P oisson Distribution Simeon
advertisement

Poisson Distribution
Simeon Poisson (1837)
Then
Approximation to the binomial
distribution when
P rfY = kg
= k!(nn! k)! k (1 )n k
n is large
k
n k
= k!(nn! k)! n
1 n
n
n
is small
= kk!
More formally, let
h
n ! for some nite > 0
1
because
! 1 and 1
as n ! 1
and show that
n!
k
(n k)!n 1
n k
n n k
n n ik
! 1 as n ! 1
e
285
Consequently, as n ! 1
n
!0
n ! !e then
Use Sterling's formula to obtain
p2nn!nne n ! 1
and p
(n k) (n k)
2(n k)(n k)
(n k)!
n
1 n n
284
!e n
Note that
with
n n
n
n ik
n!
(n k)!nk 1
!1
!0
n
h
P rfY = kg =
!1
! 1 as n ! 1
286
n k
(1 )n k
k
k
! k! e
for k = 0; 1; 2; :::
This is the probability function for a
Poisson distribution with a population
mean of .
287
Alternative derivation of the
Poisson distribution
Suppose the lifetime T of an item is
independent of its current age
Let Y = number of failures in a time
interval of length L
Then,
P r fY = k g
= P rfT1 + T2 + ::: + Tk+1 > Lg
P rfT1 + T2 + ::: + Tk > Lg
) T has an exponential
distribution with density
where
f (t) = 1e t=
Ti is the lifetime of the i th
item
for t 0; > 0
When an item fails it is immediately
replaced by a new item with the same
lifetime distribution
T1, T2, T3, .... are i.i.d. exponential
random variables with
density 1 e t=
288
289
P r T1 + T2 + ::: + Tk+1 > L
Note that
T1 + T2 + : : : + Tk 22k
2
The density function for a central
chi-square distribution with r
degrees of freedom is
1
f (x) = r=2
x(r=2) 1 e x=2
2 (r=2)
Use integration by parts to derive the
Poisson probability function
290
= P r 22k+2 > L
2
= P r 22k+2 > 2L
Z
1 k
= k+1 1
x e
2
2
(k + 1) L
=
1
2k+1 (k + 1)
+
1
Z
2L
k
= Lk e
k!
"
2xk e
2kx(k 1) e
L=
2dx
x=
21
2L
x=
#
2dx
x=
+ P r 22k > 2L
291
Then
P r (Y = k) = P r T1 + T2 + ::: + Tk+1 > L
P r (T1 + T2 + ::: + Tk > L)
= P r 22k+2 > 2L
k
= Lk e
k!
L=
2L
P r 22k >
+ P r 22k > 2L
We can also obtain
2L P r 22k >
k
= Lk e
k!
k
= e
k!
P r (Y
k) =
k j
e j =0 j !
X
= P r 22k+2 > 2
L=
This relationship can be used to do
exact tests and construct exact condence intervals for where = L= is the expected number
of failures in a time interval of length
L
293
292
The Poisson distribution bf may provide a suitable model for the distribution of
The number of occurances of a
random event in a specied
time interval
Applications of the
Poisson Distribution
Bortkiewicz (1898)
Deaths from mule kicks in the
Prussian Army
Student (W. S. Gosset) (1907)
Numbers of particles falling in
identical small regions scattered
throughout a much larger region
raindrops
oak trees in a forest
yeast particles in beer
The number of occuances of a
random event in some spatial
region
if the random events are rare and
independent.
294
295
Rutherford and Gieger (1910)
Numbers of radioactive particles
emitted in time intervals of equal
length
F. Thorndike (1926)
Number of incorrect connections
for telephone calls in a specic
time interval (Bell Labs)
W. F. Adams (1937)
Number of traÆc accidents in a
specic stretch of highway
References
Haight, F. (1967)
Handbook of the
, Wiley, New York.
Poisson Distribution
Johnson, N.L., Kotz, S. and Kemp, A.W.
(1992) Univariate Discrete Distributions, Wiley, New York.
Conditions that result in a Poisson
distribution:
Redheer (1953)
26, 185-188.
Mathematical Magazine
Walsh (1955) Operations Research, 3,
198-204.
296
297
Moments:
P r fY = kg =
mk
k!
e m
for k = 0; 1; 2; ::::
X
k
X
E (Y
m)3 = m
E (Y
m)4 = m + 3m2
Skewness:
E (Y m)3
= p1
3
=
2
m
[V (Y )]
Kurtosis:
E (Y m)4
=3+ 1
2
[V (Y )]
m
This is denoted by
Y
1
mk m
e =m
k=0 k!
k
1
V (Y ) =
(k m)2 m e m = m
k!
k=0
E (Y ) =
Distributional Properties
Y has a Poisson Distribution with
mean m = E (Y ) if
,
P oisson(m)
298
299
Normal Approximation:
Y m dist'n
! N (0; 1)
p
(2)
m
P
=1 Y
=1 mi dist'n
! N (0; 1)
P
K
K
i q i
i
P
K
i
i
=1 m
as n ! 1
as m ! 1
(3) The conditional distribution of
Y1
Y = Y..2
YK
Sums of independent Poisson
random variables:
Suppose Y1; Y2; : : : ; YK are independent
with Yi P oisson(mi), then
(1)
P
K Yi
i=1
P oisson(
P
2
3
6
6
6
6
6
4
7
7
7
7
7
5
given n = K
i=1 Yi is Mult(n; ) where
m
i = K i
for i = 1; 2; :::; K
j =1 mi
P
P
K mi )
i=1
301
300
Example: Rainfall events
Gunst, E. L., (1938) Tansactions of the
American
Society
, 384-388
of
Civil
Engineers,
103
Yi = number of storms that produced
excessive rainfall at a weather
station in a one year period,
i=1,2,...,330 stations
Number of
excessive Number
rainfall
of
events
stations
0
102
1
114
2
74
3
28
4
10
5
2
6 or more
0
330
Joint likelihood function:
2
3
Yj
330
Y
6m
e m
Y
!
j
j =1
The table indicates that 102 of the Yj 's
are zero, 114 of the Yi's are one, ...
The mle for m is
330
Y
m
^ = 1
330 j =1 j
= 1 [(102)(0) + (114)(1)
330
+(74)(2) + (28)(3)
+(10)(4) + (2)(5)]
= 1 [396]
330
= 1:2
L(m : Y1; Y2; ; ; ; Y330) =
7
5
4
X
events per station per year
302
303
An approximate (1 ) 100%
condence interval for m:
Variance of m
^ is
330
m
Yj ) =
V (m
^) = V ( 1
330 j =1
330
For large values of
k
X
X
j =1
use
This is estimated as
1:2
^
2 = m
Sm
^ 330 = 330 = :003636
= km
m
^ m N (0; 1)
m=k
^
to obtain
^
m
^ Z=2 m
k
An approximate 95% condence interval for the mean number of excessive
rainfall events is
1:2 (1:96) 1:2
330
q
v
u
u
t
Standard error for m
^ is
m
^ = :0603
Sm^ =
330
v
u
u
t
v
u
u
t
305
304
=)
(1.03, 1.32) events
An exact condence interval for m is
2(2 j Yj );1 =2 2(2+2 j Yj );=2
;
2k
2k
3
2
6
6
4
P
P
=)
2
6
4
2(792);:975
;
(2)(330)
7
7
5
2(794);:025
(2)(330)
3
2
=) 715:91 ; 873:98
660)
660
3
4
5
Are the data consistent with the
assumption that the 330 weather
stations provide a set of 330
i.i.d. Poisson counts?
Fisher's Index of Dispersion:
X2 =
7
5
k Yj
j =1
m
^
1
k Yj
1) k 1 j =1
= (k
=) [1:085; 1:324]
306
2k
m
^ 2
P
P
m
^
m
^ 2
1
when Y1, Y2, ...,Yk are i.i.d. Poisson(m)
random variables.
307
Example: Rainfall events
Goodness-of-Fit Tests
X 2 = 330:67
Create a multinomial distribution
with 330 1 = 329 df
and p-value=0.464
Each station is classied into
a category
Conclusion:
The observed numbers of excessive rainfall events at the 330
stations are consistent with an
i.i.d Poisson(m) model
Combine some categories to make
expected counts large enough
309
308
Example: Rainfall events
X1 = number of stations with zero
execessive rainfall events
X2 = number of stations with one
execessive rainfall event
X3 = number of stations with two
execessive rainfall events
X4 = number of stations with three
execessive rainfall events
X5 = number of stations with four
execessive rainfall events
X6 = number of stations with ve or
more execessive rainfall events
310
2
3
6
6
6
6
6
6
6
6
6
6
6
4
7
7
7
7
7
7
7
7
7
7
7
5
X1
X2
X = XX34
X5
X6
2
3
6
6
6
6
6
6
6
6
6
6
6
4
7
7
7
7
7
7
7
7
7
7
7
5
1
2
Mult(n; = 3 )
4
5
6
In this example, n = 330 stations and
under the i.i.d. Poison(m) model for
excessive rainfall events
j = P rfobserving a station
with exactly j excessive
rainfall eventsg
j
= m e m for j = 1; :::; 5
j!
311
Model A: i.i.d. Poisson model
6 = P rfobserving a station
with 5 or more excessive
rainfall eventsg
1 mj m
e
for j = 1; :::; 5
=
j =5 j !
X
Estimated expected counts
m
^ j 1 e m^
n^j = n
(j 1)!
for j=1, 2, 3, 4, 5 and
2
n^6 = n 1
6
4
5
X
j =1
3
^j
7
5
Model B: General alternative
Note that
mle for nj = E (Xj ) is
X
= n j = Xj
n
for j=1, 2, 3, 4, 6
1 + 2 + 3 + 4 + 5 + 6 = 1
312
Number of Expected number
excessive
of stations
rainfall
events Model B Model A
0
102
99.394
1
114
119.273
2
74
71.564
3
28
28.625
4
10
8.588
5 or more
2
2.557
330
330
313
Pearson statistic:
6 Xj n
X
^j 2
2
X =
= 0:75
n^j
j =1
with 4 df and p-value=0.945
Deviance:
1
0
6
X
X
j
2
G =2
Xj log @ A = 0:75
n^j
j =1
with 4 df and p-value=0.945
Conclusion:
314
315
data set1;
/*
This program is stored as
poisson.sas
input level count;
cards;
*/
0 102
/*
This program uses PROC IML in SAS
1 114
to test the fit of a Poisson model
2
74
against a general alternative.
It
3
28
also computes confidence intervals
4
10
for the mean of a Poisson
5
2
distribution.
run;
It is applied to
data on the number of storms with
excessive rainfall recorded at 330
weather stations.
/*
Print the data set */
(E. Grant, 1938,
TASCE, 103, 384-388)*/
proc print data=set1;
run;
317
316
/*
Use the IML procedure to compute
confidence intervals
and goodness-of-fit tests
*/
nc = nrow(x);
proc iml;
start poisson;
/*
Enter the data
/* Compute the number of categories */
/* Store category values in cc */
cc = w[ ,1];
*/
use set1;
/* Compute sample mean and variance */
read all into w;
mean = t(cc)*x/n;
var =
/* Create a column of counts */
(t(x)*(cc##2) - n*mean*mean)/n;
PRINT,,,"Sample Mean is" MEAN;
x = w[ ,2];
PRINT,"Sample Variance is" VAR;
/* Compute the total sample size */
n = sum(x);
318
319
/* Compute expected counts for the
/* Compute Pearson statistic */
iid Poisson model */
PEARSON = sum((xt-ept)##2/ept);
ep = J(nc,1,0);
ep[1,1] = n*EXP(-mean);
/* Compute likelihood ratio test */
do k = 2 to nc;
G2 = 0;
km1 = k-1;
ncf = nrow(ept);
ep[k,1] = ep[km1,1]*mean/km1;
do i = 1 to ncf;
end;
at = 0;
ep[nc,1] = ep[nc,1] + n-sum(ep);
if(xt[i,1])>0. then
/end{verbatim}
at=2*xt[i,1]*log(xt[i,1]/ept[i,1]);
g2 = g2 + at;
/* Combine categories to make each
end;
expected count larger than MB */
ept = ep;
/* Compute Fisher Deviance */
run combine;
FISHERD = n*var/mean;
320
321
dff = n-1;
PRINT "
dfp = kk - 2;
PRINT "
PVALF = 1-probchi(FISHERD,dff);
PRINT,,,"
cu
Count
Fisher Deviance =" FISHERD;
PRINT "
PRINT,,,,,,,,"Results for Fitting
Expected";
Count";
PRINT zt;
PRINT,,,"PEARSON Goodness-of-fit
Statistic =" PEARSON;
322
/*
=" DFP;
p-value =" PVALG;
PRINT "
zt=cl||cu||xt||ept;
PRINT "cl
df
PRINT "
/* Print Results */
Observed
p-value =" PVALP;
PRINT "
PVALG = 1=probchi(G2, dfp);
the Poisson Distribution";
=" DFP;
PRINT,,,"Likelihood ratio test =" G2;
PVALP = 1-probchi(PEARSON,dfp);
PRINT,"
df
df
=" DFF;
p-value =" PVALF;
Compute confidence intervals */
a = .05;
clevel = 1-a;
za = probit(1-a/2);
323
/* Use large sample normal distribution*/
*---MODULE FOR COMBINING CATEGORIES TO--;
mlower = mean - za*sqrt(mean/n);
*---KEEP ALL EXPECTED COUNTS ABOVE A SET;
mupper = mean + za*sqrt(mean/n);
*---LOWER BOUND:
MB
------------------;
print,,,"Large sample conf. intervals ";
print, " Confidence
Lower
Upper ";
print
Bound
Bound ";
"
Level
print clevel mlower mupper;
mlower = cinv(a/2,2*n*mean)/(2*n);
mupper = cinv(1-a/2,2*(n*mean+1))/(2*n);
print,,,"More accurate conf. intervals ";
print, " Confidence
Lower
Upper ";
print
Bound
Bound ";
Level
Start at the bottom of the array and
combine categories until the expected
count for the combined category
/* More accurate confidence limits */
"
/*
print clevel mlower mupper;
exceeds MB */
start combine;
mb=2;
cl = cc;
cu = cc;
xt=x;
ept=ep;
finish;
324
325
nc=nrow(cc);
ptr = ptrm1;
ptr = nc;
end;
I = J(nc, 1, .);
kk = 0;
if (ept[1] < mb) then do;
Ik = I[kk];
do until(ptr = 1);
ptrm1 = ptr - 1;
ept[Ik] = ept[Ik] + ept[1];
if (ept[ptr] < mb) then do;
xt[Ik] = xt[Ik] + xt[1];
ept[ptrm1] = ept[ptrm1] + ept[ptr];
xt[ptrm1] = xt[ptrm1] + xt[ptr];
cu[ptrm1] = cu[ptr];
cl[Ik] = cl[1];
end;
else do;
end;
kk = kk + 1;
else do;
I[kk] = 1;
kk = kk + 1;
end;
I[kk] = ptr;
end;
326
327
Obs
II = I[kk:1];
cl = cl[II];
cu = cu[II];
xt = xt[II];
ept = ept[II];
level
count
1
0
102
2
1
114
3
2
74
4
3
28
5
4
10
6
5
2
finish;
MEAN
Sample Mean is
1.2
run poisson;
VAR
Sample Variance is 1.2024242
328
329
PEARSON
PEARSON Goodness-of-fit = 0.7513034
Results for Fitting the Poisson
Distribution
DFP
Observed
Expected
Count
cl
cu
Count
0
0
102
99.39409
1
1
114
119.27291
2
2
74
71.563745
3
3
28
28.625498
4
4
10
8.5876494
5
5
2
2.5561101
df
=
4
PVALP
p-value
=
0.9448547
G2
Likelihood ratio test =
0.751497
DFP
df
=
4
PVALG
p-value =
330
0
331
Large sample confidence intervals
Confidence
Lower
Upper
Level
Bound
Bound
CLEVEL
MLOWER
MUPPER
0.95
1.0818097
1.3181903
FISHERD
Fisher
Deviance = 330.66667
DFF
df
=
329
More accurate confidence intervals
PVALF
p-value = 0.4638074
Confidence
Lower
Upper
Level
Bound
Bound
CLEVEL
MLOWER
MUPPER
0.95
332
1.0847055
1.3242132
333
combine<-function(cc, x, ep, nc, mb){
ptr <- nc
# Splus code for fitting a Poisson
I <- c()
# distribution to count data.
k <- 0
# file is stored
as
This
poisson.ssc
# First create the function "combine"
# for combining categories to keep all
# expected counts abobe a specified
# lower bound,
mb.
# Start at the bottom of the array and
# combine categories until the expected
# count for the combined category
cl<-cc
cu<-cc
while(ptr > 1) {
ptrm1 <- ptr - 1
if(ep[ptr] < mb) {
ep[ptrm1] <- ep[ptrm1] + ep[ptr]
x[ptrm1] <- x[ptrm1] + x[ptr]
cu[ptrm1]<-cu[ptr]
}
else {
k <- k + 1
# exceeds mb.
I[k] <- ptr
}
334
335
ptr <- ptrm1
# Enter the counts.
Do not skip any
}
# categoies.
Enter a zero if the
if(ep[1] < mb) {
# observed count is zero.
Ik <- I[k]
ep[Ik] <- ep[Ik] + ep[1]
x<-c(102, 114, 74, 28, 10, 2)
x[Ik] <- x[Ik] + x[1]
cl[Ik]<-cl[1]
# Compute the number of categories.
}
else {
nc<-length(x)
k <- k + 1
I[k] <- 1
# Enter the category levels.
}
II <- I[k:1]
cc<-0:(nc-1)
list(k=k, cl = cl[II], cu=cu[II],
xt = x[II], ept = ep[II])
}
# Compute the total sample size.
n<-sum(x)
336
337
# Compute mle's for the sample mean
# Combine categories to make each
# and sample variance.
# expected count at least mb.
mx<-sum(cc*x)/n
mb<-2
vx<-(sum(x*cc^2)-n*mx^2)/n
comb<-combine(cc, x, ep, nc, mb)
xt<-comb$xt
cat(" *** Sample mean is: ", mx, "\n")
ept<-comb$ept
cat(" *** Sample variance is: ", vx, "\n")
k<-comb$k
cl<-comb$cl
# Compute expected counts for the
cu<-comb$cu
# iid Poisson model.
#Compute Pearson statistic.
ep<-n*dpois(cc, mx)
ep[nc]<-ep[nc]+n-sum(ep)
PEARSON<-sum((xt-ept)^2/ept)
338
339
# Compute the G^2 statistic
#Print the results.
M<-cbind(cl, cu, xt, ept)
g2<-0
dimnames(M)<- list(NULL, c("cl", "cu",
for(i in 1:nc) {
"observed", "expected"))
at<-0
if(xt[i] > 0) {at<-2*xt[i]*
cat(" ***
log(xt[i]/ept[i])}
Results for fitting
Poisson distribution ***\n")
g2 <- g2+at}
#Compute Fisher Deviance.
FISHERD<-n*vx/mx
cat("
Pearson statistic =",PEARSON,"
cat("
df =",dfp,"\n")
cat("
p-value =",PVALP,"\n
cat("Likelihood ratio statistic=",g2,"\n")
cat("
df =",dfp,"\n")
dfp<-k-2
cat("
p-value =",PVALG,"\n
PVALF<-1-pchisq(FISHERD, dff)
cat("
Fisher deviance =",FISHERD,"
PVALP<-1-pchisq(PEARSON, dfp)
cat("
df =",dff,"\n")
PVALG<-1-pchisq(g2,dfp)
cat("
p-value =",PVALF,"\n
dff<-n-1
340
341
# Compute confidence interval for mean
a<-0.05
clevel<-1-a
# More accurate confidence limits.
#Use large sample normal distribution.
za<-qnorm(1-a/2)
mlower<-qchisq(a/2, 2*n*mx)/(2*n)
mlower<-mean-za*sqrt(mx/n)
mupper<-qchisq(1-a/2, 2+2*n*mx)/(2*n)
mupper<-mean+za*sqrt(mx/n)
cat("***Exact confidence interval***\n")
cat("***Large sample confidence interval",
"***\n")
cat(100*clevel," % confidence interval:
(", mlower, ",", mupper, ")\n")
cat(100*clevel,"% confidence interval:
(", mlower, ",", mupper, ")\n")
342
343
*** Sample mean is:
1.2
*** Sample variance is:
***
1.20242424
Results for fitting
*** Large sample confidence interval ***
95% confidence interval:
Poisson distribution ***
Pearson statistic = 0.7513
( 1.08180972473947, 1.31819027526053 )
df = 4
p-value = 0.9449
Likelihood ratio statistic = 0.7515
df = 4
p-value = 0.9448
Fisher deviance = 330.67
*** Exact confidence interval ***
95 % confidence interval:
( 1.08470554137095, 1.32421320916233 )
df = 329
p-value = 0.4638
345
344
Two-way contingency tables
with
independent Poisson counts
Model A:
Example: Bimini lizards
Likelihood Function:
Schoener (1968), Fienberg (1980) p. 36
Adult male lizards
L(m11; m12; m21; m22; Y11; Y12; Y21; Y22)
2 2 mYijij m
=
e ij
Y
!
i=1 j =1 ij
Y
Perch diameter
< 2:5 in 2:5 in
Species A
Y11; Y12; Y21; Y22 are independent
Poisson random variables
Log-likelihood:
Y11 = 63 Y12 = 102
l(m11; m12; m21; m22) =
i j log(Yij !)
P
Species B
Y21 = 24
Y
Y22 = 3
346
P
+ i j Yij log(mij )
P
P
i j mij
P
P
347
Question: Do both species of adult
male lizards have the same preference
for larger ( 2:5 in) perch diameters?
Likelihood Equations:
y
0 = @l = ij
@mij
mij
for i=1,2 and j=1,2.
1
m.l.e.'s for expected counts:
m
^ ij = Yij
i = 1; 2;
j = 1; 2
We have
Test the null hypothesis of \independence."
m m
H0 : mij = i+ +j
m++
m+j
= m++ mi+
m++ m++
where
m++ =
m
^ 11
Y11
m
^
m^ = m^ 12
= YY12 = Y
21
21
m
^ 22
Y22
2
3
2
3
6
6
6
6
6
4
7
7
7
7
7
5
6
6
6
6
6
4
7
7
7
7
7
5
mi+ =
m+j =
XX
i j
X
j
X
i
0
10
1
@
A@
A
mij
mij for i = 1; 2
mij for j = 1; 2
348
This is equivalent to
m1j
m2j
H0 :
=
m11 + m12
m21 + m22
for j=1,2
349
Note that
m m
mij = i+ +j
m++
is a function of 3 parameters. We
could use m1+; m+1; m++ because
n
Model B:
o
m+2 = m++ m+1
m2+ = m++ m1+
Y11 Y12 Y21 Y22 are independent
Poisson counts with
The parameter space for model B
has dimension
number
number
of
of
1 = 3
+
columns
rows
Yij Poisson (mij )
and
m m
mij = i+ +j
m++
for i=1,2 and j=1,2
350
0
1
0
1
B
B
@
C
C
A
B
B
@
C
C
A
351
Maximum likelihood estimation
Maximize
g(m1+; m2+; m+1; m+2; m++; 1; 2)
=
XX
i j
log(Yij !)
+ Yi+ log(mi+) +
X
X
i
j
Y+j log(m+j )
Y++ log(m++) m++
+1(m++
X
+2(m++
X
i
j
Solve the equations:
0 =
@g
= Yi+
@mi+ mi+
Y
0 = @g = +j
m+j m+j
0 =
2
j = 1; : : :
@g
= m++
@m++
Y++
1 1 2
0 = @g = m++
@1
mi+)
0 = @g = m++
@2
m +j )
1 i = 1; : : :
X
i
X
j
mi+
m+j
352
353
Solution:
m
^ i+ = Yi+
m
^ +j = Y+j
m
^ ++ = Y++
Perch diameter
< 2:5 in
2:5 in
i = 1; : : :
j = 1; : : :
m.l.e.'s for expected counts for the
independence model (model B)
m
^ m
^
m
^ ij = i+ +j
m
^ ++
Y Y
= i+ +j
Y++
column
row
total
total
=
total for
entire table
0
10
1
@
A@
A
0
1
@
A
354
Species A
m
^ 11 = 74:766 m
^ 12 = 90:234 165
Species B
m
^ 21 = 12:234 m
^ 22 = 14:766
87
X2 =
105
(Yij m
^ ij )2
= 26:2
m
^ ij
i j
XX
G2 = 2
Y
Yij log( ij ) = 24:1
m
^ ij
i j
XX
each with 1 d.f.
355
27
A unifying result:
Conclusion:
Adult males of Species B have a
stronger preference for perches with diameters less than 2.5 inches.
Tests of the null hypothesis of independence have the same formula for
two-way tables when the counts have
any of the following distributions:
Independent Poisson counts
mij 0
Single multinomial distribution
XX
i
Species B:
XX
XX
mij = n;
i
j
X
mij = n;
i
j
Yij = n
Each row is an independent multinomial (binomial) distribution
11 = 0:38
Species A: YY1+
Y21
Y2+
j
ij = 1 and
X
= 0:89
j
ij = 1 and
j
X
j
Yij = ni
Each column is an independent multinomial (binomial) distribution
X
i
ij = 1 and
X
i
mij = nj ;
X
i
Yij = nj
357
356
Log-likelihood functions with Lagrange
multipliers
In each case
H0 : mij =
Independent Poisson counts:
mi+m+j
m++
XX
i
and the m.l.e. is
and
(Xij m
^ ij )2
m
^ ij
i j
y
G2 = 2
yij log ij
m
^ ij
i j
log(n!)
+
XX
XX
i
j
XX
Yij log(mij )
i
j
mij
Single Multinomial: (mij = nij )
Y Y
m
^ ij = i+ +j
Y++
X2 =
j
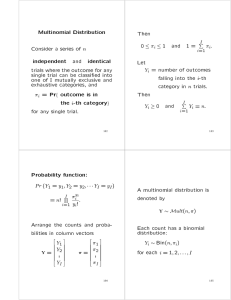
XX
log(Yij !)
0
1
@
A
XX
i
j
XX
i
j
log(Yij !) n log(n)
Yij log(mij ) + (n
XX
i
j
mij )
Independent multinomials
(binomials) for the columns
X
j
both with (I 1)(J 1) d.f.
log(nj !)
+
358
XX
i
j
XX
i
j
log(Yij !)
Yij log(mij ) +
X
j
X
j
j (nj
where mij = nj ij
nj log(nj )
X
i
mij )
359
Likelihood equations
Randomization Tests (and
the hypergeometric
distribution)
mij = mij ()
Independent Poisson Counts:
0 =
Yij
mij
XX
i
j
Single multinomial:
0 =
Yij
mij
XX
i
0 = n
j
XX
i
j
!
1
@mij
@k
Example: Vitamin C
!
@mij
@k
Examine the eectiveness of
Vitamin C as a cold
preventative.
mij
Independent multinomials:
0 =
Yij
mij
XX
i
0 = nj
j
X
i
!
j
There are 20 subjects available
for study.
@mij
@k
mij
361
360
Randomly divide the 20 subjects
into two groups, with 10 in each
group.
Randomly select one group to
receive the Vitamin C treatment
(treatment group).
Double Blind experiment:
Neither the subjects nor the people who administer the treatments and record results know
which subjects are receiving the
active treatment.
The members of the other group
(control group) are treated
with a placebo.
362
363
Results
No Cold Cold
Treated with
Vitamin C
y11 = 9
y12 = 1 10
Controls
y22 = 5 10
y21 = 5
y+1 = 14 y+2 = 6
H0 : Placebo and Vitamin C are
equally eective for preventing
colds. (independence)
HA : Vitamin C is more eective
(one-sided alternative)
364
Given the assumption that H0 is true
and all marginal totals are xed, what
is the probability of observing a table
of counts at least as inconsistant with
H0 as the observed table of counts?
146
P r(Y11 = 9) = 9201
10
9 1
5 5
= 12; 012
184; 756
= :0650
146
0
P r(Y11 = 10) = 10
20
10
10 0
4 6
1001
184; 756
= :0054
=
366
Randomization Argument:
If H0 is true, then y+1 = 14 \no colds"
and y+2 = 6 \colds" are features of the
20 subjects used in this study that cannot be changed by random assignment
of subjects to groups.
Consequently, all row totals and all column totals are \xed" quantities when
H0 is true.
Vitamin C
Placebo
No Cold
Y11
Cold
Y+1 = 14 Y+2 = 6
-
%
Y1+ = 10
Y2+ = 10
xed when H0 is true
Other counts in the 2 2 table are
determined by the value of Y11.
365
p-value = P r(Y11 9)
= P r(Y11 = 9) + P r(Y11 = 10)
= :0704
Conclusion?
Two-sided test:
H0 : Treatment and placebo are equally
eective
HA : not H0.
p value = P r(Y11 9) + P r(Y11 5)
= :1408
367
This is referred to as
Fisher's \Exact" Test
It requires an ordering of possible tables of counts with respect
to how consistent they are with
H0 relative to the specied alternative.
An appropriate ordering is not
always obvious or convenient.
368
Order tables using values of
2
G
=2
or
2
X
=
X X
i j
X X
i j
^ ij )
ij log(Yij =m
Y
(Yij
^ ij )2
m
^ ij
m
or use \exact" probabilities
(conditional on H0 is true).
Prftable of countsg
=
I (Y !) QJ (Y !)
i=1 i+
j =1 +j
Y++!(QIi=1 QJj=1 Yij !)
Q
These generally produce dierent orderings and dierent
p-values for testing H0.
370
Stay Get
Improve Same Worse
Drug A
3
6
11 20
Drug B
8
4
8 20
Drug C
10
5
5 20
21
15
24
Is the following table less consistent with H0?
4 7 9
7 3 10
10 5 5
369
Listing all tables with X 2 values equal
to or larger than X 2 for the observed table of counts becomes an
overwhelming task as the group sizes
and/or the number of response categories and/or the number of groups
increase. There are too many possible
tables.
Simulate tables of counts with.
the appropriate sets of row and
column totals and compute the
percentage with X 2 values larger
than the X 2 value for the observed
table
Use the chi-squre approximation
for the distribution of X 2 when H0
is true.
371
Example:
Incidence of Common Colds in
a double blind study involving
279 French skiers.
Large sample chi-squared test:
column row
total
total
Expected =
total for
count
entire table
L. Pauling (1971),Proc. Natl. Acad. Sciences, 68 pp 2678-2681)
Dykes & Meier (1975, JASA, 231, 10731079).
No Cold Cold
Vitamin C 122
17 139
Placebo
109
31 140
2
3
2
3
4
5
4
5
2
3
4
5
115.1 23.9
115.9 24.1
HO : Vitamin C and the
placebo are equally eective
in preventing colds
(Yij m
^ ij )2
m
^ ij
i j
= 4:81
with p-value = 0:028
X2 =
HA : Vitamin C is more eective
than the placebo
XX
373
372
/* This program is stored in the file
exact.sas
\Exact" test:
*/
DATA SET1;
INPUT ROW COL COUNT;
CARDS;
-
p value
=
f
11 122g
1 1 9
+P rfY11 109g
2 1 5
Pr Y
231 48
122 17
279
139
=
= 0:038
231 48
123 16
279
139
+
1 2 1
+ 2 2 5
run;
PROC FREQ DATA=SET1;
TABLES ROW*COL / CHISQ EXACT;
WEIGHT COUNT;
RUN;
374
375
DATA SET2;
The FREQ Procedure
INPUT ROW COL X;
Table of ROW by COL
CARDS;
1 1 3
Frequency
Percent
Row Pct
Col Pct
1 2 6
1 3 11
2 1 8
2 2 4
1
5.00
10.00
16.67
10
50.00
2
5
25.00
50.00
35.71
5
25.00
50.00
83.33
10
50.00
14
70.00
6
30.00
20
100.00
RUN;
PROC FREQ DATA=SET2;
TABLES ROW*COL / EXACT CHISQ;
WEIGHT X;
Total
9
45.00
90.00
64.29
3 1 10
3 3 5
2
1
2 3 8
3 2 5
1
Total
RUN;
377
376
Statistics for Table of ROW by COL
Statistic
DF Value
Prob
Chi-Square
1 3.8095 0.0510
Likelihood Ratio Chi-Square
1 4.0700 0.0437
Continuity Adj. Chi-Square
1 2.1429 0.1432
Mantel-Haenszel Chi-Square
1 3.6190 0.0571
Phi Coefficient
0.4364
Contingency Coefficient
0.4000
Cramer's V
0.4364
WARNING: 50% of the cells have expected counts
less than 5. Chi-Square may not be a
valid test.
Fisher's Exact Test
Cell (1,1) Frequency (F)
Left-sided Pr <= F
Right-sided Pr >= F
9
0.9946
0.0704
Table Probability (P)
Two-sided Pr <= P
0.0650
0.1409
Frequency
Percent
Row Pct
Col Pct
1
3
Total
1
3
5.00
15.00
14.29
6
10.00
30.00
40.00
11
18.33
55.00
45.83
20
33.33
2
8
13.33
40.00
38.10
4
6.67
20.00
26.67
8
13.33
40.00
33.33
20
33.33
3
10
16.67
50.00
47.62
5
8.33
25.00
33.33
5
8.33
25.00
20.83
20
33.33
21
35.00
15
25.00
24
60
40.00 100.00
Total
378
2
379
#
This code is stored in the file
#
Statistic
DF
Chi-Square
Likelihood Ratio Chi-Square
Mantel-Haenszel Chi-Square
Phi Coefficient
Contingency Coefficient
Cramer's V
Value
Prob
4 6.3643
4 6.8949
1 5.5580
0.3257
0.3097
0.2303
0.1735
0.1415
0.0184
Fisher's Exact Test
Table Probability (P)
Pr <= P
2.040E-04
0.1575
Sample Size = 60
380
exact.ssc
#
Splus has a built in function for
#
the Fisher exact test.
#
to work for any two-way table, as
#
long as the counts are not too
#
large and there are not too many
#
rows or columns in te table. The
#
manual says there can be at most
#
10 rows or 10 columns in the table,
#
but this restriction may be relaxed
#
in newer releases. It uses a
#
combination of algorithms developed
#
by Joe (1988), Cryus and Metha (1985).
#
The p-value is always for a two-sided
#
or multi-sided test.
It seems
381
> fisher.test(matrix(c(9,1,5,5),
ncol=2,byrow=T))
fisher.test(matrix(c(9,1,5,5),ncol=2,
byrow=T))
Fisher's exact test
data:
ncol = 2, byrow = T)
fisher.test(matrix(c(3, 6, 11, 8, 4,
8, 10, 5, 5),ncol=3,byrow=T))
matrix(c(9, 1, 5, 5),
p-value = 0.1409
alternative hypothesis: two.sided
fisher.test(matrix(c(3,4,5,6,7,8,9,
> fisher.test(matrix(c(3, 6, 11, 8, 4,
1,2,3,4,5,11,12,13,14,15,
8, 10, 5, 5), ncol=3,byrow=T))
16,17,1,2,3,4,5),
Fisher's exact test
ncol=12,byrow=T))
data:
matrix(c(3, 6, 11, 8, 4, 8,
10, 5, 5), ncol = 3, byrow = T)
382
383
p-value = 0.1575
alternative hypothesis: two.sided
> fisher.test(matrix(c(3,4,5,6,7,8,
9,1,2,3,4,5,11,12,13,
14,15,16,17,1,2,3,4,5),
ncol=12,byrow=T))
Fisher's Exact test is based on random
assignment of subjects to groups.
There are other \exact" tests for other
situations:
McDonald, et al. (1977)
Technometrics, 19, 145-151
Berkson (1978) Journal of
> Error in fisher.test(matrix(c(3, 4,
5, 6,7, 8, 9,..: matrix 'x'
must have at most ten rows and
ten columns.
Dumped
Statistical Planning and
Inference, 2, 27-42.
Haber (1986) Psychological
Bulletin, 2, 27-42.
Agresti (2002) Categorical Data
Analysis, Wiley, New York, 91-104.
>
384
385