Assignment 6 Solutions Stat 557 Fall 2000 Problem 1
advertisement

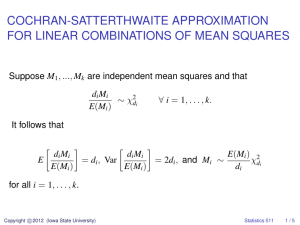
Stat 557
Fall 2000
Assignment 6 Solutions
Problem 1
(a) Yij 's are independent and Yij Poisson(mi). So the log-likelihood is
0
1
;m mY
Y
Y
e
i A
l = log @
Y
!
ij
i j
X
X
XX
= ;3 e0 1x + Yi ( + xi) ;
log(Yij !)
5
3
ij
i
=1 =1
5
5
+
5
i
i=1
+
i=1
0
1
3
i=1 j =1
(b) The mle's and corresponding standard errors are
^ = 2:878, s0 = 0:108
^ = 0:3479, s1 = 0:0593
In this case, exp(^) = 17:78 is an estimate of the mean number of colonies of TA98
salmonella at a quinoline concentration of 1.0 mg per plate. The estimate exp(^) =
141:6 indicates that the a 10-fold increase in the concentration of quinoline results in
about a 41.6 percent increase in the mean number of TA98 salmonella colonies.
^
0
^
1
(c) The mean number of colonies when the log dose of quinoline is equal to x is m = e0
Let = ( ; )T . Then,
0
1
!
@m = @m ; @m T = e0 1x; e0 1xxT = (m; mx)T
@
@ @
By the -method,
!T
!
@m
@m
^
var(m^ ) @ var() @
0
10
1
^ ) cov(^ ; ^ )
var
(
m
A@
A
= (m; mx) @
cov(^ ; ^ ) var(^ )
mx
h
i
= m var(^ ) + x var(^ ) + 2x cov(^ ; ^ )
+
0
+
1
0
0
2
Then
0
1
1
2
0
1
1
0
1
q
Sm = m^ var
^ (^ ) + x var
^ (^ ) + 2x cov
^ (^ ; ^ )
^
2
0
1
1
0
1
x.
+ 1
From the estimated model, m^ = 38:22 when x = 2:2, and
var
^ (^ ) = 0:01166;
var
^ (^ ) = 0:003518;
0
1
cov
^ (^ ; ^ ) = ;0:005766 :
0
1
Then, Sm = 2:20, and an approximate 95 percent condence interval is
^
(m^ ; z : Sm; m^ + z : Sm) = (33:91; 42:54)
0 025
^
0 025
^
Alternatively, you could rst construct a condence interval for the natural logarithm
of the mean at x = 2:2. Compute log(m^ ) = 2:878 + (0:3479)(2:2) = 3:6434, and
Slog m
(^)
q
^ (^ ) + x var
^ (^ ) + 2x cov
^ (^ ; ^ ) = 0:05756 :
= var
0
2
1
0
1
Then
log(m^ ) (1:96)Slog m
) (3:5306; 3:7562)
and an approximate 95 percent condence interval for the mean count at x = 2:2 is
(^)
(exp(3:5306); exp(3:7562))
) (34:14; 42:79) :
There is only a small dierence in these two methods in this case because the observed
counts are moderately large, but this second method would generally provide a more
accurate coverage probability than the rst method. The GENMOD procedure in SAS
uses the second method to compute condence intervals for mean responses.
(d) The deviance test of the model satisfying log(mij ) = + X against the model of the
independent Poisson counts with a dierent mean at each level of quiloline is G = 0:27,
with 5 ; 2 = 3 degrees of freedomand p-value=0.956. Assuming independent Poisson
counts, the proposed Poisson regression model can not be rejected. This is the test
you were expected to report.
0
1
2
You could also consider a second test. The deviance test of the model of the independent Poisson counts with a dierent mean at each level of quinoline against the more
general alternative that the fteen counts can all have dierent means is G = 16:98,
with 15 ; 5 = 10 degrees of freedom and p-value=0.075 . This alternative implies that
the three counts obtained at each concentration of quinoline did not come from exact
replications of the same experiment.
2
2
The sum of the G values for the previous two tests provides a deviance test of the
model satisfying log(mij ) = + X against the alternative of fteen independent
Poisson counts with potentially fteen dierent means. G = 0:27 + 16:98 = 17:25
with 15 ; 2 = 13 degrees of freedom and p-value = 0.19. Here the null hypothesis is
not rejected. If you did reject the t of the model with this test you would not know
if the model was rejected because log(mij ) = + X did not provide an adequate
description ofthe trend in the means across the quinoline concentrations, or there were
some uncontrolled background factors that prevented the three experiments at each
concentration of quinoline from being exact replicates of each other.
2
0
1
2
0
1
(e) Maximum likelihoods estimates and corresponding standard errors for the negative
binomial model are
^ = 2:8782, s0 = 0:2048.
^ = 0:3478, s0 = 0:1295.
^ = 0:0055, s = 0:0277.
The estimate of the dispersion parameter is much smaller than the standard error of the
estimate. Also, an approximate 95% condence interval of the dispersion parameter
is (0:0000; 104:9256). Hence. a zero value for the dispersion parameter is consistent
with these data, and it appears the a Poisson regression model is adequate.
0
^
1
^
^
(f) From the estimated model, log(m^ ) = 2:8782 + (:3478)(2:2) = 3:643 and m^ = 38:22
when x = 2:2, and var
^ (^ ) = 0:04196, var
^ (^ ) = 0:01678, cov
^ (^ ; ^ ) = ;0:02569.
Similar to part (c)
0
1
0
1
q
^ (^ ) + x var
^ (^ ) + 2x cov
^ (^ ; ^ ) = 0:1007
Slog m = var
and
Sm = m^
^
2
0
(^)
2
q
1
0
1
var
^ (^ ) + x var
^ (^ ) + 2x cov
^ (^ ; ^ ) = 3:85
2
0
1
0
1
An approximate 95% condence interval for the mean number of colonies when the
log-dose of quilonine equals to 2.2 is
(m^ ; z : Sm; m^ + z : Sm ) = (31:4; 46:6)
0 025
^
0 025
^
An approximate 95% condence interval with better coverage probability is constructed
by evaluating
log(m^ ) (1:96)Slog m
) (3:4456; 3:840)
(^)
3
and transforming back to the original scale
(exp(3:4456); exp(3:840))
) (31:36; 46:54) :
Condidence intervals based on the negative binomial regression model are wider than
those based on a corresponding Poisson regression model in part, because the negative
binomial regression model allows for more variation in the observed counts.
(g) Since the proposed model seems to be adequate, there is no need to serach for a better
model.
Problem 2
(a) Maximum likelihood estimates and standard errors: ^ = ;6:536, S1 = 0:9504.
^ = ;4:622, S2 = 0:7817.
^ = 0:7330, S1 = 0:1089.
^ = 0:4850, S2 = 0:08862.
1
2
^
^
1
^
2
^
(b) See Fig 1. The curves appear to underestimate the probability of at least one death
for the largest litter sizes.
(c) ^ = ;5:415, S = 0:598.
^ = 0:588, S = 0:068.
^
^
(d) The value of the deviance test of the model in part (c) against the general alternative
is G = 11:36 with 8 degrees of freedom.
The value of the deviance test of the model in part (a) against the general alternative
is G = 6:18 with 6 degrees of freedom.
Then, the value of the deviance test of the model in part (c) against the model in part
(a) is G = 11:36 ; 6:18 = 5:18 with 8 ; 6 = 2 degrees of freedom and p-value=0.075.
Therefore, the model in part (a) does not appear to provide a signicant improvement
over the model in part (c).
2
2
2
4
(e) Many models were reported. Some people t logisitc regression models with liner,
quadratic and cubic trends for litter size.. Others switched to the complimentary loglog link and t models with linear and quadratic trends for litter size. Many people
tried to t the same form of the model for each treatment, although the plot suggests
that the "curve" may bend more sharply for treatment B near 11 litters. One person
t a complimentary log-log model with a linear trend in litter size for treatment A
and a quadratic trend in litter size for treatment B. Also, the intercepts may not be
signicantly dierent for the two treatments. We will not display details for these
models, although we liked some of them as well as the following model. There are
many ways to arrive at essentially the same model (as long as you do not extrapolate
to litter sizes not included in this study).
!
ij
log 1 ; = + i X
ij
5
for (i = 1; 2; j = 1; ; 5)
The deviance test of this model against the general alternative is G = 2:717 with 7
degrees of freedom and p-value=0.91.
The estimates of the parameters and the corresponding standard errors are
= ;1:4085, s = 1:5002.
= 0:0000212, s1 = 0:00000268.
= 0:0000163, s1 = 0:00000232.
The plot of the observed death rates and the tted model versus the litter size is shown
in gure 2. By comparing gure 2 to gure 1, we can see this model appears to provide
a better t to the data than the model in part (a). It was disappointing that only a
few people bothered to show plots of their tted curves.
Some people objected to plotting a curve because litter size is discrete. You cannot
have 9.73 animals in a litter, for example, and they plotted estimated means without
showing a curve. This was okay.
2
^
1
^
2
^
#-----------------------------------------------------------------------;
#Assignment 6 Splus code;
#---------;
5
#problem 1;
#---------;
#--------------------------------------------------------------;
# A function to get the covariance matrix for the large sample ;
# approximation to the parameter estimates
;
#--------------------------------------------------------------;
f.vcov <- function(obj) {
so <- summary(obj, corr=F)
so$dispersion*so$cov.unscaled
}
# part (a)
x<-rep(c(0, 1, 1.5, 2.0, 2.5), rep(3, 5))
y<-c(12, 17, 25, 21, 26, 28, 26, 26, 35, 28, 33, 50, 34, 45, 47)
dat<-data.frame(x, y)
# part (b)
model1<-glm(y~x, family=poisson(link=log), data=dat, trace=T)
summary(model1)
# part (c)
f.vcov(model1)
x0<-2.2
m.hat<-exp(predict(model1, data.frame(x=x0)))
V<-f.vcov(model1)
s<-sqrt(t(c(m.hat, x0*m.hat))%*%V%*%c(m.hat, x0*m.hat))
z<-qnorm(0.975)
cbind(m.hat-z*s, m.hat+z*s)
# part (d)
model2<-glm(y~factor(x), family=poisson(link=log), data=dat, trace=T)
dev<-model1$dev-model2$dev
df<-model1$df-model2$df
p.value<-1-pchisq(dev, df)
6
# part (e)
library(MASS)
model.nb<-glm.nb(y~x, data=dat, control=glm.control(maxit=20))
summary(model.nb)
x0<-2.2
m.hat<-exp(predict(model.nb, data.frame(x=x0)))
V<-f.vcov(model.nb)
s<-sqrt(t(c(m.hat, x0*m.hat))%*%V%*%c(m.hat, x0*m.hat))
z<-qnorm(0.975)
cbind(m.hat-z*s, m.hat+z*s)
#---------;
#problem 2;
#---------;
#enter the data;
size<-c(7,7,8,8,9,9,10,10,11,11)
trt<-rep(c("A", "B"), 5)
none<-c(58, 75, 49, 58, 33, 45, 15, 39, 4, 5)
death<-c(16, 26, 24, 25, 33, 32, 28, 40, 29, 23)
y<-cbind(death, none)
#part (a)
a.model<-glm(y ~ trt+trt:size-1 , x=T, trace=T, family=binomial(link=logit))
summary(a.model)
#part (b)
#plot the observed death rate and the fitted death rate by the model in
#part (a) against the litter size;
x<-7:11
r<-death/(none+death)
p<-a.model$fit
r.A<-r[c(1,3,5,7,9)]
7
r.B<-r[c(2,4,6,8,10)]
p.A<-p[c(1,3,5,7,9)]
p.B<-p[c(2,4,6,8,10)]
plot(x, r.A, pch="A",
xlab="Litter Size x",
ylab="death rate (observed and fitted by model)",
main="Fig 1: problem 2, part (a))")
points(x, r.B, pch="B")
lines(x, p.A, lty=1)
lines(x, p.B, lty=2)
legend(7, 0.8, c("treatment A (fitted)", "treatment B (fitted)"), lty=1:2)
#part (c)
c.model<-glm(y ~ size , x=T, trace=T, family=binomial(link=logit))
summary(c.model)
#part (d)
dev<-c.model$dev-a.model$dev
df<-c.model$df-a.model$df
p.value<-1-pchisq(dev, df)
#part (e)
model0<-step(c.model, list(upper=~trt+size+trt:size+trt:size^2
+trt:size^3+trt:size^4+trt:size^5+trt:size^6,
lower=~size), trace=T)
summary(model0)
# === OUTPUT === #
#-----------------------------------------------;
Coefficients:
(Intercept)
Value Std. Error
t value
3.0679746 4.41900901
0.6942676
size -1.4013564 1.02420370 -1.3682399
8
trt
0.3934230 0.31851405
1.2351826
trtAI(size^2)
0.1213287 0.05865800
2.0684079
trtBI(size^2)
0.1077658 0.05856263
1.8401810
Residual Deviance: 2.433901 on 5 degrees of freedom
#-----------------------------------------------;
#Delete the insignificant term "trt";
model1<-glm(y ~ size+trt:I(size^2), x=T, trace=T,
family=binomial(link=logit))
summary(model1)
# === OUTPUT === #
#--------------------------------------------------;
Coefficients:
(Intercept)
Value Std. Error
t value
3.2420054 4.39779743
0.7371884
size -1.4278875 1.01897369 -1.4012997
trtAI(size^2)
0.1172089 0.05816256
2.0151945
trtBI(size^2)
0.1132818 0.05812053
1.9490851
Residual Deviance: 3.976015 on 6 degrees of freedom
#----------------------------------------------------;
#Delete the insignificant term "size";
model2<-glm(y ~ trt:I(size^2), x=T, trace=T, family=binomial(link=logit))
summary(model2)
# === OUTPUT === #
#---------------------------------------------------;
Coefficients:
Value
Std. Error
t value
(Intercept) -2.92150241 0.312069865 -9.361693
trtAI(size^2)
0.03614524 0.004149556
8.710626
trtBI(size^2)
0.03223410 0.003967145
8.125265
p.value<-1-pchisq(result2$dev, result2$df)
9
Residual Deviance: 5.966825 on 7 degrees of freedom
#--------------------------------------------------;
#a
better model;
model3<-glm(y ~ trt:I(size^5), x=T, trace=T, family=binomial(link=logit))
summary(model3)
p.value<-1-pchisq(model3$dev, model3$df)
# === OUTPUT === #
#----------------------------------------------------;
Coefficients:
Value
Std. Error
t value
(Intercept) -1.40850852077 1.500262e-001 -9.388414
trtAI(size^5)
0.00002120519 2.687591e-006
7.890036
trtBI(size^5)
0.00001629306 2.320851e-006
7.020298
Residual Deviance: 2.717231 on 7 degrees of freedom
#----------------------------------------------------;
#plot the observed death rate and the fitted death rate by model 3
#against the litter size;
p<-model3$fit
p.A<-p[c(1,3,5,7,9)]
p.B<-p[c(2,4,6,8,10)]
plot(x, r.A, pch="A",
xlab="Litter Size x",
ylab="death rate (observed and fitted by model)",
main="Fig 2: problem 2, part (e)")
points(x, r.B, pch="B")
lines(x, p.A, lty=1)
lines(x, p.B, lty=2)
legend(7, 0.8, c("treatment A (fitted)", "treatment B (fitted)"), lty=1:2)
10
! [-0.1in,0in][9.8in,11in] c:5576f00s.p2.g1.ps
11