0.57500 0.65000 0.72500
0.50000 0.42500 0.50000 0.57500 0.65000 0.57500
0.50000 0.42500 0.50000 0.57500 0.65000 0.57500
0.50000 0.42500 0.50000 0.57500 0.50000 0.57500
0.50000 0.42500 0.50000 0.57500 0.50000 0.57500
0.50000 0.42500 0.50000 0.57500 0.50000 0.42500
0.50000 0.42500 0.50000 0.57500 0.50000 0.42500
0.50000 0.42500 0.50000 0.42500 0.50000 0.57500
0.50000 0.42500 0.50000 0.42500 0.50000 0.57500
0.50000 0.42500 0.50000 0.42500 0.50000 0.42500
0.50000 0.42500 0.50000 0.42500 0.50000 0.42500
0.50000 0.42500 0.50000 0.42500 0.35000 0.42500
0.50000 0.42500 0.50000 0.42500 0.35000 0.42500
0.50000 0.42500 0.50000 0.42500 0.35000 0.27500
0.50000 0.42500 0.50000 0.42500 0.35000 0.27500
0.50000 0.42500 0.35000 0.42500 0.50000 0.57500
0.50000 0.42500 0.35000 0.42500 0.50000 0.57500
0.50000 0.42500 0.35000 0.42500 0.50000 0.42500
0.50000 0.42500 0.35000 0.42500 0.50000 0.42500
0.50000 0.42500 0.35000 0.42500 0.35000 0.42500
0.50000 0.42500 0.35000 0.42500 0.35000 0.42500
0.50000 0.42500 0.35000 0.42500 0.35000 0.27500
0.50000 0.42500 0.35000 0.42500 0.35000 0.27500
0.50000 0.42500 0.35000 0.27500 0.35000 0.42500
0.50000 0.42500 0.35000 0.27500 0.35000 0.42500
0.50000 0.42500 0.35000 0.27500 0.35000 0.27500
0.50000 0.42500 0.35000 0.27500 0.35000 0.27500
0.50000 0.42500 0.35000 0.27500 0.20000 0.27500
0.50000 0.42500 0.35000 0.27500 0.20000 0.27500
0.50000 0.42500 0.35000 0.27500 0.20000 0.12500
0.50000 0.42500 0.35000 0.27500 0.20000 0.12500
ED50
.94197
.84680
.79205
.72710
.74862
.69260
.66042
.61250
.71345
.66042
.63027
.58767
.60290
.56233
.53750
.49227
.68397
.63125
.60290
.56233
.57665
.53750
.51268
.47082
.55280
.51268
.48732
.44720
.46250
.42178
.39522
.34197
.65802
.60477
.57823
.53750
.55280
.51268
.48732
.44720
.52918
.48733
.46250
.42335
.43768
.39710
.36875
.31602
.61602
.66875
.39710
.43768
.42335
.46250
.18733
.22918
.44120
.48733
.33733
.25280
.23750
.27822
.00477
.05802
104
The fixed doses and estimated ED50 for 64 possible outcomes
when L=6 for RM1Cc (n=1, c=.001) design.
RM1C0
S
X1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
1
1
1
0
0
0
0
0 0
0 1
1 0
1 1
1
1 0 0
1 0 1
1
1
0
1
1
1
0
0
0
0
1
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1 0
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
1
0
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0 0 0
0 0 1
0 1 0
0
1
1
1 0 0
1 0 1
1
1
0
1
1
1
X2
X3
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50075
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.50050 0.50025
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49975
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
0.50000 0.49950 0.49925
X4
0.50092
0.50092
0.50092
0.50092
0.50092
0.50092
0.50092
0.50092
0.50058
0.50058
0.50058
0.50058
0.50058
0.50058
0.50058
0.50058
0.50042
0.50042
0.50042
0.50042
0.50042
0.50042
0.50042
0.50042
0.50008
0.50008
0.50008
0.50008
0.50008
0.50008
0.50008
0.50008
0.49992
0.49992
0.49992
0.49992
0.49992
0.49992
0.49992
0.49992
0.49958
0.49958
0.49958
0.49958
0.49958
0.49958
0.49958
0.49958
0.49942
0.49942
0.49942
0.49942
0.49942
0.49942
0.49942
0.49942
0.49908
0.49908
0.49908
0.49908
0.49908
0.49908
0.49908
0.49908
X5
X6
0.50104 0.50114
0.50104 0.50114
0.50104 0.50094
0.50104 0.50094
0.50079 0.50089
0.50079 0.50089
0.50079 0.50069
0.50079 0.50069
0.50071 0.50081
0.50071 0.50081
0.50071 0.50061
0.50071 0.50061
0.50046 0.50056
0.50046 0.50056
0.50046 0.50036
0.50046 0.50036
0.50054 0.50064
0.50054 0.50064
0.50054 0.50044
0.50054 0.50044
0.50029 0.50039
0.50029 0.50039
0.50029 0.50019
0.50029 0.50019
0.50021 0.50031
0.50021 0.50031
0.50021 0.50011
0.50021 0.50011
0.49996 0.50006
0.49996 0.50006
0.49996 0.49986
0.49996 0.49986
0.50004 0.50014
0.50004 0.50014
0.50004 0.49994
0.50004 0.49994
0.49979 0.49989
0.49979 0.49989
0.49979 0.49969
0.49979 0.49969
0.49971 0.49981
0.49971 0.49981
0.49971 0.49961
0.49971 0.49961
0.49946 0.49956
0.49946 0.49956
0.49946 0.49936
0.49946 0.49936
0.49954 0.49964
0.49954 0.49964
0.49954 0.49944
0.49954 0.49944
0.49929 0.49939
0.49929 0.49939
0.49929 0.49919
0.49929 0.49919
0.49921 0.49931
0.49921 0.49931
0.49921 0.49911
0.49921 0.49911
0.49896 0.49906
0.49896 0.49906
0.49896 0.49886
0.49896 0.49886
ED50
0.50122
0.50106
0.50102
0.50086
0.50097
0.50081
0.50077
0.50061
0.50089
0.50073
0.50069
0.50053
0.50064
0.50047
0.50044
0.50027
0.50073
0.50056
0.50053
0.50036
0.50047
0.50031
0.50027
0.50011
0.50039
0.50023
0.50019
0.50003
0.50014
0.49998
0.49994
0.49978
0.50022
0.50006
0.50002
0.49986
0.49997
0.49981
0.49978
0.49961
0.49989
0.49973
0.49969
0.49953
0.49964
0.49948
0.49944
0.49928
0.49973
0.49956
0.49953
0.49936
0.49948
0.49931
0.49928
0.49911
0.49939
0.49923
0.49919
0.49903
0.49914
0.49898
0.49894
0.49878
105
The fixed doses and estimated ED50 for 64 possible outcomes
when L=6 for RM1Cc (n=1, c=.2) design.
RM1C2
S
X1
0
0
0
0
1
1
1
0
0
1
1
0
0
1
0
1
1
0
1
0
1
0
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
1 1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
1
0 0 0
0 0 1
0 1 0
0 1 1
1 0 0
1 0 1
1
1
0
1
1
1
0 0 0
0 0 1
0 1 0
0 1 1
1 0 0
1 0 1
1
0
1 1
1
1
X2
X3
x4
X5
X6
ED50
0.50000 0.60000 0.65000 0.68333 0.70833 0.72833 0.74500
0.50000 0.60000 0.65000 0.68333 0.70833 0.72833 0.71167
0.50000 0.60000 0.65000 0.68333 0.70833 0.68833 0.70500
0.50000 0.60000 0.65000 0.68333 0.70833 0.68833 0.67167
0.50000 0.60000 0.65000 0.68333 0.65833 0.67833 0.69500
0.50000 0.60000 0.65000 0.68333 0.65833 0.67833 0.66167
0.50000 0.60000 0.65000 0.68333 0.65833 0.63833 0.65500
0.50000 0.60000 0.65000 0.68333 0.65833 0.63833 0.62167
0.50000 0.60000 0.65000 0.61667 0.64167 0.66167 0.67833
0.50000 0.60000 0.65000 0.61667 0.64167 0.66167 0.64500
0.50000 0.60000 0.65000 0.61667 0.64167 0.62167 0.63833
0.50000 0.60000 0.65000 0.61667 0.64167 0.62167 0.60500
0.50000 0.60000 0.65000 0.61667 0.59167 0.61167 0.62833
0.50000 0.60000 0.65000 0.61667 0.59167 0.61167 0.59500
0.50000 0.60000 0.65000 0.61667 0.59167 0.57167 0.58833
0.50000 0.60000 0.65000 0.61667 0.59167 0.57167 0.55500
0.50000 0.60000 0.55000 0.58333 0.60833 0.62833 0.64500
0.50000 0.60000 0.55000 0.58333 0.60833 0.62833 0.61167
0.50000 0.60000 0.55000 0.58333 0.60833 0.58833 0.60500
0.50000 0.60000 0.55000 0.58333 0.60833 0.58833 0.57167
0.50000 0.60000 0.55000 0.58333 0.55833 0.57833 0.59500
0.50000 0.60000 0.55000 0.58333 0.55833 0.57833 0.56167
0.50000 0.60000 0.55000 0.58333 0.55833 0.53833 0.55500
0.50000 0.60000 0.55000 0.58333 0.55833 0.53833 0.52167
0.50000 0.60000 0.55000 0.51667 0.54167 0.56167 0.57833
0.50000 0.60000 0.55000 0.51667 0.54167 0.56167 0.54500
0.50000 0.60000 0.55000 0.51667 0.54167 0.52167 0.53833
0.50000 0.60000 0.55000 0.51667 0.54167 0.52167 0.50500
0.50000 0.60000 0.55000 0.51667 0.49167 0.51167 0.52833
0.50000 0.60000 0.55000 0.51667 0.49167 0.51167 0.49500
0.50000 0.60000 0.55000 0.51667 0.49167 0.47167 0.48833
0.50000 0.60000 0.55000 0.51667 0.49167 0.47167 0.45500
0.50000 0.40000 0.45000 0.48333 0.50833 0.52833 0.54500
0.50000 0.40000 0.45000 0.48333 0.50833 0.52833 0.51167
0.50000 0.40000 0.45000 0.48333 0.50833 0.48833 0.50500
0.50000 0.40000 0.45000 0.48333 0.50833 0.48833 0.47167
0.50000 0.40000 0.45000 0.48333 0.45833 0.47833 0.49500
0.50000 0.40000 0.45000 0.48333 0.45833 0.47833 0.46167
0.50000 0.40000 0.45000 0.48333 0.45833 0.43833 0.45500
0.50000 0.40000 0.45000 0.48333 0.45833 0.43833 0.42167
0.50000 0.40000 0.45000 0.41667 0.44167 0.46167 0.47833
0.50000 0.40000 0.45000 0.41667 0.44167 0.46167 0.44500
0.50000 0.40000 0.45000 0.41667 0.44167 0.42167 0.43833
0.50000 0.40000 0.45000 0.41667 0.44167 0.42167 0.40500
0.50000 0.40000 0.45000 0.41667 0.39167 0.41167 0.42833
0.50000 0.40000 0.45000 0.41667 0.39167 0.41167 0.39500
0.50000 0.40000 0.45000 0.41667 0.39167 0.37167 0.38833
0.50000 0.40000 0.45000 0.41667 0.39167 0.37167 0.35500
0.50000 0.40000 0.35000 0.38333 0.40833 0.42833 0.44500
0.50000 0.40000 0.35000 0.38333 0.40833 0.42833 0.41167
0.50000 0.40000 0.35000 0.38333 0.40833 0.38833 0.40500
0.50000 0.40000 0.35000 0.38333 0.40833 0.38833 0.37167
0.50000 0.40000 0.35000 0.38333 0.35833 0.37833 0.39500
0.50000 0.40000 0.35000 0.38333 0.35833 0.37833 0.36167
0.50000 0.40000 0.35000 0.38333 0.35833 0.33833 0.35500
0.50000 0.40000 0.35000 0.38333 0.35833 0.33833 0.32167
0.50000 0.40000 0.35000 0.31667 0.34167 0.36167 0.37833
0.50000 0.40000 0.35000 0.31667 0.34167 0.36167 0.34500
0.50000 0.40000 0.35000 0.31667 0.34167 0.32167 0.33833
0.50000 0.40000 0.35000 0.31667 0.34167 0.32167 0.30500
0.50000 0.40000 0.35000 0.31667 0.29167 0.31167 0.32833
0.50000 0.40000 0.35000 0.31667 0.29167 0.31167 0.29500
0.50000 0.40000 0.35000 0.31667 0.29167 0.27167 0.28833
0.50000 0.40000 0.35000 0.31667 0.29167 0.27167 0.25500
106
The fixed doses and estimated ED50 for 64 possible outcomes
when L=6 for RM1Cc (n=1, c=.408) design.
RM1C4
S
X1
0
0
0
0
1
1
1
0
0
1
1
0
0
1
0
1
1
0
1
0
1
0
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
0
0
0
0 0
0 1
1 0
1 1
1 0 0
1 0 1
1
0
1
1 1 1
0 0 0
0
0
0
1
0 1
1 0
1 1
0 0
1 0 1
1
1
0
1
1
1
0
0
0
0
0 0
0 1
1 0
1 1
1
0
0
1 0
1
1
1
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
1
1
1
0
0
0
0
1
1
1
0
0
1
1
0
0
1
0
0
1
1
1
0
0
0
0
1
1
0 0
0 1
1 0
1 1
0 0
0 1
1
1
0
1
1
1
0
1
0
1
0
1
x2
x3
X4
x5
x6
ED50
0.50000 0.70408 0.80612 0.87415 0.92517 0.96599 1.00000
0.50000 0.70408 0.80612 0.87415 0.92517 0.96599 0.93197
0.50000 0.70408 0.80612 0.87415 0.92517 0.88435 0.91837
0.50000 0.70408 0.80612 0.87415 0.92517 0.88435 0.85034
0.50000 0.70408 0.80612 0.87415 0.82313 0.86395 0.89796
0.50000 0.70408 0.80612 0.87415 0.82313 0.86395 0.82993
0.50000 0.70408 0.80612 0.87415 0.82313 0.78231 0.81633
0.50000 0.70408 0.80612 0.87415 0.82313 0.78231 0.74830
0.50000 0.70408 0.80612 0.73810 0.78912 0.82993 0.86395
0.50000 0.70408 0.80612 0.73810 0.78912 0.82993 0.79592
0.50000 0.70408 0.80612 0.73810 0.78912 0.74830 0.78231
0.50000 0.70408 0.80612 0.73810 0.78912 0.74830 0.71429
0.50000 0.70408 0.80612 0.73810 0.68707 0.72789 0.76190
0.50000 0.70408 0.80612 0.73810 0.68707 0.72789 0.69388
0.50000 0.70408 0.80612 0.73810 0.68707 0.64626 0.68027
0.50000 0.70408 0.80612 0.73810 0.68707 0.64626 0.61224
0.50000 0.70408 0.60204 0.67007 0.72109 0.76190 0.79592
0.50000 0.70408 0.60204 0.67007 0.72109 0.76190 0.72789
0.50000 0.70408 0.60204 0.67007 0.72109 0.68027 0.71429
0.50000 0.70408 0.60204 0.67007 0.72109 0.68027 0.64626
0.50000 0.70408 0.60204 0.67007 0.61905 0.65986 0.69388
0.50000 0.70408 0.60204 0.67007 0.61905 0.65986 0.62585
0.50000 0.70408 0.60204 0.67007 0.61905 0.57823 0.61224
0.50000 0.70408 0.60204 0.67007 0.61905 0.57823 0.54422
0.50000 0.70408 0.60204 0.53401 0.58503 0.62585 0.65986
0.50000 0.70408 0.60204 0.53401 0.58503 0.62585 0.59184
0.50000 0.70408 0.60204 0.53401 0.58503 0.54422 0.57823
0.50000 0.70408 0.60204 0.53401 0.58503 0.54422 0.51020
0.50000 0.70408 0.60204 0.53401 0.48299 0.52381 0.55782
0.50000 0.70408 0.60204 0.53401 0.48299 0.52381 0.48980
0.50000 0.70408 0.60204 0.53401 0.48299 0.44218 0.47619
0.50000 0.70408 0.60204 0.53401 0.48299 0.44218 0.40816
0.50000 0.29592 0.39796 0.46599 0.51701 0.55782 0.59184
0.50000 0.29592 0.39796 0.46599 0.51701 0.55782 0.52381
0.50000 0.29592 0.39796 0.46599 0.51701 0.47619 0.51020
0.50000 0.29592 0.39796 0.46599 0.51701 0.47619 0.44218
0.50000 0.29592 0.39796 0.46599 0.41497 0.45578 0.48980
0.50000 0.29592 0.39796 0.46599 0.41497 0.45578 0.42177
0.50000 0.29592 0.39796 0.46599 0.41497 0.37415 0.40816
0.50000 0.29592 0.39796 0.46599 0.41497 0.37415 0.34014
0.50000 0.29592 0.39796 0.32993 0.38095 0.42177 0.45578
0.50000 0.29592 0.39796 0.32993 0.38095 0.42177 0.38776
0.50000 0.29592 0.39796 0.32993 0.38095 0.34014 0.37415
0.50000 0.29592 0.39796 0.32993 0.38095 0.34014 0.30612
0.50000 0.29592 0.39796 0.32993 0.27891 0.31973 0.35374
0.50000 0.29592 0.39796 0.32993 0.27891 0.31973 0.28571
0.50000 0.29592 0.39796 0.32993 0.27891 0.23810 0.27211
0.50000 0.29592 0.39796 0.32993 0.27891 0.23810 0.20408
0.50000 0.29592 0.19388 0.26190 0.31293 0.35374 0.38776
0.50000 0.29592 0.19388 0.26190 0.31293 0.35374 0.31973
0.50000 0.29592 0.19388 0.26190 0.31293 0.27211 0.30612
0.50000 0.29592 0.19388 0.26190 0.31293 0.27211 0.23810
0.50000 0.29592 0.19388 0.26190 0.21088 0.25170 0.28571
0.50000 0.29592 0.19388 0.26190 0.21088 0.25170 0.21769
0.50000 0.29592 0.19388 0.26190 0.21088 0.17007 0.20408
0.50000 0.29592 0.19388 0.26190 0.21088 0.17007 0.13605
0.50000 0.29592 0.19388 0.12585 0.17687 0.21769 0.25170
0.50000 0.29592 0.19388 0.12585 0.17687 0.21769 0.18367
0.50000 0.29592 0.19388 0.12585 0.17687 0.13605 0.17007
0.50000 0.29592 0.19388 0.12585 0.17687 0.13605 0.10204
0.50000 0.29592 0.19388 0.12585 0.07483 0.11565 0.14966
0.50000 0.29592 0.19388 0.12585 0.07483 0.11565 0.08163
0.50000 0.29592 0.19388 0.12585 0.07483 0.03401 0.06803
0.50000 0.29592 0.19388 0.12585 0.07483 0.03401 0.00000
107
The fixed doses and estimated ED50 when L=6 for RM2Cc (n=2,
c=.001,
.25) designs.
RM2C0
s
x1
X2
x3
0
0
0
1
0
1
2
0
1
1
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50050
0.50050
0.50050
0.50050
0.50050
0.50050
0.50050
0.50050
0.50050
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.49950
0.49950
0.49950
0.49950
0.49950
0.49950
0.49950
0.49950
0.49950
0.50075
0.50075
0.50075
0.50050
0.50050
0.50050
0.50025
0.50025
0.50025
0.50025
0.50025
0.50025
0.50000
0.50000
0.50000
0.49975
0.49975
0.49975
0.49975
0.49975
0.49975
0.49950
0.49950
0.49950
0.49925
0.49925
0.49925
1 2
2 0
2
1
2 2
0 0
0 1
0 2
1 0
1
1
1 2
2 0
2
2
0
0
0
1
1
2
1
1
0
1
2
0
1 2
2 0
2 1
2
2
ED50
0.50092
0.50075
0.50058
0.50067
0.50050
0.50033
0.50042
0.50025
0.50008
0.50042
0.50025
0.50008
0.50017
0.50000
0.49983
0.49992
0.49975
0.49958
0.49992
0.49975
0.49958
0.49967
0.49950
0.49933
0.49942
0.49925
0.49908
RM2C2
0
0
s
X1
x2
0
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.62500
0.62500
0.62500
0.62500
0.62500
0.62500
0.62500
0.62500
0.62500
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.37500
0.37500
0.37500
0.37500
0.37500
0.37500
0.37500
0.37500
0.37500
1
0 2
1
0
1
1
1
2
2 0
2 1
2 2
0 0
0
0
1
1
1
1
0
2
1
2
2
0
2
1
2
2
0
0
0 1
0 2
1
0
1
1
2
2
2
2
1
0
1
2
x3
0.68750
0.68750
0.68750
0.62500
0.62500
0.62500
0.56250
0.56250
0.56250
0.56250
0.56250
0.56250
0.50000
0.50000
0.50000
0.43750
0.43750
0.43750
0.43750
0.43750
0.43750
0.37500
0.37500
0.37500
0.31250
0.31250
0.31250
ED50
0.72917
0.68750
0.64583
0.66667
0.62500
0.58333
0.60417
0.56250
0.52083
0.60417
0.56250
0.52083
0.54167
0.50000
0.45833
0.47917
0.43750
0.39583
0.47917
0.43750
0.39583
0.41667
0.37500
0.33333
0.35417
0.31250
0.27083
108
The fixed doses and estimated ED50 when L=6 for RM2Cc (n=2,
c=.5454) design.
RM2C5
X1
00
00
00
01
01
0
0
2
1
02
02
02
10
10
10
1
1
1
2
0
1
0
1
2
0
1
2
0
11
11
12
12
12
20
20
20
1
2
2
0
1
21
21
1
2
0
0
1
2
1
2
2 2 0
22
22
1
2
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
x2
x3
0.77273 0.90909
0.77273 0.90909
0.77273 0.90909
0.77273 0.77273
0.77273 0.77273
0.77273 0.77273
0.77273 0.63636
0.77273 0.63636
0.77273 0.63636
0.50000 0.63636
0.50000 0.63636
0.50000 0.63636
0.50000 0.50000
0.50000 0.50000
0.50000 0.50000
0.50000 0.36364
0.50000 0.36364
0.50000 0.36364
0.22727 0.36364
0.22727 0.36364
0.22727 0.36364
0.22727 0.22727
0.22727 0.22727
0.22727 0.22727
0.22727 0.09091
0.22727 0.09091
0.22727 0.09091
ED50
1.00000
0.90909
0.81818
0.86364
0.77273
0.68182
0.72727
0.63636
0.54545
0.72727
0.63636
0.54545
0.59091
0.50000
0.40909
0.45455
0.36364
0.27273
0.45455
0.36364
0.27273
0.31818
0.22727
0.13636
0.18182
0.09091
0.00000
109
The fixed doses and estimated ED50 when L=6 for RM3Cc (n=3,
c=.001,
.333,
.667) designs.
RM3C0
0
0
0
0
1
0
1
2
3
0
1
1
1
1
2
2
2
2
3
3
3
3
2
3
0
1
2
3
0
1
2
3
xl
x2
ED50
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50050
0.50050
0.50050
0.50050
0.50017
0.50017
0.50017
0.50017
0.49983
0.49983
0.49983
0.49983
0.49950
0.49950
0.49950
0.49950
0.50075
0.50058
0.50042
0.50025
0.50042
0.50025
0.50008
0.49992
0.50008
0.49992
0.49975
0.49958
0.49975
0.49958
0.49942
0.49925
RM3C3
0
0
0
0
1
1
0
1
2
3
0
1
1
2
1 3
2 0
2
1
2
2
3
3
3
3
2
3
0
1
2
3
x1
x2
ED50
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.66667
0.66667
0.66667
0.66667
0.55556
0.55556
0.55556
0.55556
0.44444
0.44444
0.44444
0.44444
0.33333
0.33333
0.33333
0.33333
0.75000
0.69444
0.63889
0.58333
0.63889
0.58333
0.52778
0.47222
0.52778
0.47222
0.41667
0.36111
0.41667
0.36111
0.30556
0.25000
RM3C6
0
0
0
0
1
0
1
2
3
0
1
1
1
2
1 3
2 0
2 1
2
2
2 3
3 0
3
1
3 2
3 3
x1
x2
ED50
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.83333
0.83333
0.83333
0.83333
0.61111
0.61111
0.61111
0.61111
0.38889
0.38889
0.38889
0.38889
0.16667
0.16667
0.16667
0.16667
1.00000
0.88889
0.77778
0.66667
0.77778
0.66667
0.55556
0.44444
0.55556
0.44444
0.33333
0.22222
0.33333
0.22222
0.11111
0.00000
110
The fixed doses and estimated ED50 when L=6 for RM6Cc (n=6,
c=.001,
1) designs.
.5,
RM6C0
s
xl
ED50
O
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50050
0.50033
0.50017
0.50000
0.49983
0.49967
0.49950
1
2
3
4
5
6
RM6C5
O
1
2
3
4
5
6
xl
ED50
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.75000
0.66667
0.58333
0.50000
0.41667
0.33333
0.25000
RM6C10
s
xl
ED50
O
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
0.50000
1.00000
0.83333
0.66667
0.50000
0.33333
0.16667
0.00000
1
2
3
4
5
6