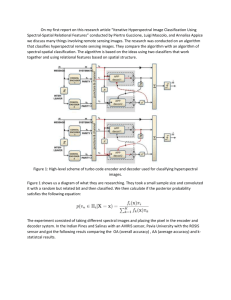
This report was taken from the: Prepared by
advertisement

This report was taken from the: Final Report on ITD Contributions to the Healthy Forest Project Prepared by Institute for Technology Development with input from Science Systems and Applications, Inc. Grant #NNS07AA21G October 1, 2009 1 Hyperspectral Data of Invasive Species and Sudden Oak Death The Institute for Technology Development (ITD) has collected and analyzed hyperspectral data using its patented hyperspectral sensor for demonstrating how the use of high resolution airborne hyperspectral imagery could improve early detection of anomalies in forest conditions of surveyed lands. Specifically, ITD engaged in two separate case studies. The first study investigated hyperspectral imaging as a means to improve the pre-visual detection of Sudden Oak Death (SOD) syndrome, while the second focused on the identification of invasive plant species that threaten forests of the Eastern United States. The following two subsections are summarized from the documents “Detection, Mapping, and Monitoring of Sudden Oak Death Using Hyperspectral Imagery” (Copenhaver, et. al.) and “Identification of Invasive Plant Species in the Daniel Boone National Forest Using Hyperspectral Imagery” (Copenhaver, et. al.2008). These compilations are intended to be an initial draft of what could be a US Forest Service General Technical Report publication. Sudden Oak Death Detection Case Study Sudden oak death (SOD) caused by the invasive non-native pathogen Phytophthora ramorum, is the name given to an epidemic tree disease, first detected in 1995. This pathogen is an Oomycete – not a fungus. It is in the water mold family and related to certain marine algae. It is lethal to a number of oak species: coast live oak (Quercus agrifolia), California black oak (Q. kelloggii), and Shreve oak (Q. parvula var. shrevei).and tanbark oak (Lithocarpus densiflorus) (McPherson et al., 2001). The disease is currently known to exist in the coastal ranges in California and several locations in southern Oregon (Kelly, 2001). Since first detected, over one million individual overstory trees of tanoak, California black oak, and coastal live oak have died due to SOD infection (California Oak Mortality Task Force, 2007a). The symptoms of P. ramorum infection vary among plant species and symptoms are not necessarily consistent within any given species. One characteristic of the infection in oak species is the sudden simultaneous leaf death on a major stem or of the entire tree, thus, the term sudden oak death. Infected trees may exhibit ‘bleeding‘ of sap from apparently intact bark or through cankers that develop on the tree from near the soil level up to as high as 4 m (California Oak Mortality Task Force, 2007b). Several other organisms are often associated with P. ramorum infection, including ambrosia beetles (Monarthrum scutellare and M. dentiger) and the saprophytic fungus Hypoxylon thuorsianum (Goheen et al., 2006). Black, dome-shaped fruiting bodies may appear near the P. ramorum canker and on other portions of the bole of the tree (Garbelotto et al., 2001). At present, the only definitive ways to diagnose a SOD infection in a tree are by culturing the pathogen or by amplifying the DNA using polymerase chain reaction (PCR) techniques (Kelly, 2001). Since it was first detected, SOD has had a significant economic impact. At a cost of more than $15 million, over 1.6 million plants were investigated by state agencies and USDA-APHIS for SOD. In one Southern California nursery, over one million camellias valued at nearly $9 million, were destroyed because of SOD infection (Alexander, 2005). California exports nearly half its nursery production (i.e., plants and flowers) to markets in other states or countries, while Oregon exports an estimated 90% if its production. Canada is a major market for that nursery stock and initially, Canada shut its markets to most plant stock from the states of Oregon and California. Without reopened market access, Oregon nurseries alone faced losses in sales to Canada of $15 to $20 million. Costs also appear in the form of quarantines and regulations. Of the funds available to address the epidemic, California has dispersed over $1 million dollars to SOD infected counties for the removal 2 of trees that pose a threat to life, property, and/or public works (Cole, 2001). In Oregon, more than $1.5 million has been spent in an attempt to eradicate the pathogen from forests. There are many examples of the use of remotely sensed imagery to monitor overstory forest health parameters. Although a majority of the work has focused on coniferous forests, remote sensing has also been utilized to investigate the health and structure of hardwood forests (Boyer et al., 1998; Everitt et al., 1999; Gong et al., 1999). High resolution, multispectral imagery has been used for the identification of SOD in the coastal Western US forests (Kelly, 2001; Kelly, 2003; Kelly et al., 2004). The seasonality of image acquisition is a critical consideration for accurate monitoring of this disease. It appears that late spring through summer is the ideal time for data acquisition. The specific months within this time frame may vary from year to year, but optimal imagery must be acquired after complete leaf expansion in the oaks and before California buckeye (Aesculus californica) begins its summer deciduous process. When buckeye begins to change color, it can appear very similar to SOD-affected trees when viewed from a remote location. In some areas of California, buckeye may begin changing in late June (Kelly, 2003). Sudden Oak Death has several characteristics that make an integrated monitoring approach that combines remote sensing with fieldwork ideal. Generally, as trees with the disease die, the entire crown changes dramatically from healthy green to light brown over a relatively short period of time. The change from green to light brown, however, may be more subtle with the canopy progressively yellowing over a period from months to years. After canopy change has occurred, the brown leaves can stay attached to the branches for months, and in areas where SOD is advanced, the affected trees display spatial clumping, with diseased trees likely to be clumped together. This pattern can result in dramatic spectral reflectance changes across broad areas (Kelly, 2001). High spatial resolution remotely sensed data has provided mixed results in mapping and monitoring SOD. Kelly (2003) observed that SOD has spectral reflectance shift across the visible to NIR range as the disease progresses. They found, however, that automated spectral classification methods alone do not provide high accuracies (i.e., 68.8%). None-the-less, classifications using the visible and NIR bands combined with Tasseled Cap “greenness” and “wetness” components improved overall accuracy to 81.7%. Further work (Kelly et al., 2004) evaluated supervised, unsupervised, and “hybrid” classification methods for discriminating dead and dying tree crowns (as a result of SOD) from the surrounding forest mosaic. They determined that unsupervised and supervised methods provided overall classification accuracies of 66.1-78.8% and 73.5-87.9%, respectively. They indicated that use of hybrid methods, which incorporated both unsupervised and supervised techniques, resulted in overall accuracies of 93-95%. The Western Wildland Environmental Threat Assessment Center (WWETAC) in Prineville, OR was designated to coordinate U.S. Forest Service (USFS) efforts in the western U.S. to reduce the likelihood of forest disturbances, mitigate the effects of disturbances, improve information access through centralization, and improve tracking of forest risk and disturbance over space and time. Another center, located in Asheville, NC has a similar directive, with a focus on threats to Eastern forests. One high priority overarching goal of the threat assessment centers is the development of an integrated, national Early Warning System (EWS) to identify, detect, and rapidly respond to forest environmental threats. Although the operational requirements for this EWS are still evolving within the USFS, it is envisioned to be a multi-source, multi-scale tool that provides broad area, high frequency reconnaissance; detailed location-specific analysis using higher-resolution imagery and ground data; and predictive modeling of potential and emerging forest threats. 3 The overall goal of the ITD SOD case study was to evaluate the use of hyperspectral imagery for improving early warning system capability as it relates to pre-visual detection of SOD syndrome. Specific objectives are to: 1) collect representative ground data from forest species infected and not infected with SOD; 2) perform two hyperspectral image acquisitions over the study area; 3) develop a classification product that separates tanoak and other host species (when applicable) from other forest species; 4) determine the degree of separability between SOD infected and non-infected species and at what stage of development SOD may first be identified with hyperspectral imagery; 5) identify spectral locations most useful for identifying SOD; and 6) classify and map the extent of SOD in the study areas using multiple analysis techniques. After the September 2006 working meeting in Portland, Oregon, a project was established to evaluate the use of remotely sensed imagery for the detection and mapping of SOD. With the assistance of the USFS, project collaborators were identified and two study sites selected: one in Humboldt County, California; the other in Curry County, Oregon (Figure 1). Both areas had and continue to have significant amounts of SOD. Both study areas contained numerous highly vulnerable tanoak trees and experienced widespread yet variable occurrences of SOD. Figure 1. Location of SOD Hyperspectral Case Study. Hyperspectral imagery was acquired using ITD‘s RDACS-H4 hyperspectral sensor on two occasions over both study areas and ground data were collected by the project collaborators and ITD for use in all subsequent analyses. The data was converted to at-sensor radiance, was atmospherically calibrated, and also georectified. To address the project‘s objectives, a number of statistical and image processing techniques were utilized to compute necessary cover type and SOD detection products. All analyses efforts focused on techniques that may be integrated into the EWS, once it becomes operational. Because of the nature of the work being performed, the conducted analysis relied on a tiered approach. First, analysis was performed to separate the species of interest (i.e., tanoak) from other forest land cover. Second, analysis was performed to identify SOD occurrence within the tanoak species cover type. In addition to identifying the tanoak species cover, the hyperspectral data was also preliminarily analyzed for its ability to identify and map the locations of other SOD host species such as California Bay Laurel. 4 To determine if statistical differences existed between the spectral reflectance data for infected and non-infected trees and other native land cover, a mixed model (Littell et al., 1996) was utilized. The mixed model analysis was used to characterize the spectral differences between forest land cover/species types as well as the various symptoms expressed by SOD. For each combination of land cover types and/or vegetation health, comparisons were made using the ESTIMATE statements in the MIXED procedure in the SAS System Version 9.0 (SAS Institute, Cary, NC). The analysis was performed on each image date as well as on the reflectance differences that were observed between the two dates. For the image classifications, most of the analysis was performed using the Iterative Self-Organizing Data Analysis Technique (ISODATA) unsupervised classification algorithm (Tou and Gonzalez, 1974). The ISODATA algorithm was utilized to break the imagery into a large number of classes (at least 60 initial cluster classes). After the initial cluster development, the resulting classes were labeled and merged to identify tanoak and/or other species with SOD using statistical techniques and ground data collected by the project team. If shadows were prevalent in the data, they were labeled as such using unsupervised techniques. Accuracy assessments were performed using a portion of the ground data that was set aside prior to the start of analysis. Because of the less diverse species composition of the relevant forest types found in Oregon, a majority of the analyses was focused on that study site. This was done with the hope that some of the spectral signatures developed from the Oregon study site will be useful in identifying species of interest as well as SOD occurrence at the California site. Ground data points were collected by the project cooperators and by ITD staff during visits to the study sites. The points that were collected not only recorded the locations of SOD, but also recorded the locations of other species (e.g., alder, Douglas fir, madrone). At the California study site a total of 321 points were collected on the ground; while at the Oregon site, a total of 281 points were collected. Additional ground points were obtained from the OakMapper online database of known SOD infections in coastal California. From 2001 to 2007, nearly 1000 points of known SOD infections (in tanoak only) were obtained and utilized in the analysis of the temporal MODIS datasets provided by NASA SSC and SSAI. The hyperspectral datasets provided excellent results for the identification and mapping of forest land cover types, especially of the SOD host species. SOD detection, however, presented more of a challenge and was only possible in the later, terminal stages of infection. The estimated classification accuracies for the forest types for the study sites are summarized in Tables 1 and 2. Table 1. Estimated forest land cover classification accuracies for the Oregon study site. Class Tanoak Alder Douglas fir Clearing/burn area Sparse vegetation Overall Accuracy 90 96 93 97 94 94 5 Table 2. Estimated forest land cover classification accuracies for the California study site. Class California redwood Tanoak California bay laurel White oak Madrone Coast live oak Douglas fir Overall Accuracy 93 79 81 73 85 73 89 81 To identify SOD occurrence, the previously classified tanoak cover were segmented into additional classes and then further processed to identify trees potentially infected with SOD. On several flight lines of the Oregon study site, some of the tanoak had been treated with herbicide approximately five weeks prior to image data collection. Thus, an attempt was also made to define a class that contained the tanoaks that were treated with herbicide. Results from a portion of flight line OR-11 in Oregon are shown in Figure 2. The classification shows the surrounding healthy vegetation as well as the dead tanoak and the tanoak that are declining from the herbicide treatment. Accuracy assessment results (Table 3) indicate very good accuracies for the mapping of the dead tanoak and the declining tanoak. Overall accuracies were typically above 85% for the classifications identifying tanoak that died as a result of SOD infection. 6 Figure 2. Identification of dead tanoak (from SOD) and declining tanoak from a prior application of herbicide to help control the spread of SOD. Table 3. Estimated classification accuracies for detection of dead and declining tanoak in Oregon. Class Advanced SOD Declining tanoak (herbicide treated) Overall Accuracy 90 83 81 For the California study site, it was expected that accuracies for the identification of SOD would be lower than those observed in Oregon. The overall accuracies for the identification of SOD were typically around 80%, agreeing well with the results Kelly et al. (2004) obtained from their study. Figure 3 shows the results of an unsupervised ISODATA classification performed on flight line CA 9 (July 2007). The zoom area shown in the map indicates two instances where SOD was identified via the ground data and in the imagery. 7 Figure 3. Unsupervised classification products from flight line CA-9 in the California study area. In addition to the hyperspectral datasets, ASTER and MODIS satellite data were also obtained. MODIS 250 meter vegetation phenology datasets for the June -July time frame of 2007 corresponded to the collection period of the hyperspectral datasets. The coarser spatial resolution provided by ASTER and the time-series MODIS products proved largely ineffective in discriminating between healthy and SOD infected tanoak trees (accuracies ≤ 60%). The ASTER data at 15 to 30 meter spatial resolution, however, was useful for mapping forest land cover types, especially at the Oregon study site. Despite the larger spatial extent covered by the satellite image datasets, the coarser spatial resolution of the data resulted in significantly lower accuracies than those obtained using airborne hyperspectral imagery. This is due in part to the small patch sizes of the SOD-infected forests. For this particular application, the Multi-Temporal / Multi-Resolution (MT/MR) approach may prove difficult to implement as the acreage of disturbance would need to be substantial for detection with moderate to coarse spatial resolution satellite datasets. Alternative datasets with higher spatial resolution and future sensors may increase the feasibility of this application. The classification methodology used in this work developed by this effort has been described in detail in the report titled, “Detection, Mapping, and Monitoring of Sudden Oak Death Using Hyperspectral Imagery” and should aid 8 project collaborators in their ongoing efforts to mitigate the impact of SOD in coastal California and Oregon (Copenhaver et al., 2008). In summary, the RDACS-H4 hyperspectral datasets provided excellent results for the identification and mapping of forest land cover types in the study areas, especially of those that are host species for SOD. SOD detection, however, presented more of a challenge and was only possible in the later stages of infection. From the two dates of imagery acquired (October 2006 and July 2007), the July imagery proved to be the most useful in discriminating between SOD-infected and healthy tanoak trees (accuracies ≥ 80%). Since the completion of this project, there have been new journal publications on the use of high spatial resolution hyperspectral remote sensing for SOD detection, including work by Pu et al., (2008a, 2008b). The latter publications also provide positive findings on the feasibility of using airborne hyperspectral remote sensing for aiding SOD mitigation. Case Study on Invasive Plant Species Detection This subsection regards results of an ITD case study on the use of high spatial resolution airborne hyperspectral image data for detecting targeted invasive plant species that occur in forests of the eastern US. Invasive plant species cause serious ecological disturbances making monitoring and mapping important. There are thousands of invasive plant species that occur in the Unites States many of which pose problems to native forests. Non-indigenous weeds are spreading and invading approximately 700,000 hectares of US wildlife habitat per year (Babbitt, 1998). According to a statement from U.S. Department of the Interior (1998, 2000), weeds have infested 100 million acres in the U.S., spread at 14 percent per year – consuming 4,600 acres of Federal lands per day. According to a research report in 2000, the annual damage caused by invasive plants is $34 billion and at least 5000 introduced plant species have escaped and currently exist in natural U.S. ecosystems (Pimentel, 2000). According to a 1998 conservative estimate count, there are 5000 alien plant species, 350 of which experts say are serious and dangerous invaders. Farmers and ranchers spend more than $5 billion just for controlling weeds and losses exceed $7 billion (Babbitt, 1998). Traditional, field-based mapping and monitoring of the extent and density of invasive plant infestation presents several challenges, including inaccessibility of areas for field sampling, extent and density, and budget constraints on field sampling and monitoring (Jakubauskas, 2002). Early discovery and mapping of invasive plants distribution can reduce cost, improve efficiency and reduce time required in eradication efforts (Pimentel, 2000). Several remote sensing technologies have shown potential for mapping invasive species occurrences at small to broad regional scales with greater accuracy and precision than field based methods. Aerial photographs have been effective in mapping a few weeds like leafy spurge (Everitt, 1996). However, the major disadvantage of this approach is that it relies on the nonnative plant possessing visually detectable unique characteristics at the observed time of year. Processing of the imagery via aerial photo interpretation requires extensive manual labor (Everitt, 1996; Anderson, 1993) and only small spatial areas can be collected (Underwood et al., 2003). Researchers have also tried using high resolution multispectral digital imagery (Carson, 1995) to map invasions. Hyperspectral remote sensing provides high-resolution spectral data and the potential for remote discrimination between subtle differences in ground cover types. In some cases, hyperspectral data from space, air, or in-situ has been used to identify occurrence of targeted invasive forest, shrub, and herbaceous plant species. In these examples, advanced imaging techniques were often used to aid or enable the detection process. For example, Underwood et al. (2003) evaluated three methods and 9 hyperspectral imagery to detect invasive plants, namely, minimum noise fraction (MNF), continuum removal, and band ratio indices for mapping iceplant and jubata grass. Validation with field sampled data showed high mapping accuracy for all three methods. Another example regards the use of hyperspectral imaging technology for the detection of kudzu. By using the Haar discrete wavelet transform (DWT) to extract pertinent features from hyperspectral signatures, a detection system was implemented and tested for practical use of kudzu detection (Li, 2001). The study used a hand held ASD spectroradiometer (Hatchell, 1999) to do spectral analysis and reported 100% accuracy (overall) in detection of kudzu. Williams et al. (2002) used AVIRIS airborne hyperspectral imagery to map leafy spurge using a mixture tuned matched filter (MTMF). Overall performance of the MTMF for estimating the percent canopy cover of leafy spurge for all sites was good (r2 = 0.69) with better performance in prairie areas (r2 = 0.79) and poorer performance occurring on wooded sites (r2 = 0.57). These results demonstrated that in open canopies with leafy spurge in the understory, the spectral signature is sufficiently distinct to be detectable. Imagery from the Hyperion hyperspectral sensor on board the EO-1 spacecraft was used to estimate and map the occurrence of Chinese tallow during senescence (Ramsey 2001). Using a sub-pixel extraction model, five characteristic spectra that adequately represented the site-specific and also the corresponding Hyperion imaged canopy reflectance spectra data sets were extracted. Ramsey (2001) also employed Landsat ETM+ data to detect Chinese Tallow based on identification of subtle spectral anomalies (Ramsey, 2001). Invasive species such as leafy spurge and spotted knapweed randomly distributed in a non-uniform environment were mapped using hyperspectral imagery obtained using the Probe-1 airborne sensor. An overall accuracy of 86% was obtained using the Breiman Cutler classification method (Lawrence, 2006). In a project aimed towards early detection of tamarisk, remotely sensed data products such as natural color, color-infrared, and hyperspectral imagery were collected in the summer season. The results verified that such imagery could be used for mapping tamarisk during the growing season but early detection would require sophisticated analysis techniques (Laes and Maus, 2006). Additionally, extensive research has been carried out for the detection of cogon grass using sophisticated analysis techniques such as wavelet transform (Mathur, 2002), best-bases selection, decision-fusion (Venkataraman, 2005), and projection pursuit (Lin, 2004; West, 2006). Case study discussed in this subsection was done in collaboration with the Eastern Forest Environmental Threat Assessment Center (EFETAC) in Asheville, NC. EFETAC was designated the lead for developing and implementing a forest threat Early Warning System (EWS). EFETAC is responsible for aiding U.S. Forest Service (USFS) efforts to identify, assess, and mitigate forest threats in the eastern United States. In doing so, this group works to mitigate the effects and reduce the likelihood of forest disturbances, improve information access through centralization, and improve tracking of forest risk and disturbance over space and time. The Western Forest Threat center, located in Prineville, OR has a similar directive, with a focus on threats to western forests. The overarching goal of the threat assessment centers is the development of an integrated, national EWS to identify, detect, and rapidly respond to forest environmental threats. Although the operational requirements for the EWS are still evolving within the USFS, it is envisioned to be a multi-source, multi-scale tool that provides broad area, high frequency reconnaissance; detailed location-specific analysis using high-resolution imagery and ground data; and predictive modeling of potential and emerging forest threats. The overall goal of this invasive species study was to evaluate the use of airborne hyperspectral imagery for improving early warning system capability as it relates to the identification and mapping 10 invasive species in a forest ecosystem. Specific objectives were to: 1) collect representative ground data from native and invasive species in the study area; 2) collect hyperspectral imagery over the study area; 3) determine the degree of separability between species and identify spectral locations most useful for species discrimination; 4) classify and map the extent of invasive species using image classification techniques; and 5) evaluate the use of advanced analysis techniques (i.e., wavelet analysis) for mapping invasive species. The study regarded a few targeted invasive plant species. The study of all non-native invasive plants in the Eastern U.S. is beyond the scope of this project. Based on discussions with the project collaborators, several species were identified that are particularly harmful in Eastern US forests. This study focused on autumn olive, multiflora rose, ailanthus, Chinese silvergrass, and mimosa. The hyperspectral imagery was collected in portions of the Daniel Boone National Forest located in Eastern Kentucky. This location was chosen by the EFETAC because of the large number of invasive species present, especially in areas surrounding reclaimed mine lands, highways, and railroad rightof-ways. Ground points were collected using a differentially-corrected global positioning receiver (DGPS) and a ruggedized handheld field computer on four different occasions. The ground data were collected as points, lines, or polygons for every species identified and were used to provide training as well as accuracy assessment points. Remotely sensed imagery was collected using the ITD RDACS-H4 hyperspectral sensor. The RDACS-H4 is an updated version of the sensor described by Mao (2000) and collects data from 294nm to 1173-nm at 6-nm bandwidths for a total of 150 bands. Imagery was collected at a spatial resolution of one (1) meter. The image footprint from the sensor covers an area of 1600 × 5700 meters. Two data acquisitions were carried out: one took place 21 Oct. 2006 and the other on June 2007. Image acquisition dates were determined by Forest Service personnel in cooperation with ITD and were timed to match phenological periods of optimal separability between invasive and native species such as green-up, flowering or senescence. During the October data collection mission, imagery was obtained over 17 flight lines and during the June data collection mission, imagery was obtained over 7 flight lines. The number was reduced to cover flight lines for which adequate ground data existed and also to provide time to acquire 0.5-m resolution imagery over many of the study sites. Flight lines were spread throughout the locations within the forest with known invasive weed problems (Figure 4). The data was converted to at-sensor radiance, was atmospherically calibrated, and also georectified. 11 Figure 4. Map of the June 2007 flight lines for the Daniel Boone National Forest. Unsupervised classification categorizes the images into a specified number of spectral classes according to image statistics and pixel values (Campbell, 2002). The ISODATA (Iterative SelfOrganizing Data Analysis Technique) method initially assigns pixels into spectral classes based on statistics. The new centroids are found for each class and the pixels are then re-allocated to the closest centroid if it has changed position. This process is repeated until the change threshold that is determined by the analyst is met (Campbell, 2002). The change threshold was set at 95 percent leaving only five percent of the pixels acceptable to be able to be changed for the classification. The statistical settings are also determined by the analyst, for this dataset a standard deviation of 1.0 was used and a minimum class distance of 3.0 was used. Smaller values for the class standard deviation and class distance allow for less variance within each class. A total of 50 spectral classes were utilized for the DBNF dataset. Image data with spectral bands ranging from 400 to 819nm were utilized for all classifications. Imagery was spectrally subset from the original 150 bands to 72 bands because of noise in the data below 400 nm and spectral bleeding occurring in the near-infrared (NIR). To prepare the image for use in the ISODATA unsupervised classification, all pixel values were multiplied by 1000. This was done to provide a greater dynamic range for unsupervised classification with the ISODATA algorithm. This analysis utilized the information provided by the ground truth information collected for the project. The points collected for each site were overlaid on the georeferenced and calibrated hyperspectral datasets. The regions of interest (ROIs) were identified on the imagery as points, buffered polygons, irregular polygons, and lines. To perform the End Member analysis, a spectral library must first be built. This is done using the Spectral library builder tool available in ENVI 4.2. The tool uses the previously identified ROIs to build a spectrum unique to a particular species. The reliability of the spectrum is based on the number of training points. For example, if 150 pixels are used to build a spectrum autumn olive, the 12 spectrum would be more reliable than if a spectrum was built using only 15 pixels. Figure 5 shows the spectra created from flight line 10. In this example, the image consisted of 72 bands and 15 different types of vegetation were identified from the ground points. The study area in flight line 10 consists primarily of a reclaimed strip mine and the invasive plant species of interest here are autumn olive and bush honeysuckle. Figure 5. Spectral library of End Member spectra of relevant native and invasive plant species. After the spectral library has been constructed, it is then saved as an End Member collection file. This is an ASCII file generated by ENVI and contains information about the unique set of spectra that were collected. This file is then utilized in the classification process at a later stage. The wavelet transform is a digital signal processing technique that can be used for scalar analysis of hyperspectral reflectance curves. This signal processing tool is used in analysis to differentiate invasive species from neighboring native species as well as to differentiate one invasive from another. The unsupervised ISODATA classification was performed on four different flight lines within the Daniel Boone National Forest during the time period of June 2007. The reason these four flight lines were chosen was because of the abundance of ground points that were collected within each flight line. The DOQQ and information from the ground data collection missions in the area aided in determining the appropriate land cover type each of the spectral classes represented. The ISODATA unsupervised classification was utilized for the Daniel Boone National Forest dataset because of the relatively diverse species matrix observed in the study area. The focus of this project was identification of invasive species, which is why there was more ground truth points collected within the invasive land cover types, while the native species land cover types were not as heavily collected. This fact made supervised classification difficult because each land cover class needs numerous points for it to be classified. Of the different flight lines, Flight line 7 had a larger amount of ground truth points and this is exhibited by the highest overall classification accuracy (85%) (Figure 6) (Table 4). Another reason that flight line 7 might have higher classification accuracy is because the ground truth points were collected within a larger land area (i.e., greater diversity). Most of the ground points within the flight lines were all collected within a small area, whereas with flight line 7 the ground points collected covered almost the entire length of the flight line. Also, when looking at the individual land cover accuracies for flight line 7, the most accurate classification was the ailanthus, which had the most pixels available to be classified. The number of pixels used for the accuracy assessment could also 13 result in misleading classification accuracies for some of the land cover classes, particularly the native species. When investigating the number of pixels that were utilized to evaluate the classification, several species had less than 10 pixels. Thus, some bias (either positive or negative) may exist when accuracies were computed. Figure 6. Overview of the ISODATA classification from flight line 7 imagery acquired 12, June 2007. In this image, there are more roads and urban areas and more ground truth points that were collected. Table 4. Estimated accuracy of ISODATA classification of targeted invasive plant species – computed from flight line 7. Class Ailanthus Native Overall Accuracy Producer Accuracy % 82.8 96.6 Consumer Accuracy % 99.3 49.1 84.8 The End Member classification technique was applied to the hyperspectral imagery collected in the Daniel Boone National Forest (DBNF) on 21 Oct. 2006 and 12 June 2007. The spectral library developed from the ground data was used to classify the flight lines of interest for each collection. The classification image in Figure 7 shows the different types of land cover identified in the flight line 7 by the classification algorithm. An accuracy assessment was performed on each classification and is shown in Table 5 for flight line 7. 14 Figure 7. The overview of the End Member classification for flight line 7 (atmospherically calibrated method) from 12 June 2007 imagery collection. Table 5. Estimated classification accuracies of targeted invasive species from classification of flight 7 data acquired 12 June 2007. Class Calibration Method Ailanthus Overall Accuracy User Accuracy % Emp. Line Atmospheric 97.8 96.5 Emp. Line 59 Producer Accuracy % Emp. Line Atmospheric 59 72 Atmospheric 56 The End Member classification was implemented on the imagery collected in the Daniel Boone National Forest so that the spectral library developed using the ground data collected in one flight line may be used for other flight lines with limited ground data. This made it possible for various land covers to be collected during ground data collection. This method was very useful in classification of flight lines where only a few ground points were collected, (i.e., flight line 17). The spectral library for flight line 10 was used together with the spectral library created for flight line 17, which made the objective of applying the End Member technique successful. Since the goal of the project was to identify invasive species, individual class accuracies of the invasive species provide a clearer picture of the effectiveness of this method of analysis. The overall accuracy could be misleading since the distribution of ground points collected for all types of landcover in a particular flight line was not evenly distributed. Ailanthus has been effectively identified in flight lines 7 and 17 with very good user accuracies. Autumn olive was best identified in flight line 10, especially for the October data collection. Bush honeysuckle has not been identified effectively in flight line 10 mainly due to the fact that there were very few training pixels. This was observed in the drastic producer/user accuracy difference. The wavelet analysis model was applied to flight lines 10 and 17 from the hyperspectral imagery collected in October 2006 and flight lines 3, 7, 10 and 18 from the June 2007 imagery were analyzed using the model that was developed for the analysis in this project. Both the Haar and db1 wavelets 15 were applied to the imagery. Analysis I consistently performed better than Analysis II. Hence, the results from Analysis I are reported for June imagery analysis whereas results from both analyses are shown for the October imagery. The wavelet analysis model has been applied on data from both date imagery collections- October 2006 and June 2007. This model was applied to parts of the imagery neighboring areas from where ground points were selected. The probability of finding invasive plants in a small area of the imagery was explored here. These small areas were selected around regions that would be revisited during the June ground data collection. Large areas of the imagery were not analyzed done due to time restraints posed by the model execution for the entire imagery. The accuracy assessment of bush honeysuckle was better using this method than any of the other analysis methods for the October imagery. However, bush honeysuckle could not be considered for analysis for the June imagery since the number of pixels available for analysis was too small to use for training or testing the classifier. Autumn olive was identified with high accuracies in flight line 10 in October; however analysis of June imagery yielded very poor results reconfirming the conclusion that autumn olive is better discriminated at the end of growth season in fall than in spring. The frost that set in early spring of 2007 could have also affected the growth pattern thus leading to such poor accuracies. This was isolated only to flight line 10 and not flight line 3 where autumn olive was identified with very high accuracies. Wavelet analysis was successful in identifying Chinese silvergrass in flight lines 3 and 18 whereas End Member classification had failed. Ailanthus was identified in flight line 7 in the June imagery with high accuracies whereas analysis of flight line 17 from the October imagery did not yield extremely good results. Hyperspectral imaging sensor technology has paved the way for the remote sensing community to carry out detailed intensive research on target recognition applications. The purpose of this project was to use hyperspectral imagery as an operational, diagnostic tool to analyze forest threats. To achieve this objective, an end-to-end process that includes 1) four ground data collections, 2) two hyperspectral imagery collections, 3) preprocessing of data, 4) analysis of processed imagery using ISODATA, End Member, and Wavelet analysis techniques were carried out. The unsupervised ISODATA classification technique was implemented on the hyperspectral dataset since there were various species of vegetation present in the forest and limited ground truth points collected for certain land cover types. Ailanthus (flight line 7) was classified accurately using this analysis, but results for the other invasive species were not as accurate. It could be either that the ISODATA classification technique was not successful for classification of the hyperspectral imagery or that the number of pixels used for the accuracy assessment led to misleading classification accuracy. The End Member analysis technique was mainly implemented so that the spectral library created using the ground data collected in one flight line may be used for other flight lines with limited ground data. The limited number of ground points available for each class for both invasive and native species led to the choice of this analysis technique. Hence, the objective of implementing was achieved. The classification accuracies of invasive species such as autumn olive and ailanthus were good in the flight lines of interest, but the classification of Chinese silvergrass and bush honeysuckle were not impressive. A few reasons for this occurrence could be either 1) the spectral signature created for the invasive species was not accurate; or 2) spectral separability of a given invasive species from other invasive or native species nearby was not possible due to very similar spectra. 16 The Wavelet analysis technique was implemented to analyze minor variances in the spectra of the invasive species and used features extracted from the minor variances to identify the presence of invasive species. The wavelet analysis produced increased classification accuracies for all flight lines except for flight line 10 (June collection). The classification accuracy for Chinese silvergrass increased significantly in flight lines 3 as well as 18 from below 50% to over 75%. Bush honeysuckle could not be classified using the wavelet analysis model due to the lack of training data. The spectral-spatial resampling approach provided insight about the possibility of invasive plant identification using lower resolution commercial imagery. The accuracies and the reliability of both the wavelet and end member methods dropped significantly. Therefore, a multi-tier approach to identify invasive species that have been identified in this project with high accuracies would probably be a difficult task, at least given the techniques and data tried in this case study. In summary, the feasibility of using airborne high spatial resolution hyperspectral imagery to detect occurrences of targeted selective invasive plant species was demonstrated and validated for an area of concern within the Daniel Boone National Forest. This is but one case study and more are desirable to demonstrate and assess feasibility for additional invasive plant threats to native forests. This potential has also been demonstrated previously for other invasive plants, though both its research and operational use in regard to Federal, State, and private forest land surveys would require additional investment of the USFS. Public Dissemination of ITD Hyperspectral Research Findings Efforts were also made to identify suitable conferences for public dissemination of the Healthy Forest project‘s hyperspectral case studies. It was determined that the following conferences would be highly relevant: 1) American Society for Photogrammetry and Remote Sensing Annual Conference (www.asprs.org) 2) Society of Photographic Instrumentation Engineers (SPIE) Conferences (spie.org/) International Geoscience & Remote Sensing Symposium (IGARSS) (http://www.igarss08.org/) 3) USDA Forest Service Remote Sensing Conference (www.fs.fed.us/eng/rsac/) Tree Care Industry Exposition (http://www.treecareindustry.org/public/meetings_tci_expo.htm) 4) Pacific Agriculture Show (www.agricultureshow.net/) – relevant to SOD only References References are given below in 4 subsections with regard to SOD, invasive plant species, and hyperspectral plant stress detection. SOD Alexander, J. 2005. Review of Phytophthora ramorum in European and North American nurseries. [Online]. Available at http://www.fs.fed.us/psw/publications/documents/psw_gtr196/psw_gtr196_001c_01Alexander. pdf (verified 26 Feb. 2007). Sudden oak death 2nd Science Symposium: The state of our knowledge, Monterey, CA. 17 Boyer, M., J. Miller, M. Belanger, and E. Hare. 1988. Senescence and spectral reflectance in leaves of northern pin oak (Quercus palustris Muenchh.). Remote Sens. of Environ., 25:71-87. California Oak Mortality Task Force. 2007a. Sudden oak death [Online]. Available at http://nature.berkeley.edu/comtf/index.html (accessed 26 Feb. 2007; verified 01 Mar. 2007). Univ. of California, Berkeley. California Oak Mortality Task Force. 2007b. Sudden oak death: Plant symptoms [Online]. Available at http://nature.berkeley.edu/comtf/html/plant_symptoms.html (accessed 26 Feb. 2007; verified 01 Mar. 2007). Univ. of California, Berkeley. Cole, E.F. 2001. The relentless spread of sudden oak death: Tracking a mysterious killer. California Coast and Ocean 17(3):1-4. Everitt, J.D., D. Escobar, D. Appel, W. Riggs, and M. Davis. 1999. Using airborne digital imagery for detecting oak wilt disease. Plant Dis. 83(6):502-505. Copenhaver, K., J. Fridgen, K. Hellrung, and S. Venkataraman, 2008, Detection, Mapping, and Monitoring of Sudden Oak Death Using Hyperspectral Imagery : Final Report for 2008. ITD Report to NASA. Gao, B.C., K.B. Heidelbrecht, and A.F.H. Goetz. 1993. Derivation of scaled surface reflectances from AVIRIS data. Remote Sens. Environ. 44:165‐178. Garbelotto, M., P. Svihra, and D. Rizzo. 2001. Sudden oak death syndrome fells three oak species. California Agric. 55(1):9. Goheen, E.M., E. Hansen, A. Kanaskie, N. Osterbauer, J. Parke, J. Pscheidt, and G. Chastagner. 2006. Sudden oak death and Phytophthora ramorum: A guide for forest managers, Christmas tree growers, and foresttree nursery operators in Oregon and Washington. Publication EM 8877. Oregon State Univ. Ext. Service, Corvallis. Gong, P., X. Mei, G.S. Bignig, and Z. Zhang. 1999. Monitoring oak woodland change using digital photogrammetry. J. of Remote Sens. 3(4):285‐289. ITT Visual Information Solutions. 2007. ENVI User‘s Guide. ITT Visual Information Solutions, Inc. Boulder, CO. Jensen, J.R. 1996. Introductory digital image processing: A remote sensing perspective. 2nd ed. Prentice Hall, Upper Saddle River, NJ. Kelly, M.N. 2001. Monitoring sudden oak death in California using high resolution imagery [Online]. Available at http://nature.berkeley.edu/comtf/pdf/Bibliography/kelly2002a.pdf (verified 26 Feb. 2007). 5th Symp. on Oak Woodlands: Oaks in California‘s changing landscape, San Diego, CA. Kelly, M.N. 2003. Remote sensing of sudden oak death using ADAR imagery. In Proc. 9th Forest Service Rem. Sens. App. Conference: Rapid delivery of remote sensing products, Amer. Soc. Photogram. Remote Sens., San Diego, CA. 18 Kelly, M.N., D. Shaari, Q. Guo, and D. Liu. 2004. A comparison of standard and hybrid classifier methods for mapping hardwood mortality in areas affected by sudden oak death. Photogram. Engr. and Remote Sens. 70(11):1229-1239. Lange, N., B.P. Carlin, and A.E. Gelfand. 1992. Hierarchical Bayes Models for the progression of HIV infection using longitudinal CD4 T‐Cell numbers (with discussion). J. Amer. Stat. Assoc. 87:615-632. Littell, R.C., G.A. Milliken, W.W. Stroup, and R.D. Wolfinger. 1996. SAS system for mixed models. SAS Institute, Cary, NC. Mao, C. 2000. Hyperspectral focal plane scanning: An innovative approach to airborne and laboratory pushbroom hyperspectral sensing. p. 424-428. In Proc. Geospatial Info. in Agric. And Forestry, Vol. 1. ERIM Int‘l, Ann Arbor, MI. McPherson, D.L. Wood, A.J. Storer, N. Maggi Kelly, and R.B. Standiford. 2001. Sudden oak death, a new forest disease in California. Integrated Pest Mngmt. Rev., 6:243‐246. Montes, M.J. and B.C. Gao. 2004. Tafkaa_6S: An atmospheric correction algorithm for the land. Remote Sens. Div., Naval Research Laboratory, Washington, D.C. Pu, R., N. M. Kelly, Q. Chen and P. Gong, 2008, Spectroscopic determination of health levels of Coast Live Oak (Quercus agrifolia) Leaves, Geocarto International. 23(1):3-20. Pu, R., M. Kelly, G. L. Anderson and P. Gong, 2008, Using CASI hyperspectral imagery to detect mortality and vegetation stress associated with a new hardwood forest disease, PE&RS. 74(1):65-75. Tou, J.T. and R.C. Gonzalez. 1974. Pattern recognition principles. Addison‐Wesley, Reading, MA. Invasive Plant Species Copenhaver, K., J. Fridgen, K. Hellrung, S. Venkataraman, Identification of Invasive Species in the Daniel Boone National Forest Using Hyperspectral Imagery: Final Report. 2008. Report delivered to NASA Anderson, G.L., J.H. Everitt, A.J. Richardson, and D.E. Escobar. 1993. Using satellite data to map false broomweed (Ericameria austrotexana) infestation on south Texas rangelands. Weed Tech. 7:865-871. Babbit, B. 1998. Statement on invasive alien species [Online]. Available at ttp://www.hear.org/articles/babbit9804/ (verified 26 Feb. 2007). Science in Wildland Seed Management Symposium, Denver, CO, Bureau of Land Management. Bruce, L.M., Cheriyadat, A., Burns, M. 2003. Wavelets: Getting Perspective, IEEE Potentials, 22(2): 24-27. Burrus, R.A., Gopinath, H. Guo, Introduction to Wavelets and Wavelet Transforms: A Primer, Prentice-Hall, Inc., New Jersey, 1998. 38 19 Campbell, J.B., (2002). Introduction to Remote Sensing, Guilford Press, New York, NY, 3rd edition. Carson, H.W., L.W. Lass, and R.H. Callihan. 1995. Detection of yellow hawkweek with high resolution multispectral digital imagery. Weed Tech. 9:477-483. Department of the Interior. 2000. Invasive species fact sheet [Online]. Available at http://www.nps.gov/plants/alien/pubs/doiinvsp.htm (verified 27 Feb. 2007). ENVI Staff, ENVI User‘s Guide (1997). Better Solutions Consulting, LLC., Lafayette, CO 614 pp. Everitt, J.H., D.E. Escobar, M.A. Alaniz, M.R. Davis, and J.V. Richerson. 1996. Using spatial information technologies to map Chinese tamarisk (Tamarix chinensis) infestations. Weed Sci. 44:194-201. Gao, B.C., K.B. Heidelbrecht, and A.F.H. Goetz. 1993. Derivation of scaled surface reflectances from AVIRIS data. Remote Sens. Environ. 44:165-178. Gonzalez, R. and Woods, R. Digital Image Processing, 2nd edition, Prentice Hall Upper Saddle River New Jersey, 2001. Hatchell, D.C. 1999. Analytical Spectral Devices, Inc. (ASD) Technical Guide (3rd ed.). Jakubauskas, M.E., D.L. Peterson, S.W. Campbell, F. deNoyelles, Jr., S.D. Campbell, and D. Penny. 2002. Mapping and monitoring invasive aquatic plant obstructions in navigable waterways using satellite multispectral imagery. In Proc. Pecora 15/Land Satellite Information IV. Laes , D. and P. Maus. 2006. Tamarisk remote sensing inventory and assessment [Online]. Available at http://www.fs.fed.us/eng/rsac/documents/0046-TIP1.pdf (verified 01 Feb. 2007). U.S. Forest Service, Salt Lake City, UT. Lawrence, R.L., S.D. Wood, and R.L. Sheley. 2006. Mapping invasive plants using hyperspectral imagery and Breiman Cutler classifications (Randomforest). Rem. Sens. Environ. 100(3):356-362. Li, J., L.M. Bruce, J.D. Byrd, and J. Barnett. 2001. Automated detection of Pueraria montana (kudzu) through Haar analysis of hyperspectral reflectance data. Proc. IEEE Geoscience and Remote Sensing Symposium, 5:2247-2249 Lin, H. and L. M. Bruce. 2004. Projection pursuits for dimensionality reduction of hyperspectral signals in target recognition applications. IEEE Proc. International Geoscience and Remote Sensing Symposium, 2: 960-963. Mallat, S.G. 1998. A wavelet tour of signal processing. Academic Press, San Diego. Mao, C. 2000. Hyperspectral focal plane scanning: An innovative approach to airborne and laboratory pushbroom hyperspectral sensing, Proc. Geospatial Information in Agriculture and Forestry. 1: 424-428. Mathur, A., L.M. Bruce, and J. Byrd. 2002. Discrimination of subtly different vegetative species via hyperspectral data. Proc. IEEE Geoscience andRemote Sensing Symposium, 2: 805-807. 20 Montes, M.J. and B.C. Gao. 2004. Tafkaa_6S: An atmospheric correction algorithm for the land. Remote Sens. Div., Naval Research Laboratory, Washington, D.C. Pimentel, D., L. Lach, R. Zuniga, and D. Morrison. 2000. Environmental and economic costs of nonindigenous species in the United States. Bioscience. 50(1):53-65. Ramsey III, E., G. Nelson, A. Rangoonwala, and R. Ehrlich. 2001. Mapping the invasive species Chinesetallow [Online]. Available at http://eo1.gsfc.nasa.gov/new/validationReport/Technology/Documents/Tech.Val.Report/ Science_Summary_Ramsey_Rev%202.pdf (accessed 15 Jan. 2007; verified 26 Feb. 2007). NASAGoddard Spaceflight Center, Greenbelt, MD. Richards, J.A. and Jia, X. Remote Sensing Digital Image Analysis, 3rd ed., New York: Springer, 1999. Schlapfer D., Schaepman M., and Itten K.I., 1998: PARGE. Parametric Geocoding Based on GCPCalibrated Auxiliary Data. Descour and Shen (Eds.), SPIE , Imaging Spectrometry IV, San Diego, 3438: 334-344. Smith, G.M. and E.J., Milton, 1999. The use of the empirical line method to calibrate remotely sensed data to reflectance. International Journal of Remote Sensing. 20(13): 2653-2662. Underwood, E., S. Ustin, and D. DiPietro. 2003. Mapping nonnative plants using hyperspectral imagery. Rem. Sens. of Environ. 86:150–161. Venkataraman, S. and L.M. Bruce. 2005. Hyperspectral dimensionality reduction via localized discriminant bases. Proc. IEEE Geoscience and Remote Sensing Symp. 2:1245- 1248 West, T.R. 2006. Hyperspectral dimensionality reduction via sequential parametric projection pursuits for automated invasive species target recognition. Master‘s Thesis. Mississippi State Univ., Starkville. 40 Williams, A.P. and E.R. Hunt Jr. 2002. Estimation of leafy spurge cover from hyperspectral imagery using mixture tuned matched filtering. Rem. Sens. of Environ. 82:446–456. Hyperspectral Plant Stress Detection Carter, G., and A. Knapp, 2001. Leaf optical properties in higher plants: linking spectral characteristics to stress and chlorophyll concentration. American Journal of Botany 88(4): 677-684. Carter, G. A., M. Seal, and T. Haley. 1998. Airborne detection of Southern Pine Beetle activity using key spectral bands. Canadian Journal of Forest Research 28:1040-1045. Govender, M., K. Chetty, and H. Bulcock, 2006. A review of hyperspectral remote sensing and its application in vegetation and water resource studies. Water SA 33 (2): 145-151. Hallett, R., Pontius, J., Martin, M., and L. Plourde. 2008. The practical utility of hyperspectral remote sensing for early EAB detection. In: Emerald Ash Borer Research and Development Meeting; 21 October 24-25, 2007; Pittsburg, PA. Morgantown, WV: Forest Health Technology Enterprise Team: 67-68. Jackson, S., G. Raber, J. Griffith, and M. Graves, 2008. Concepts for sensor data fusion to detect vegetation stress and implications of ecosystem health following Hurricane Katrina. ERDC TNSWWRP-08-06. 15pp. Jones, C., P. Weckler, N. Maness, M. Stone, and R. Jayasekara, 2004. Estimating water stress in plants using hyperspectral imagery. Paper number 043065, 2004 ASAE Annual Meeting. Lawrence, R., and M. Labus, 2003. Early detection of Douglas-Fir beetle infestation with subcanopy resolution hyperspectral imagery. Western Journal of Applied Forestry 18 (3): 1-5. Mutanga, O., R. Ismail, F. Ahmet, and L. Kumar, 2007. Using insitu hyperspectral remote sensing to disciminate pest attacked pine forests in South Africa. 2007 Proceedings of the Asian Association on Remote Sensing. Pontius, J.; Martin, ME, and Hallett, RA. 2005a. Using AVIRIS to assess hemlock abundance and early decline in the Catskills, New York. Remote Sensing of Environment. 2005; 97:163-173. Pontius, J.; Hallett, R. A., and Martin, M. E. 2005b. Assessing hemlock decline using hyperspectral imagery: signature analysis, indices comparison and algorithm development. Journal of Applied Spectroscopy. 2005; 59 (6): 836-843. Pontius, J.; Hallett, R. A., and Martin, M. E. 2006. Foliar chemistry linked to hemlock woolly adelgid success and hemlock susceptibility. Environmental Entomology 35(1):112-120. Pontius, J., Martin, M., Plourde, L. and R. Hallett. 2008. Ash Decline Assessment in Emerald Ash Borer infested regions: a test of tree-level, hyperspectral technologies. Remote Sensing of Environment, 112, 5: 2665-2676. Prather, J., E. Aumack, T. Sisk, H. Hampton, B. Dickson, and Y. Xu, 2004. An Assessment of Various Definitions of the Wildland-Urban Interface (WUI): Implications for Management in a Southwestern Landscape. ESA Meeting Presentation, August 1-8. Pu, R., P. Gong, G. Biging, and M. Larrieu, 2003. Extraction of red edge optical parameters from hyperion data for estimation of forest leaf area index. IEEE Transactions on Geoscience and Remote Sensing 41 (4): 916-921. Pu, R., N. M. Kelly, Q. Chen and P. Gong, 2008, Spectroscopic determination of health levels of Coast Live Oak (Quercus agrifolia) Leaves, Geocarto International. 23(1):3-20. Pu, R., M. Kelly, G. L. Anderson and P. Gong, 2008, Using CASI hyperspectral imagery to detect mortality and vegetation stress associated with a new hardwood forest disease, PE&RS. 74(1):65-75. Shafri, H., M. Salleh, and A. Ghiyamat, 2006. Hyperspectral remote sensing of vegetation using red edge position techniques. American Journal of Applied Sciences 3 (6): 1864-1871. 43 22 Turner, W., S. Spector, N. Gardiner, M. Fladeland, E. Sterling, and M. Steininger, 2003. Remote Sensing for Biodiversity science and conservation. Trends in Ecology and Evolution 18: 306-314. Zarco-Tejada, P., 2000. Hyperspectral remote sensing of closed forest canopies: estimation of chlorophyll fluorescence and pigment content. Doctoral Dissertation, Graduate Programme in Earth and Space Science, York University, Toronto, Ontario, Canada. 224 pp. Zarco-Tejada, P., J. Miller, G. Mohammed, T. Noland, and P. Sampson, 2002. Vegetation stress detection through Chlorophyll a + b estimation and fluorescence effects on hyperspectral imagery. J. Environ. Qual. 31: 1433-1441. 23