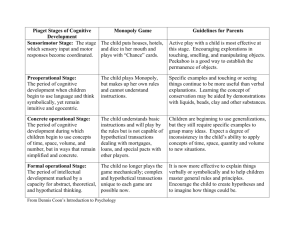
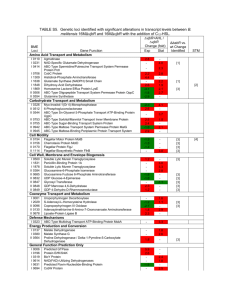
Table S1: 16S rRNA gene sequence for CL500-11-LM reconstructed by... Closest match in GenBank Reconstructed sequence (CL500-11-LM)
advertisement

Table S1: 16S rRNA gene sequence for CL500-11-LM reconstructed by EMIRGE. Closest match in GenBank Uncultured Crater Lake bacterium CL500-11 16S ribosomal RNA gene (AF316759) Reconstructed sequence (CL500-11-LM) GTTTGATCCTGGCTCAGGATGAACGCTGGCGGCGTGCCTAATACATGCAAGTCGAACGAA GAGTGTAGCAATATGCTCTTAGTGGCGAACGGGTGAGTAACACGTGGGTGACCTACCCCA AAGTGTGGAATAACTCCTCGAAAGAGGTGCTAATACCGCATGTGGTCCTAGAGATTAGAG GCTTAGGGACTAAAGACTTCGGTCGCTTTGGGAGGGGCCCGCGGCCCATCAGCTAGTTGG TGAGGTAATGGCTCACCAAGGCAGTGACGGGTAGGGGACTTGAGAGGGTGACCCCCCACA CTGGAACTGAAACACGGTCCAGACACCTACGGGTGGCAGCAGTAGGGAATATTGGTCAAT GGGCGAAAGCCTGAACCAGCAACGCCGCGTGCGCGATGAAGGCCTTCGGGTCGTAAAGCG CTTTTGGGAGGGATGAAAATGACAGTACCTCCCGAATAAGGATCGGCTAACTACGTGCCA GCAGCCGCGGTAAGACGTAGGATCCAAGCGTTATCCGGAATTACTGGGCGTAAAGGGCGT GTAGGAGGTTGGGCAAGTCGGCCATGAAAGCTCCCGGCTCAACTGGGAGAGGCTGGTCGA TACTGCCTGGCTAGAGGGCAAGAGAGGGAGGTGGAATTCCCGGTGTAGTGGTGAAATGCG TAGATATCGGGAGGAACACCAGTGGCGAAGGCGGCCTCCTGGCTTGTACCTGACTCTGAA ACGCGAAAGCATGGGTAGCAAACAGGATTAGAAACCCTGGTAGTCCATGCCATAAACGAT GTCAACTGGGTGTCGGCGAAGTAAAATTTGTCGGTGCCGAAGCTAACGCGTTAAGTTGAC CGCCTGAGGACTACGATCGCAAGATTAAAACTCAAAGGAATTGACGGGGACCCGCACAAG CAGCGGAGCGTGTGGTTTAATTCGAGGCTACGCGAAGAACCTTACCAGGGTTTGACATTC ATGTAGTAGGGAAGCGAAAGCGGACCGACCCTTCGGGGAGCATGAACAGGTGATGCATGG CTGTCGTCAGCTCGTGCCGTGAGGTGTTCGGTTAAGTCCGTTAACGAGCGCAACCCTTGT CGCATGTTACACGTGTCATGCGAGACTGCCGGTGTCAAACCGGAGGAAGGTGGGGACGAC GTCAAGTCAGCATGTCCTTTATATCCTGGGCTACACACACGCTACAATGGCCGGGACAAT GGGTTGCCAAGCCGCGAGGCGGAGCCAATCCCTCAAACCCGGTCTCAGTTCGGATTGTAG GCTGCAACTCGCCTACATGAAGTTGGAGTTGCTAGTAACCGCGCGTCAGCACAGTGCGGT GAATACGTTCCCGGGTCTTGTACACACCGCCCGTCACGTCATGGGAGCTGGCAATGCCTG AAGCCGGTGGCCTAACCGCAAGGAAGGAGCCGTCTAGGGCAGGGTCGGTGACTGGGACGA AGTCGTAACAAGGTAGCAGTACCGGAA Table S2. Assembly statistics. Number of contigs, their total length, N50, and largest contig length from (a) the Joint Genome Institute assemblies for metagenomic data generated from the 0.22-3 µm fraction of water samples collected from the deep waters at the off-shore station of Lake Michigan in spring, summer, and fall (Sp13.BD.MM110.DD, Su13.BD.MM110.DN, and Fa13.BD.MM110.DN, respectively), (b) GAM-NGS merging of these three assemblies, (c) the Chloroflexi ESOM bin and its constituent subbins identified by MaxBin, (d) the CL500-11-LM bin identified by mmetagenome, and (e) the manually curated CL500-11-LM bin. Assembly Joint Genome Institute Sp13.BD.MM110.DD Su13.BD.MM110.DN Fa13.BD.MM110.DN Contigs Length (bp) N50 (bp) Max (bp) 60,737 5,333 50,283 141,586,820 10,092,395 111,363,949 2,595 1,911 2,423 131,701 24,845 73,994 67,811 173,739,793 3022 131701 869 198 453 210 3,352,252 540,165 2,371,731 417,901 5,973 55,999 Maxbin 1 Maxbin 2 Maxbin 3 Chloroflexi CL500-11-LM (mmgenome) 503 2,585,961 7,247 44,001 Chloroflexi CL500-11-LM curated 398 2,248,119 7,673 44,001 gam-ngs assembly Chloroflexi bin (ESOM) Table S3. checkM analysis. Genome completeness and purity for the original and curated CL500-11-LM sequence bin, as well as for its closest fully sequenced isolate. Contamination” is the proportion of markers present at larger copy number than expected, while “Strain heterogeneity” is the share of contamination that can be explained by strain-variable regions included in the assembly (> 95% sequence identity between redundant marker gene copies). Genome CL500-11-LM CL500-11-LM curated A. thermophila Marker lineage k__Bacteria k__Bacteria k__Bacteria marker gene occurrence 0 1 2 3 4 5+ 22 124 17 0 0 0 22 137 4 0 0 0 9 148 6 0 0 0 Completeness Contamination Strain heterogeneity 84.46 % 84.46 % 93.64 % 13.64 % 3.64 % 4.73 % 94.12 % 75.00 % 0% Table S4: CL500-11-LM genes that support an aerobic heterotrophic lifestyle. Locus_tag Product Name A. Aerobic phosphorylation Ga0063436_12234 NADH-quinone oxidoreductase, B subunit Ga0063436_12235 NADH:ubiquinone oxidoreductase 49 kD subunit 7 Ga0063436_12236 Formate hydrogenlyase subunit 6/NADH:ubiquinone oxidoreductase 23 kD subunit (chain I) B. Glycolysis Ga0063436_100611 glyceraldehyde-3-phosphate dehydrogenase (NAD+) Ga0063436_103915 6-phosphofructokinase Ga0063436_10843 DhnA-type fructose-1,6-bisphosphate aldolase Ga0063436_111617 bifunctional phosphoglucose/phosphomannose isomerase Ga0063436_11275 pyruvate kinase Ga0063436_12043 phosphoglycerate mutase Ga0063436_12687 Phosphoglycerate kinase Ga0063436_12691 enolase B. Tricarboxylic acid cycle Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide acyltransferase (E2) component, and related enzymes Ga0063436_113722 Pyruvate/2-oxoglutarate dehydrogenase complex, dehydrogenase (E1) component, eukaryotic type, Ga0063436_113723 beta subunit Ga0063436_113724 pyruvate dehydrogenase E1 component, alpha subunit Ga0063436_12068 dihydrolipoamide dehydrogenase Ga0063436_101414 aconitase Ga0063436_11053 succinyl-CoA synthetase, alpha subunit Ga0063436_11054 succinyl-CoA synthetase, beta subunit Ga0063436_121110 isocitrate dehydrogenase (NADP) Ga0063436_13091 Pyruvate:ferredoxin oxidoreductase and related 2-oxoacid:ferredoxin oxidoreductases, beta subunit Ga0063436_13092 2-oxoacid:acceptor oxidoreductase, alpha subunit Ga0063436_10852 Aspartate ammonia-lyase Ga0063436_12117 succinate dehydrogenase subunit B Ga0063436_12118 succinate dehydrogenase subunit A Ga0063436_12119 Succinate dehydrogenase, hydrophobic anchor subunit Ga0063436_121113 citrate synthase Ga0063436_10633 malate synthase EC 1.6.5.3 1.6.5.3 1.6.5.3 1.2.1.12 2.7.1.11 4.1.2.13 5.3.1.8 / 5.3.1.9 2.7.1.40 5.4.2.12 4.2.1.11 2.3.1.12 1.2.4.1 1.2.4.1 1.8.1.4 4.2.1.3 6.2.1.5 6.2.1.5 1.1.1.42 1.2.7.3 1.2.7.3 4.2.1.2 1.3.5.1 1.3.5.1 2.3.3.1 2.3.3.9 Table S5: Gene annotation and read recruitment data for CL500-11-LM. Locus Tag Product Name COG Pfams Metatranscriptomics (number of reads recruited - offshore station) Sp.SN Ga0063436_10011 pfam00120 pfam03951 pfam03710 pfam08335 pfam00005 pfam12848 pfam00211 pfam00672 E Ga0063436_10022 L-glutamine synthetase (EC 6.3.1.2) Glutamate-ammonia ligase adenylyltransferase/GlnD PIIuridylyltransferase ATPase components of ABC transporters with duplicated ATPase domains Adenylate cyclase, family 3 (some proteins contain HAMP domain) Ga0063436_10023 Bacteriorhodopsin R Ga0063436_10024 Phytoene dehydrogenase and related proteins Q pfam01036 pfam01266 pfam13450 Ga0063436_10411 Phytoene/squalene synthetase I Ga0063436_10412 phytoene desaturase Q Ga0063436_10413 Arabinose efflux permease Ga0063436_10414 Ga0063436_10415 Ga0063436_10012 Ga0063436_10021 R Sp.DD Su.DCMD Su.DN Fa.DN 319 956 375 75 705 953 8 172 25 0 211 325 250 686 122 88 587 570 418 812 174 150 338 442 1582 4191 582 480 1206 1850 148 389 66 21 67 162 pfam00494 84 401 18 16 71 163 pfam01593 145 694 52 10 133 236 G pfam05977 34 116 24 4 90 166 FAD dependent oxidoreductase - pfam12831 170 335 150 3 447 487 Arabinose efflux permease G pfam07690 84 173 60 8 204 269 Ga0063436_10416 CDP-alcohol phosphatidyltransferase - pfam01066 18 80 10 12 50 95 Ga0063436_10417 GTP cyclohydrolase I (EC 3.5.4.16) H pfam01227 47 100 26 36 72 112 Ga0063436_10041 - 10 3 7 0 10 13 C pfam03450 pfam00111 pfam01799 20 22 15 18 77 87 Ga0063436_10043 CO dehydrogenase flavoprotein C-terminal domain Aerobic-type carbon monoxide dehydrogenase, small subunit CoxS/CutS homologs Asp-tRNAAsn/Glu-tRNAGln amidotransferase A subunit and related amidases J pfam01425 10 140 28 10 117 271 Ga0063436_10044 hypothetical protein - 0 0 3 0 6 43 Ga0063436_10045 CoA binding domain/CoA-ligase - pfam00549 pfam02629 pfam13607 36 41 75 10 118 236 Ga0063436_10046 Protein of unknown function (DUF1116) - pfam06545 61 205 51 18 204 325 Ga0063436_10047 R 318 143 37 492 632 C pfam04199 pfam01315 pfam02738 183 Ga0063436_10048 Predicted metal-dependent hydrolase Aerobic-type carbon monoxide dehydrogenase, large subunit CoxL/CutL homologs 172 305 150 53 500 705 Ga0063436_10051 glutamate 5-kinase - pfam00696 19 94 18 4 149 223 Ga0063436_10052 - - 36 84 30 41 207 239 Ga0063436_10053 hypothetical protein Predicted oxidoreductases (related to aryl-alcohol dehydrogenases) C pfam00248 99 201 80 57 391 499 Ga0063436_10054 Acyl carrier protein IQ pfam00550 19 36 27 6 43 34 Ga0063436_10055 Acyl-CoA synthetases (AMP-forming)/AMP-acid ligases II IQ pfam00501 Ga0063436_10056 hypothetical protein - - Ga0063436_10061 AhpC/TSA antioxidant enzyme - Ga0063436_10062 Arabinose efflux permease Ga0063436_10063 Uncharacterized conserved protein Ga0063436_10064 Ga0063436_10065 Ga0063436_10066 Ga0063436_10042 T Sp.SD 78 299 75 21 323 438 213 534 119 40 554 707 pfam13911 54 437 7 18 57 79 G pfam07690 254 670 21 10 55 120 S - 14 301 12 16 24 78 Phytoene dehydrogenase and related proteins Q pfam13450 58 636 12 22 52 187 hypothetical protein - - 10 266 18 4 36 93 Alpha/beta hydrolase family Predicted hydrolases or acyltransferases (alpha/beta hydrolase superfamily) Response regulators consisting of a CheY-like receiver domain and a winged-helix DNA-binding domain His Kinase A (phospho-acceptor) domain/Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase - pfam12695 23 177 12 16 28 68 R pfam12697 pfam00072 pfam00486 pfam00512 pfam02518 23 71 2 0 22 40 43 66 16 64 100 162 41 71 41 79 98 163 Major Facilitator Superfamily glyceraldehyde-3-phosphate dehydrogenase (NAD+) (EC 1.2.1.12) - 15 47 14 63 39 53 G pfam07690 pfam00044 pfam02800 12 40 37 81 38 82 - - 0 0 11 12 6 20 IQR 139 183 39 36 141 175 P pfam00106 pfam01476 pfam12849 1266 790 236 4577 1588 1922 P pfam00528 145 228 64 340 322 427 P pfam00528 57 65 29 81 109 195 Ga0063436_100617 hypothetical protein Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) phosphate ABC transporter substrate-binding protein, PhoT family (TC 3.A.1.7.1) phosphate ABC transporter membrane protein 1, PhoT family (TC 3.A.1.7.1) phosphate ABC transporter membrane protein 2, PhoT family (TC 3.A.1.7.1) phosphate ABC transporter ATP-binding protein, PhoT family (TC 3.A.1.7.1) P pfam00005 32 41 41 45 111 121 Ga0063436_10071 methylthioadenosine phosphorylase (EC 2.4.2.28) F pfam01048 37 163 26 4 145 194 Ga0063436_10072 Glycosyltransferases involved in cell wall biogenesis M pfam00535 23 158 24 0 117 204 Ga0063436_10067 Ga0063436_10068 Ga0063436_10069 Ga0063436_100610 Ga0063436_100611 Ga0063436_100612 Ga0063436_100613 Ga0063436_100614 Ga0063436_100615 Ga0063436_100616 TK - Ga0063436_10073 transcriptional attenuator, LytR family - pfam03816 13 72 24 12 130 169 Ga0063436_10074 hypothetical protein - - 69 326 46 30 231 693 Ga0063436_10075 hypothetical protein - - 6 103 4 0 78 114 Ga0063436_10076 hypothetical protein - 47 64 16 20 206 205 Ga0063436_10077 cell division protein FtsZ D pfam00091 pfam12327 84 163 90 22 424 652 Ga0063436_10078 hypothetical protein - - 54 171 46 34 239 481 Ga0063436_10079 - - 146 374 114 26 596 912 V 136 22 4 79 94 M pfam00005 pfam02687 pfam12704 66 Ga0063436_100711 hypothetical protein ABC-type antimicrobial peptide transport system, ATPase component ABC-type transport system, involved in lipoprotein release, permease component 151 637 84 30 435 554 Ga0063436_100712 L-aminopeptidase/D-esterase EQ pfam03576 72 296 62 9 268 400 Ga0063436_100713 Predicted permeases R pfam01925 8 30 32 3 105 151 Ga0063436_100714 ADP-ribosylglycohydrolase O pfam03747 8 80 20 0 164 175 Ga0063436_100715 Smr domain - pfam01713 22 14 10 6 47 70 Ga0063436_100716 HemN C-terminal domain - pfam06969 16 30 22 2 53 112 Ga0063436_100717 Radical SAM superfamily - pfam04055 84 70 30 34 140 180 Ga0063436_100718 hypothetical protein - 169 83 84 7 384 385 Ga0063436_11331 Glycosyltransferase M 226 499 159 44 978 1472 Ga0063436_11332 Glycosyltransferase M 91 295 104 12 447 715 Ga0063436_11333 Glycosyl transferases group 1/Methyltransferase domain - pfam00534 pfam13414 pfam13432 pfam14559 pfam00534 pfam14559 pfam12847 pfam13524 25 170 67 28 360 372 Ga0063436_11334 Protein kinase domain - 36 62 20 0 58 84 Ga0063436_11335 Glycosyltransferase M pfam00069 pfam00534 pfam00535 54 69 44 42 275 354 Ga0063436_11336 hypothetical protein - - 8 16 16 0 95 126 Ga0063436_11337 Tetratricopeptide repeat - 0 0 0 0 0 4 Ga0063436_11338 M 48 95 20 8 156 352 2291 3518 1493 879 8907 10937 667 1637 665 299 2875 3906 Ga0063436_113311 Glycosyltransferases involved in cell wall biogenesis Bacterial flagellin N-terminal helical region/Bacterial flagellin Cterminal helical region Bacterial flagellin N-terminal helical region/Bacterial flagellin Cterminal helical region Flagellar hook-associated protein 2 N-terminus/Flagellar hookassociated protein 2 C-terminus - pfam13432 pfam00535 pfam13414 pfam13432 pfam00669 pfam00700 pfam00669 pfam00700 pfam02465 pfam07195 169 491 195 33 910 1471 Ga0063436_13821 Flagellar hook-associated protein 2 C-terminus - pfam07195 27 110 31 26 134 289 Ga0063436_13822 FlaG protein pfam03646 201 185 115 68 528 720 Ga0063436_13823 Flagellin-specific chaperone FliS NU O - 104 105 58 43 258 355 Ga0063436_13824 Sortase family - pfam04203 44 222 12 14 269 423 Ga0063436_13825 hypothetical protein - 124 111 54 36 273 335 Ga0063436_13826 flagellar basal-body rod protein FlgC N pfam00460 pfam06429 80 105 72 64 302 392 Ga0063436_13827 NU 48 61 22 215 342 260 738 273 78 1198 1437 Ga0063436_13829 Flagellar motor switch protein N pfam02049 pfam01514 pfam08345 pfam01706 pfam14841 pfam14842 65 Ga0063436_13828 flagellar hook-basal body complex protein FliE flagellar basal-body M-ring protein/flagellar hook-basal body protein (fliF) 182 268 196 72 757 1060 Ga0063436_138210 Flagellar biosynthesis/type III secretory pathway protein NU 68 357 58 32 317 652 Ga0063436_138211 type III secretion system ATPase, FliI/YscN (EC 3.6.3.15) 77 216 60 32 406 567 Ga0063436_138212 flagellar export protein FliJ NU NO U pfam02108 pfam00006 pfam02874 pfam02050 35 79 12 10 93 231 Ga0063436_138213 hypothetical protein - - 5 27 4 0 57 84 Ga0063436_138214 Transglycosylase SLT domain - pfam01464 25 114 40 2 124 269 Ga0063436_138215 flagellar basal-body rod protein FlgB N pfam00460 24 97 21 14 186 201 Ga0063436_138216 Flagellar hook-length control protein FliK - pfam02120 22 130 40 14 150 293 Ga0063436_138217 Flagellar hook capping protein - N-terminal region - 133 195 82 34 251 331 Ga0063436_138218 flagellar hook-basal body protein N pfam03963 pfam00460 pfam06429 pfam07559 200 298 168 109 948 1022 Ga0063436_138219 hypothetical protein - - 96 266 64 6 286 449 Ga0063436_138220 carbon storage regulator, CsrA T pfam02599 44 194 85 12 245 538 Ga0063436_100710 Ga0063436_11339 Ga0063436_113310 - NU Ga0063436_138221 Uncharacterized protein conserved in bacteria S pfam02623 93 306 82 58 457 587 Ga0063436_138222 Flagellin and related hook-associated proteins N 124 419 174 40 713 830 Ga0063436_138223 - 175 559 225 88 1057 1769 Ga0063436_10101 flagellar hook-associated protein FlgK DnaB-like helicase N terminal domain/DnaB-like helicase C terminal domain - pfam00669 pfam00460 pfam06429 pfam00772 pfam03796 159 270 65 59 359 430 Ga0063436_10102 Uncharacterized conserved protein S pfam02577 280 500 243 78 773 895 Ga0063436_10103 ADP-ribose pyrophosphatase F pfam00293 73 459 82 49 462 697 Ga0063436_10104 ribosomal protein L7/L12 J pfam00542 634 888 426 207 2246 2495 Ga0063436_10105 LSU ribosomal protein L10P J pfam00466 205 812 158 80 1159 1363 Ga0063436_10106 ribosomal protein L1, bacterial/chloroplast J 326 827 196 54 1146 1298 Ga0063436_10107 LSU ribosomal protein L11P J pfam00687 pfam00298 pfam03946 294 622 208 102 1021 1245 Ga0063436_10108 transcription antitermination protein nusG K pfam02357 318 756 189 179 979 1297 Ga0063436_10109 preprotein translocase, SecE subunit, bacterial U pfam00584 98 196 64 35 361 447 Ga0063436_101011 ribosomal protein L33, bacterial type J pfam00471 197 417 163 43 755 776 Ga0063436_10111 Nucleotidyl transferase Predicted kinase related to galactokinase and mevalonate kinase - 50 64 35 5 129 167 R pfam00483 pfam00288 pfam08544 51 105 58 6 153 170 GDP-D-mannose dehydratase Pyruvate/2-oxoglutarate dehydrogenase complex, dehydrogenase (E1) component, eukaryotic type, alpha subunit Pyruvate/2-oxoglutarate dehydrogenase complex, dehydrogenase (E1) component, eukaryotic type, beta subunit Predicted pyridoxal phosphate-dependent enzyme apparently involved in regulation of cell wall biogenesis M pfam01370 34 14 20 8 96 139 C 48 113 74 6 186 172 C pfam00676 pfam02779 pfam02780 58 143 34 14 103 126 M pfam01041 44 99 58 10 104 183 - pfam01370 3 75 7 20 25 66 Ga0063436_10121 NAD dependent epimerase/dehydratase family haloacid dehalogenase superfamily, subfamily IA, variant 1 with third motif having Dx(3-4)D or Dx(3-4)E R 26 204 32 2 157 249 Ga0063436_10122 molybdenum cofactor biosynthesis protein A, bacterial H 38 234 77 28 369 403 Ga0063436_10123 Mismatch repair ATPase (MutS family) L pfam13419 pfam04055 pfam06463 pfam13353 pfam00488 pfam05192 36 123 35 28 218 320 Ga0063436_10131 Activator of Hsp90 ATPase homolog 1-like protein - 41 130 19 0 47 110 Ga0063436_10132 Predicted integral membrane protein S 13 208 55 33 213 339 Ga0063436_10133 FAD/FMN-containing dehydrogenases C pfam08327 pfam10028 pfam10035 pfam01565 pfam02913 132 125 73 40 274 399 Ga0063436_10141 Peptidase family M20/M25/M40 - pfam01546 14 68 16 6 87 120 Ga0063436_10142 E 264 66 40 320 371 E pfam02274 pfam01546 pfam07687 52 Ga0063436_10143 N-Dimethylarginine dimethylaminohydrolase Acetylornithine deacetylase/Succinyl-diaminopimelate desuccinylase and related deacylases 64 254 104 32 287 405 Ga0063436_10144 dTDP-4-dehydrorhamnose reductase (EC 1.1.1.133) M pfam04321 62 222 52 20 214 301 Ga0063436_10145 Predicted glycosyltransferases R pfam00535 20 67 10 0 118 197 Ga0063436_10146 Glycosyltransferases involved in cell wall biogenesis M pfam00535 24 64 44 10 61 207 Ga0063436_10147 RNase HII (EC 3.1.26.4) L pfam01351 0 74 2 0 50 57 Ga0063436_10148 Glycosyltransferase M pfam00534 15 78 13 4 91 94 Ga0063436_10149 Capsular polysaccharide biosynthesis protein M - 38 161 38 14 183 248 Ga0063436_101410 Capsular polysaccharide biosynthesis protein M pfam02706 100 244 76 25 326 351 Ga0063436_101411 capsular exopolysaccharide family D pfam13614 89 180 57 31 258 381 Ga0063436_101412 glucose-1-phosphate thymidylylransferase, long form M pfam00483 84 275 71 21 287 310 Ga0063436_101413 hypothetical protein - 28 114 46 25 145 166 Ga0063436_101414 aconitase (EC 4.2.1.3) C pfam00330 pfam00694 Ga0063436_101415 SnoaL-like domain - Ga0063436_101416 (p)ppGpp synthetase, RelA/SpoT family TK Ga0063436_101417 molybdenum cofactor synthesis domain Ga0063436_101418 Predicted membrane protein Ga0063436_101419 Ga0063436_101420 Ga0063436_101421 Ga0063436_10112 Ga0063436_10113 Ga0063436_10114 Ga0063436_10115 Ga0063436_10116 Ga0063436_10117 233 654 257 154 941 1328 135 492 138 104 757 912 112 134 46 14 290 308 H pfam12680 pfam02824 pfam04607 pfam13291 pfam13328 pfam00994 pfam03453 pfam03454 35 88 16 18 197 159 S pfam07884 96 229 137 24 280 431 hypothetical protein - - 215 266 138 77 493 634 S1 RNA binding domain - pfam00575 475 796 326 115 1249 1539 Nucleoside-diphosphate-sugar epimerases MG pfam01370 64 242 90 38 453 518 Ga0063436_101422 GDP-mannose 4,6-dehydratase M 60 173 65 27 319 418 G pfam01370 pfam00408 pfam02878 pfam02879 pfam02880 Ga0063436_101423 Phosphomannomutase Ga0063436_101424 hydrolase, TatD family 148 204 114 79 484 706 L pfam01026 91 423 42 4 260 407 Ga0063436_101425 Ga0063436_101426 Geranylgeranyl pyrophosphate synthase H pfam00348 172 844 58 61 270 503 Starch synthase catalytic domain - pfam08323 28 131 14 11 46 85 Ga0063436_10151 O-antigen ligase like membrane protein - pfam13425 30 132 16 8 162 197 Ga0063436_10152 hypothetical protein - - 66 275 78 18 458 614 Ga0063436_10153 Predicted membrane protein (DUF2142) - pfam09913 36 73 52 20 252 253 Ga0063436_10154 hypothetical protein - - 164 304 106 60 641 755 Ga0063436_10155 Carbohydrate binding domain - 93 173 119 88 432 386 Ga0063436_10156 Excinuclease ABC subunit C L pfam02018 pfam01541 pfam02151 pfam08459 pfam12826 51 305 137 16 307 464 Ga0063436_10157 murein biosynthesis integral membrane protein MurJ R pfam03023 63 109 36 12 137 227 Ga0063436_10158 Methyltransferase domain - pfam08241 58 71 14 2 185 256 Ga0063436_10159 LPXTG-site transpeptidase (sortase) family protein - pfam04203 51 289 101 10 285 404 Ga0063436_101510 hypothetical protein - 89 176 42 38 139 229 Ga0063436_101511 J 31 447 35 13 184 341 Ga0063436_101512 bacterial peptide chain release factor 1 (bRF-1) protein-(glutamine-N5) methyltransferase, release factorspecific pfam00472 pfam03462 J pfam05175 44 316 34 31 118 181 Ga0063436_101513 Zn-dependent hydrolases, including glyoxylases R pfam00753 202 425 127 59 528 731 Ga0063436_10161 hypothetical protein - - 4 0 4 19 18 17 Ga0063436_10162 hypothetical protein - 61 87 54 14 84 125 Ga0063436_10171 DNA-directed DNA polymerase III (polc) L pfam02811 pfam07733 pfam14579 64 199 90 16 375 548 Ga0063436_10172 ADP-ribose pyrophosphatase F pfam00293 42 141 59 62 196 259 Ga0063436_10173 LysM domain - pfam01476 92 308 85 6 344 578 Ga0063436_10174 hypothetical protein - - 12 40 12 0 64 139 Ga0063436_10175 Uncharacterized low-complexity proteins S pfam00805 62 237 96 46 379 589 Ga0063436_10176 L-threonine synthase (EC 4.2.3.1) E pfam00291 54 385 43 10 452 685 Ga0063436_10177 hypothetical protein - - 11 13 13 0 16 48 Ga0063436_10178 Periplasmic component of the Tol biopolymer transport system U pfam07676 85 262 88 44 354 661 Ga0063436_10179 Short-chain dehydrogenases of various substrate specificities R pfam00106 33 138 20 2 211 322 Ga0063436_101710 Carbonic anhydrase P pfam00194 86 352 67 38 376 609 Ga0063436_101711 Calx-beta domain - 93 167 113 33 477 511 Ga0063436_101712 E 39 140 22 12 190 360 Ga0063436_101713 Choline dehydrogenase and related flavoproteins Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) pfam03160 pfam00732 pfam05199 IRQ pfam00106 11 61 18 0 75 178 Ga0063436_101714 hypothetical protein - - 69 156 75 16 265 306 Ga0063436_101715 hypothetical protein ABC-type dipeptide/oligopeptide/nickel transport systems, permease components ABC-type dipeptide/oligopeptide/nickel transport systems, permease components - 0 14 2 0 12 19 EP pfam00528 pfam12911 7 119 37 34 130 179 PE pfam00528 86 122 73 38 270 460 E pfam00496 789 1769 857 378 4338 4621 KE pfam00155 26 158 35 14 140 246 Ga0063436_101720 ABC-type dipeptide transport system, periplasmic component Transcriptional regulators containing a DNA-binding HTH domain and an aminotransferase domain (MocR family) and their eukaryotic orthologs Diadenosine tetraphosphate (Ap4A) hydrolase and other HIT family hydrolases FGR pfam01230 74 155 39 26 160 270 Ga0063436_101721 Glycerophosphoryl diester phosphodiesterase C 34 126 32 16 102 117 Ga0063436_101722 alpha-galactosidase (EC:3.2.1.22) G pfam03009 pfam02056 pfam11975 12 47 30 5 121 144 Ga0063436_101723 GMP synthase - Glutamine amidotransferase domain F 24 122 48 20 211 228 Ga0063436_101724 DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) L pfam00117 pfam01149 pfam06827 pfam06831 12 171 22 4 119 268 Ga0063436_101725 Adenosine/AMP deaminase - pfam00962 58 77 33 16 218 287 Ga0063436_101726 Adenosine deaminase SecA preprotein cross-linking domain/SecA Wing and Scaffold domain/SEC-C motif F pfam00962 pfam01043 pfam02810 93 281 112 74 551 656 222 681 170 79 573 833 Ga0063436_101716 Ga0063436_101717 Ga0063436_101718 Ga0063436_101719 Ga0063436_101727 - pfam07516 pfam07517 Ga0063436_101728 SecA DEAD-like domain - 24 201 28 12 74 141 FE pfam07517 pfam13793 pfam14572 Ga0063436_101729 ribose-phosphate pyrophosphokinase Ga0063436_101730 LAO/AO transport system ATPase 97 229 47 10 214 388 E pfam03308 56 48 15 4 142 252 Ga0063436_101731 Ga0063436_101732 methylmalonyl-CoA mutase C-terminal domain I pfam02310 127 395 186 22 544 533 heat-inducible transcription repressor HrcA K pfam01628 183 494 121 35 307 379 Ga0063436_101733 Molecular chaperone GrpE (heat shock protein) O pfam01025 137 421 120 59 232 328 Ga0063436_101734 chaperone protein DnaK O 242 917 183 134 772 1147 Ga0063436_101735 chaperone protein DnaJ O pfam00012 pfam00226 pfam00684 pfam01556 156 474 84 64 359 529 Ga0063436_101736 ribosomal protein L11 methyltransferase J pfam06325 57 150 21 36 179 264 Ga0063436_10191 FOG: GAF domain T - 68 336 72 12 275 570 Ga0063436_10192 Uncharacterized conserved protein S pfam06240 64 304 74 4 259 380 Ga0063436_10193 Molybdopterin biosynthesis enzyme H - 94 472 87 18 517 817 Ga0063436_10194 - pfam04250 94 205 48 37 348 599 G pfam00528 44 148 71 68 260 364 Ga0063436_10202 Protein of unknown function (DUF429) carbohydrate ABC transporter membrane protein 1, CUT1 family (TC 3.A.1.1.-) carbohydrate ABC transporter membrane protein 2, CUT1 family (TC 3.A.1.1.-) G pfam00528 90 235 91 31 356 595 Ga0063436_10211 Methylase involved in ubiquinone/menaquinone biosynthesis H 16 112 10 19 67 87 Ga0063436_10212 Glycosyltransferase M pfam08241 pfam00534 pfam13579 74 117 20 10 117 212 Ga0063436_10213 conserved hypothetical protein - pfam03706 92 75 47 12 164 235 Ga0063436_10221 hypothetical protein - - 11 50 17 2 83 132 Ga0063436_10222 hypothetical protein - - 104 95 42 18 248 346 Ga0063436_10223 FeS assembly ATPase SufC O pfam00005 1021 1791 342 197 1348 1324 Ga0063436_10224 Iron-regulated ABC transporter membrane component SufB O pfam01458 2459 4178 1124 521 3791 4250 Ga0063436_10225 FeS assembly protein SufD O pfam01458 695 1637 330 76 1353 1308 Ga0063436_10226 cysteine desulfurases, SufSfamily E pfam00266 244 569 104 57 329 422 Ga0063436_10227 C pfam01592 119 203 59 29 220 244 Ga0063436_10228 SUF system FeS assembly protein, NifU family Ferredoxin subunits of nitrite reductase and ring-hydroxylating dioxygenases PR pfam00355 77 137 23 18 156 248 Ga0063436_10229 Aldo/keto reductase family - pfam00248 36 103 17 14 57 66 Ga0063436_10231 Glycosyl hydrolase family 1 - pfam00232 16 174 18 14 132 214 Ga0063436_10232 hypothetical protein - - 4 0 0 4 6 35 Ga0063436_10233 FR47-like protein - 18 147 12 12 63 173 Ga0063436_10234 J 36 308 94 23 340 390 R 4 12 14 26 48 54 Ga0063436_10241 asparaginyl-tRNA synthetase (EC 6.1.1.22) Membrane protein involved in the export of O-antigen and teichoic acid carbohydrate ABC transporter substrate-binding protein, CUT1 family (TC 3.A.1.1.-) pfam08445 pfam00152 pfam01336 pfam01943 pfam14667 G pfam01547 2871 4062 1887 839 11493 10902 Ga0063436_10242 carbamate kinase (EC 2.7.2.2) E 80 91 100 47 161 233 Ga0063436_10243 E 167 302 139 40 436 494 Ga0063436_10244 putative selenium metabolism hydrolase Coenzyme F420-dependent N5,N10-methylene tetrahydromethanopterin reductase and related flavindependent oxidoreductases pfam00696 pfam01546 pfam07687 C pfam00296 174 338 135 20 436 454 Ga0063436_13055 Hydroxypyruvate isomerase G 16 180 42 24 174 217 Ga0063436_13054 Glutamate dehydrogenase/leucine dehydrogenase E 44 204 94 74 343 517 Ga0063436_13053 Acyl-CoA dehydrogenases I pfam01261 pfam00208 pfam02812 pfam00441 pfam02770 pfam02771 69 144 54 31 227 400 Ga0063436_13051 Septum formation initiator - pfam04977 69 350 80 73 291 443 Ga0063436_10251 G pfam00294 30 136 29 4 124 323 Ga0063436_10252 hexose kinase, 1-phosphofructokinase family Ribulose-5-phosphate 4-epimerase and related epimerases and aldolases G 61 134 56 15 311 451 Ga0063436_10253 rRNA methylase, putative, group 3 J 43 226 16 8 203 360 Ga0063436_10254 cysteinyl-tRNA synthetase (EC 6.1.1.16) J pfam00596 pfam00588 pfam08032 pfam01406 pfam09190 122 395 69 27 416 672 Ga0063436_10255 Integral membrane protein (PIN domain superfamily) R pfam01938 146 307 57 15 459 615 Ga0063436_10261 MOSC domain - 25 46 38 0 127 84 Ga0063436_10262 histidyl-tRNA synthetase (EC 6.1.1.21) J pfam03473 pfam03129 pfam13393 39 288 86 10 375 441 Ga0063436_10201 Ga0063436_10235 Ga0063436_10265 hypothetical protein - - 10 0 14 36 34 46 Ga0063436_10273 glycine cleavage system H protein E pfam01597 153 369 69 21 291 286 Ga0063436_10274 glycine dehydrogenase (decarboxylating) E pfam02347 582 1544 363 78 1117 1392 Ga0063436_10281 Ppx/GppA phosphatase family - pfam02541 44 30 24 0 84 117 Ga0063436_10282 phosphohistidine phosphatase, SixA T pfam00300 26 0 20 8 55 123 Ga0063436_10283 Uncharacterized conserved protein S pfam05235 52 123 48 16 164 220 Ga0063436_10291 O-antigen ligase like membrane protein - pfam13425 14 57 16 4 75 159 Ga0063436_10292 hypothetical protein - - 42 108 32 26 124 116 Ga0063436_10301 Polysaccharide deacetylase - pfam01522 26 26 10 12 55 36 Ga0063436_10302 hypothetical protein - - 99 159 52 75 186 219 Ga0063436_10303 hypothetical protein - 181 505 84 61 447 576 Ga0063436_10304 DNA-directed RNA polymerase subunit beta' (EC 2.7.7.6) K pfam00623 pfam04983 pfam04997 pfam04998 pfam05000 3244 7166 2029 1184 9394 12161 Ga0063436_10305 Superfamily II DNA and RNA helicases LKJ pfam00270 107 451 93 41 574 687 Ga0063436_10306 DNA gyrase/topoisomerase IV, subunit A - pfam00521 8 4 12 19 55 83 Ga0063436_10311 Glycine/serine hydroxymethyltransferase E pfam00464 139 349 152 11 358 625 Ga0063436_10312 hypothetical protein - 0 6 2 0 20 47 Ga0063436_10313 tRNA synthetases class I (I, L, M and V)/Anticodon-binding domain of tRNA/Leucyl-tRNA synthetase, Domain 2 - 262 192 153 74 540 711 Ga0063436_10314 SOS regulatory protein LexA KT pfam00133 pfam08264 pfam13603 pfam00717 pfam01726 115 429 88 28 383 507 Ga0063436_10315 Transcriptional regulator, effector-binding domain/component KT 8 0 0 8 35 28 Ga0063436_10321 hydroxylysine kinase/5-phosphonooxy-L-lysine phospho-lyase apoenzyme - 51 Ga0063436_10331 hypothetical protein - Ga0063436_10332 valyl-tRNA synthetase (EC 6.1.1.9) Ga0063436_10333 Ga0063436_10341 pfam00202 pfam01551 pfam01636 202 54 16 269 346 25 22 28 4 145 107 J pfam00133 pfam08264 pfam10458 105 336 107 30 516 586 Prolyl oligopeptidase family - pfam00326 11 44 18 0 95 109 hypothetical protein - - 62 0 22 12 37 45 Ga0063436_10342 hypothetical protein - - 54 114 57 38 204 175 Ga0063436_10343 hypothetical protein - - 139 122 47 50 182 165 Ga0063436_10344 Aminotransferase class-III - pfam00202 108 281 56 13 290 393 Ga0063436_10351 NAD-dependent aldehyde dehydrogenases C pfam00171 54 450 145 55 455 675 Ga0063436_10352 Acetyl esterase (deacetylase) Q 78 83 65 17 209 397 Ga0063436_10353 Superfamily II DNA and RNA helicases LKJ pfam05448 pfam00270 pfam00271 273 623 140 54 399 608 Ga0063436_10354 hypothetical protein - - 11 22 6 13 34 37 Ga0063436_10361 - - 2 128 12 0 43 119 O pfam02628 pfam07664 pfam07670 60 365 42 8 149 252 Ga0063436_10363 hypothetical protein Uncharacterized protein required for cytochrome oxidase assembly Ferrous iron transport protein B C terminus/Nucleoside recognition 222 367 246 102 2315 2143 Ga0063436_10364 Ferrous iron transport protein B - 142 317 94 72 1253 1070 Ga0063436_10365 Mn-dependent transcriptional regulator K pfam02421 pfam02742 pfam04023 195 371 168 172 1422 1528 Ga0063436_10366 hypothetical protein - - 0 59 14 10 54 69 Ga0063436_10367 Periplasmic component of the Tol biopolymer transport system U 155 374 148 46 369 630 Ga0063436_10368 Peptidase family M23/LysM domain - pfam07676 pfam01476 pfam01551 109 389 122 42 530 747 Ga0063436_10369 Endonuclease IV (EC 3.1.21.-) Acetylornithine deacetylase/Succinyl-diaminopimelate desuccinylase and related deacylases Predicted hydrolases or acyltransferases (alpha/beta hydrolase superfamily) L 63 120 59 6 276 441 E pfam01261 pfam01546 pfam07687 45 138 16 6 216 312 R pfam12697 30 22 32 15 80 141 pyridoxal-phosphate dependent TrpB-like enzyme 2-keto-4-pentenoate hydratase/2-oxohepta-3-ene-1,7-dioic acid hydratase (catechol pathway) R pfam00291 342 842 276 198 2115 2637 Q pfam01557 117 220 112 36 788 957 MG pfam01370 94 339 99 46 696 1077 Ga0063436_103615 Nucleoside-diphosphate-sugar epimerases amino acid ABC transporter ATP-binding protein, PAAT family (TC 3.A.1.3.-) E pfam00005 123 300 100 45 423 664 Ga0063436_103616 Uncharacterized protein conserved in bacteria S pfam09996 106 447 191 66 623 779 Ga0063436_10362 Ga0063436_103610 Ga0063436_103611 Ga0063436_103612 Ga0063436_103613 Ga0063436_103614 - Ga0063436_103617 Uncharacterized protein conserved in archaea S pfam02596 41 250 50 8 163 240 Ga0063436_103618 H pfam00994 20 129 21 28 130 162 Ga0063436_103619 molybdenum cofactor synthesis domain ABC-type Mn2+/Zn2+ transport systems, permease components P pfam00950 34 57 22 2 116 153 Ga0063436_103620 ABC-type Mn/Zn transport systems, ATPase component P pfam00005 59 248 85 16 210 386 Ga0063436_103621 - 53 17 8 45 61 G pfam01297 pfam01256 pfam03853 5 Ga0063436_10391 Periplasmic solute binding protein family yjeF C-terminal region, hydroxyethylthiazole kinaserelated/yjeF N-terminal region 109 430 54 23 559 937 Ga0063436_10392 YtxH-like protein - pfam12732 184 523 63 17 521 800 Ga0063436_10393 hypothetical protein - - 30 224 15 25 211 239 Ga0063436_10394 - - 12 152 38 12 119 177 J pfam03054 144 336 61 8 357 467 Ga0063436_10396 hypothetical protein tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase (EC 2.1.1.61) Cysteine sulfinate desulfinase/cysteine desulfurase and related enzymes E pfam00266 76 326 53 26 326 520 Ga0063436_10397 hypothetical protein - - 37 111 41 4 128 202 Ga0063436_10398 Beta-lactamase superfamily domain - pfam13483 38 74 45 10 203 213 Ga0063436_10399 - pfam13483 39 105 21 0 83 85 Ga0063436_103910 Beta-lactamase superfamily domain Carbohydrate-binding module 48 (Isoamylase N-terminal domain) - 19 24 20 4 70 66 Ga0063436_103911 alpha-1,4-glucan:alpha-1,4-glucan 6-glycosyltransferase G pfam02922 pfam00128 pfam02806 39 132 34 14 107 134 Ga0063436_103912 LysM domain - 78 150 68 20 180 295 Ga0063436_103913 folylpolyglutamate synthase/dihydrofolate synthase H pfam01476 pfam02875 pfam08245 14 94 40 28 210 265 Ga0063436_103914 FecR family protein - pfam04773 355 859 236 170 997 1201 Ga0063436_103915 6-phosphofructokinase (EC 2.7.1.11) G pfam00365 71 239 83 32 307 566 Ga0063436_103916 TPR repeat - pfam13414 88 226 41 18 244 307 Ga0063436_103917 hypothetical protein - - 54 339 36 0 303 426 Ga0063436_103921 LysM domain - 172 231 101 30 394 414 Ga0063436_103922 DNA polymerase III, subunit gamma and tau L pfam01476 pfam12169 pfam13177 152 570 142 58 596 839 Ga0063436_103923 DNA-binding protein, YbaB/EbfC family S 58 88 38 20 141 205 Ga0063436_103924 DNA replication and repair protein RecR L pfam02575 pfam02132 pfam13662 51 284 54 22 256 348 Ga0063436_103925 ribosomal protein S6 J pfam01250 189 334 177 122 749 958 Ga0063436_103926 single stranded DNA-binding protein (ssb) L pfam00436 112 307 108 63 558 543 Ga0063436_103927 ribosomal protein S18 J 85 161 51 8 280 468 Ga0063436_103928 - 88 277 73 64 428 530 Ga0063436_10421 SurA N-terminal domain/PPIC-type PPIASE domain Molybdopterin-binding domain of aldehyde dehydrogenase/Aldehyde oxidase and xanthine dehydrogenase, a/b hammerhead domain pfam01084 pfam13616 pfam13624 - pfam01315 pfam02738 154 843 67 27 211 430 Ga0063436_10422 PPOX class probable F420-dependent enzyme, Rv2061 family - - 47 212 18 13 77 87 Ga0063436_10423 hypothetical protein - - 48 154 21 8 56 59 Ga0063436_10424 Predicted ring-cleavage extradiol dioxygenase R pfam12681 8 42 17 0 26 60 Ga0063436_10425 hypothetical protein - - 2 0 25 0 0 25 Ga0063436_10431 Bacterial extracellular solute-binding proteins, family 5 Middle - pfam00496 14 35 31 30 161 200 Ga0063436_10432 Glycine/D-amino acid oxidases (deaminating) E pfam01266 28 57 46 8 136 202 Ga0063436_10433 E pfam00496 6780 10732 5252 2521 23555 22413 Ga0063436_10434 ABC-type dipeptide transport system, periplasmic component ABC-type dipeptide/oligopeptide/nickel transport systems, permease components EP 194 472 190 128 954 885 Ga0063436_10441 iron (metal) dependent repressor, DtxR family K 79 340 60 56 265 413 Ga0063436_10442 Membrane carboxypeptidase (penicillin-binding protein) M pfam00528 pfam01325 pfam02742 pfam04023 pfam00905 pfam00912 109 333 124 18 445 644 Ga0063436_10443 Predicted phosphoesterase R pfam12850 12 104 35 42 62 140 Ga0063436_10444 Methyltransferase domain - pfam08241 8 74 38 22 112 127 Ga0063436_10451 YjeF-related protein N-terminus - pfam03853 36 53 36 14 43 91 Ga0063436_10452 YHYH protein - pfam14240 75 174 69 28 200 245 Ga0063436_10454 hypothetical protein - - 113 202 93 38 331 401 Ga0063436_10455 hypothetical protein - - 200 673 157 88 906 1101 Ga0063436_10456 putative regulatory protein, FmdB family - pfam09723 48 85 34 8 82 131 Ga0063436_10457 Pentapeptide repeats (8 copies) - pfam00805 56 362 80 56 202 303 Ga0063436_10458 ABC-type amino acid transport/signal transduction systems, ET pfam00497 904 1408 860 345 4295 4387 Ga0063436_10395 periplasmic component/domain Ga0063436_10459 amine acid ABC transporter, permease protein, 3-TM region, His/Glu/Gln/Arg/opine family E pfam00528 36 130 57 20 296 388 Ga0063436_10661 Secreted and surface protein containing fasciclin-like repeats M 101 536 121 9 415 645 Ga0063436_10662 Uncharacterized conserved protein S pfam02469 pfam13369 pfam13371 122 428 39 45 213 389 Ga0063436_10461 hypothetical protein - - 103 475 165 52 551 826 Ga0063436_10462 FecR family protein - pfam04773 520 1670 375 180 2129 2838 Ga0063436_10463 DNA polymerase III, delta subunit (EC 2.7.7.7) L pfam06144 28 122 58 4 153 259 Ga0063436_10464 hypothetical protein - 2 72 2 0 37 134 Ga0063436_10465 replication restart DNA helicase PriA L pfam00270 pfam00271 12 127 16 0 72 183 Ga0063436_10471 Uncharacterised protein family UPF0066 - pfam01980 11 71 9 17 39 68 Ga0063436_10473 Arabinose efflux permease G pfam07690 16 165 44 2 199 295 Ga0063436_10474 16S rRNA m(7)G-527 methyltransferase (EC 2.1.1.170) M pfam02527 58 105 54 20 146 347 Ga0063436_10475 Arabinose efflux permease G pfam07690 54 129 35 8 121 210 Ga0063436_10476 Esterase/lipase R 3 31 12 0 42 81 Ga0063436_10484 translation elongation factor TU J pfam12697 pfam00009 pfam03143 pfam03144 2570 4396 1538 902 8501 9004 Ga0063436_10485 ribosomal protein S10, bacterial/organelle J pfam00338 265 501 143 82 848 1070 Ga0063436_10486 LSU ribosomal protein L3P J pfam00297 268 600 171 113 1290 1350 Ga0063436_10487 50S ribosomal protein L4, bacterial/organelle J pfam00573 268 505 156 98 761 786 Ga0063436_10488 Ribosomal protein L23 J pfam00276 157 566 121 22 608 814 Ga0063436_10489 hypothetical protein - - 30 121 36 19 178 284 Ga0063436_10491 flagellar hook-basal body protein N - 208 234 144 40 617 777 Ga0063436_10492 Secreted and surface protein containing fasciclin-like repeats M pfam02469 361 575 283 130 1381 1939 Ga0063436_10493 hypothetical protein - 0 18 4 0 15 43 Ga0063436_10494 RNA polymerase sigma factor, FliA/WhiG family K pfam04539 pfam04542 pfam04545 151 312 74 52 498 573 Ga0063436_10495 hypothetical protein - - 26 54 15 0 56 139 Ga0063436_10496 Flagellar GTP-binding protein N pfam00448 18 216 22 2 230 363 Ga0063436_10497 Flagellar biosynthesis pathway, component FlhA NU pfam00771 79 424 156 39 550 957 Ga0063436_10498 Flagellar biosynthesis pathway, component FlhB NU pfam01312 89 334 112 38 475 634 Ga0063436_10499 Flagellar biosynthesis pathway, component FliR NU pfam01311 25 251 53 0 181 303 Ga0063436_104910 flagellar biosynthetic protein FliQ NU pfam01313 9 75 21 6 77 145 Ga0063436_104911 flagellar biosynthetic protein FliP NU pfam00813 59 97 45 24 186 265 Ga0063436_104912 Flagellar biosynthesis protein, FliO - pfam04347 36 224 32 8 334 566 Ga0063436_104913 flagellar motor switch protein FliN NU 153 480 122 101 730 924 Ga0063436_104914 Flagellar motor switch protein N pfam01052 pfam01052 pfam02154 398 744 285 196 1399 1353 Ga0063436_104915 Flagellar basal body-associated protein N 200 335 151 65 844 1131 Ga0063436_104916 Flagellar motor protein N 192 538 193 8 737 1011 Ga0063436_104917 Flagellar motor protein N pfam03748 pfam00691 pfam13677 pfam00691 pfam13677 177 615 164 84 786 1077 Ga0063436_104918 Flagellar motor component N pfam01618 324 1434 304 180 1448 1851 Ga0063436_104919 N 145 67 41 275 322 358 492 216 48 1147 1349 Ga0063436_104921 PAS domain S-box - 145 520 113 75 777 1102 Ga0063436_104922 Signal transduction histidine kinase T pfam06289 pfam00072 pfam00486 pfam00072 pfam00512 pfam02518 pfam13426 pfam13492 pfam00512 pfam02518 115 Ga0063436_104920 Uncharacterized protein, possibly involved in motility Response regulators consisting of a CheY-like receiver domain and a winged-helix DNA-binding domain 70 200 99 14 346 572 Ga0063436_104923 hypothetical protein - - 38 116 16 2 109 209 Ga0063436_104924 Uncharacterized low-complexity proteins S pfam00805 245 558 243 137 1014 1346 Ga0063436_104925 - 228 100 34 433 595 P pfam00027 pfam02254 pfam07885 159 Ga0063436_104926 Cyclic nucleotide-binding domain Kef-type K+ transport systems, predicted NAD-binding component 217 590 234 94 925 1380 Ga0063436_10501 Inorganic H+ pyrophosphatase - pfam03030 527 1500 494 298 2886 2728 Ga0063436_10502 integral membrane protein, TerC family P pfam03741 130 378 57 98 319 340 TK Ga0063436_10503 Arabinose efflux permease G pfam07690 27 33 34 27 86 165 Ga0063436_10504 hypothetical protein - - 0 35 13 0 41 63 Ga0063436_10505 hypothetical protein - - 4 16 9 0 22 61 Ga0063436_10506 S-layer homology domain - pfam00395 143 354 107 10 346 627 Ga0063436_10511 - 343 320 103 948 843 F pfam00856 pfam01725 pfam13177 234 Ga0063436_10512 SET domain non-canonical purine NTP pyrophosphatase, RdgB/HAM1 family 56 81 24 26 181 277 Ga0063436_10513 Predicted esterase R pfam02230 63 185 40 58 154 304 Ga0063436_10514 hypothetical protein - 39 156 35 9 102 127 Ga0063436_10515 TIGR01777 family protein R pfam08338 pfam13460 235 618 135 59 360 353 Ga0063436_10516 Arabinose efflux permease G pfam07690 6 88 24 2 67 108 Ga0063436_10517 hypothetical protein - - 39 132 29 10 120 255 Ga0063436_10518 Deoxynucleoside kinases F pfam01712 16 140 26 18 107 173 Ga0063436_10519 hypothetical protein - - 24 106 19 8 109 172 Ga0063436_105110 Protein involved in cellulose biosynthesis (CelD) M pfam13480 16 130 50 24 143 213 Ga0063436_12241 deoxyribose-phosphate aldolase F pfam01791 63 102 68 20 209 296 Ga0063436_12242 Aldehyde dehydrogenase family - pfam00171 8 23 14 6 22 35 Ga0063436_10531 hypothetical protein probable selenium-dependent hydroxylase accessory protein YqeC selenium-dependent molybdenum hydroxylase system protein, YqeB family - - 72 146 68 39 127 190 R pfam12804 21 156 44 4 151 259 - - 30 59 21 0 79 125 - 0 6 12 0 28 48 C pfam01315 pfam02738 126 355 172 48 603 840 C pfam01799 54 162 46 2 140 221 Ga0063436_10541 hypothetical protein Aerobic-type carbon monoxide dehydrogenase, large subunit CoxL/CutL homologs Aerobic-type carbon monoxide dehydrogenase, small subunit CoxS/CutS homologs Acetyltransferases, including N-acetylases of ribosomal proteins J pfam13302 16 146 33 35 150 153 Ga0063436_10542 Cyclic nucleotide-binding domain - pfam00027 108 256 116 59 296 448 Ga0063436_10543 putative TIM-barrel protein, nifR3 family J pfam01207 132 421 81 0 247 316 Ga0063436_10544 Uncharacterized conserved protein S - 43 83 31 12 175 174 Ga0063436_10545 HEAT repeats - pfam13646 1 53 4 0 46 85 Ga0063436_10551 methyltransferase, FkbM family - pfam05050 63 74 46 28 186 190 Ga0063436_10552 Glycosyltransferase M pfam00534 85 160 61 34 313 430 Ga0063436_10553 hypothetical protein - 1 0 9 6 46 23 Ga0063436_10554 Glycosyltransferase M 53 270 61 2 274 498 Ga0063436_10561 K 14 219 34 6 138 198 Ga0063436_10562 iron (metal) dependent repressor, DtxR family Predicted metal-dependent protease of the PAD1/JAB1 superfamily pfam00534 pfam13692 pfam01325 pfam02742 pfam04023 R pfam14464 50 415 68 60 266 310 Ga0063436_10563 Predicted membrane protein S pfam02660 59 318 63 76 257 465 Ga0063436_10564 hypothetical protein - - 17 75 19 0 108 201 Ga0063436_10565 NO U - 28 219 17 0 117 200 Ga0063436_10566 hypothetical protein Type II secretory pathway, prepilin signal peptidase PulO and related peptidases pfam01478 6 85 8 0 41 141 Ga0063436_10567 hypothetical protein - 16 87 14 2 71 176 Ga0063436_10568 G 22 138 6 2 88 179 Ga0063436_10569 Sugar kinases, ribokinase family Phosphopantothenate-cysteine ligase (EC 6.3.2.5)/Phosphopantothenoylcysteine decarboxylase (EC 4.1.1.36) pfam00294 pfam10343 H pfam02441 pfam04127 36 252 42 10 227 347 Ga0063436_105610 pantothenate kinase, type III K pfam03309 18 97 34 7 120 229 Ga0063436_105611 M pfam01380 73 184 34 38 235 361 Ga0063436_105612 Predicted phosphosugar isomerases Predicted hydrolases or acyltransferases (alpha/beta hydrolase superfamily) R pfam12697 18 41 2 8 34 94 Ga0063436_105613 Uncharacterized protein conserved in bacteria S 17 41 14 0 48 44 Ga0063436_105614 Hemolysins and related proteins containing CBS domains R 66 283 84 16 303 482 Ga0063436_105615 Hemolysins and related proteins containing CBS domains R 112 444 74 38 312 613 Ga0063436_10571 cytochrome c oxidase, subunit II C pfam04342 pfam00571 pfam01595 pfam03471 pfam00571 pfam01595 pfam03471 pfam00034 pfam00116 pfam02790 394 1331 189 100 806 879 Ga0063436_10532 Ga0063436_10533 Ga0063436_10534 Ga0063436_10535 Ga0063436_10536 Ga0063436_10572 Heme/copper-type cytochrome/quinol oxidases, subunit 1 C pfam00115 869 1707 299 192 1238 1537 Ga0063436_10573 hypothetical protein - - 159 303 40 39 290 310 Ga0063436_10574 Heme/copper-type cytochrome/quinol oxidase, subunit 3 C pfam00510 172 536 87 44 301 402 Ga0063436_10575 Predicted membrane protein S pfam09678 81 393 34 67 235 242 Ga0063436_10576 Uncharacterized conserved protein S pfam02104 73 403 67 0 116 233 Ga0063436_10577 protoheme IX farnesyltransferase O pfam01040 72 315 50 18 145 222 Ga0063436_10581 - pfam11258 25 144 24 9 201 218 Ga0063436_10582 Protein of unknown function (DUF3048) LL-diaminopimelate aminotransferase apoenzyme (EC 2.6.1.83) E pfam00155 68 168 55 27 183 245 Ga0063436_10591 pyrimidine 5'-nucleotidase R pfam13419 0 73 21 0 58 111 Ga0063436_10592 - pfam13180 20 81 24 0 94 136 Ga0063436_10601 PDZ domain Inhibitor of the KinA pathway to sporulation, predicted exonuclease R 12 57 26 8 63 145 Ga0063436_10602 FAD/FMN-containing dehydrogenases C pfam00929 pfam01565 pfam02913 84 154 90 0 346 432 Ga0063436_10603 - 24 24 14 86 43 O pfam07883 pfam02625 pfam13478 38 Ga0063436_10604 Cupin domain Xanthine and CO dehydrogenases maturation factor, XdhC/CoxF family 27 122 26 10 147 227 Ga0063436_10605 Protein containing von Willebrand factor type A (vWA) domain R pfam05762 39 227 56 18 251 403 Ga0063436_10606 AAA domain (dynein-related subfamily) - pfam07728 141 210 86 50 291 322 Ga0063436_10607 Ala-tRNA(Pro) hydrolase (EC 3.1.1.-) S 10 77 49 6 179 378 Ga0063436_10608 acetyl-coenzyme A synthetase (EC 6.2.1.1) I pfam04073 pfam00501 pfam13193 180 437 280 90 1423 1875 Ga0063436_10609 - 268 113 59 584 984 451 815 331 74 2364 3723 1646 2594 1722 403 8750 11782 C pfam00501 pfam00941 pfam03450 pfam01315 pfam02738 pfam00111 pfam01799 185 Ga0063436_106012 AMP-binding enzyme Aerobic-type carbon monoxide dehydrogenase, middle subunit CoxM/CutM homologs Aerobic-type carbon monoxide dehydrogenase, large subunit CoxL/CutL homologs Aerobic-type carbon monoxide dehydrogenase, small subunit CoxS/CutS homologs 205 382 364 90 1439 2142 Ga0063436_106013 Uncharacterized protein conserved in bacteria (DUF2062) - pfam09835 18 44 24 46 102 90 Ga0063436_106014 Esterase/lipase I pfam12695 8 47 45 28 94 223 Ga0063436_106015 Dihydrodipicolinate synthase/N-acetylneuraminate lyase EM pfam00701 16 80 56 12 140 192 Ga0063436_13501 NADH(P)-binding - 10 72 17 10 82 91 Ga0063436_13502 Glycosyltransferase M pfam13460 pfam00534 pfam13579 41 83 34 10 105 167 Ga0063436_13503 Glycosyltransferases involved in cell wall biogenesis M pfam00535 18 48 10 0 60 90 Ga0063436_13504 Glycosyl transferases group 1 - pfam00534 34 107 66 18 115 135 Ga0063436_10611 Glycosyl transferases group 1 - pfam00534 0 25 0 0 3 2 Ga0063436_10612 hypothetical protein - - 18 80 31 38 114 107 Ga0063436_10613 M pfam00534 58 262 71 33 213 371 Ga0063436_10614 Glycosyltransferase Membrane protein involved in the export of O-antigen and teichoic acid R pfam01943 24 66 42 20 83 146 Ga0063436_10615 Glycosyl transferases group 1 - pfam00534 20 98 22 0 91 92 Ga0063436_10621 nucleoside-binding protein R pfam02608 3274 3963 2195 951 12737 12507 Ga0063436_10622 ABC transporter - pfam00005 105 168 96 33 446 452 Ga0063436_10631 E pfam01810 42 40 11 10 113 70 Ga0063436_10632 Putative threonine efflux protein Predicted oxidoreductases (related to aryl-alcohol dehydrogenases) C pfam00248 35 68 48 24 133 204 Ga0063436_10633 malate synthase (EC 2.3.3.9) C pfam01274 102 191 88 28 521 585 Ga0063436_10634 agmatinase (EC 3.5.3.11) E pfam00491 32 209 39 38 202 311 Ga0063436_10635 Xaa-Pro aminopeptidase E 68 439 83 22 321 415 Ga0063436_10636 thioredoxin-disulfide reductase O pfam00557 pfam00070 pfam07992 104 236 98 40 429 513 Ga0063436_10637 hypothetical protein - - 41 68 22 0 100 83 Ga0063436_10638 - 261 42 40 168 214 HE pfam00389 pfam02826 47 Ga0063436_10641 hypothetical protein Phosphoglycerate dehydrogenase and related dehydrogenases 94 76 70 43 258 395 Ga0063436_10642 ParB-like nuclease domain - pfam02195 40 78 32 12 134 212 Ga0063436_10651 hypothetical protein - 117 425 142 42 522 621 Ga0063436_10652 L 185 512 172 122 709 1025 Ga0063436_10653 chromosomal replication initiator protein DnaA Transcriptional regulators containing a DNA-binding HTH domain and an aminotransferase domain (MocR family) and their eukaryotic orthologs pfam00308 pfam08299 KE pfam00155 114 195 74 16 252 382 Ga0063436_10654 3-dehydroquinate dehydratase (EC 4.2.1.10) E pfam01220 114 323 72 41 325 358 Ga0063436_106010 Ga0063436_106011 C C Ga0063436_10655 3-dehydroquinate synthase (EC 4.2.3.4) E Ga0063436_10656 F Ga0063436_10657 O-6-methylguanine DNA methyltransferase Permeases of the drug/metabolite transporter (DMT) superfamily pfam01202 pfam01761 pfam01035 pfam02805 pfam12833 GER Ga0063436_10658 Major Facilitator Superfamily - Ga0063436_10671 Sugar kinases, ribokinase family Ga0063436_10672 Ga0063436_10673 20 223 48 4 261 378 10 113 20 0 64 180 pfam00892 22 333 28 18 138 223 pfam07690 44 67 11 27 99 113 G pfam00294 53 65 45 20 158 228 ribokinase G pfam00294 132 572 139 75 482 825 Fucose dissimilation pathway protein FucU G pfam05025 81 129 133 4 287 333 Ga0063436_10674 D-aminopeptidase E pfam04951 80 119 87 4 193 221 Ga0063436_10675 Nucleoside 2-deoxyribosyltransferase - pfam05014 1 58 8 22 63 80 Ga0063436_10676 ABC-type multidrug transport system, ATPase component V pfam00005 181 441 47 56 124 192 Ga0063436_10677 hypothetical protein - - 10 50 2 18 12 8 Ga0063436_10678 ABC-2 type transporter - pfam01061 32 488 26 36 47 172 Ga0063436_10679 Arabinose efflux permease G 68 261 51 20 202 281 Ga0063436_10681 pseudouridine synthase J 90 213 86 28 397 465 Ga0063436_10682 cytidylate kinase F pfam07690 pfam00849 pfam01479 pfam01553 pfam02224 32 256 40 38 220 375 Ga0063436_10683 Fe-S oxidoreductase, related to NifB/MoaA family C pfam04459 64 131 72 40 259 296 Ga0063436_10684 Predicted membrane protein S pfam00892 10 80 7 0 84 112 Ga0063436_10691 hypothetical protein - - 88 97 65 16 250 324 Ga0063436_10692 Sugar transferases involved in lipopolysaccharide synthesis M pfam02397 31 71 21 0 94 95 Ga0063436_10693 CoA-binding domain - pfam13727 14 110 30 16 125 159 Ga0063436_10694 Glycosyltransferases, probably involved in cell wall biogenesis M pfam13641 4 179 32 22 165 278 Ga0063436_14152 Uncharacterised protein family (UPF0104) - pfam03706 41 93 48 46 213 244 Ga0063436_14151 hypothetical protein - - 49 57 28 24 175 145 Ga0063436_10701 hypothetical protein - - 94 183 72 14 327 460 Ga0063436_10702 probable rRNA maturation factor YbeY R 8 12 10 0 38 44 Ga0063436_10703 HDIG domain - pfam02130 pfam01966 pfam07698 39 103 26 18 116 113 Ga0063436_10711 Dolichyl-phosphate-mannose-protein mannosyltransferase - 65 233 44 34 309 338 Ga0063436_10712 putative hydrolase, CocE/NonD family R 102 433 146 34 680 1008 Ga0063436_10713 arginase (EC 3.5.3.1) E 75 202 44 14 335 482 Ga0063436_10714 O-succinylbenzoate synthase (EC 4.2.1.113) MR 51 203 53 18 106 240 Ga0063436_13462 3-hydroxyacyl-CoA dehydrogenase I Ga0063436_13461 acetyl-CoA acetyltransferases I Ga0063436_10721 N-methylhydantoinase A/acetone carboxylase, beta subunit EQ Ga0063436_10722 N-methylhydantoinase B/acetone carboxylase, alpha subunit Thiamine pyrophosphate-requiring enzymes [acetolactate synthase, pyruvate dehydrogenase (cytochrome), glyoxylate carboligase, phosphonopyruvate decarboxylase] EQ Ga0063436_10725 Glycine/D-amino acid oxidases (deaminating) Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) Ga0063436_10726 Ga0063436_10731 pfam13231 pfam02129 pfam08530 pfam00491 pfam02746 pfam13378 pfam00378 pfam00725 pfam02737 173 763 164 88 621 900 pfam00108 pfam01968 pfam05378 79 452 66 4 334 483 21 73 40 20 143 138 30 30 16 8 52 98 EH pfam02538 pfam00205 pfam02775 pfam02776 23 106 32 0 52 112 E pfam01266 14 59 27 6 53 75 IQR pfam13561 2 4 6 0 18 79 hypothetical protein - - 24 0 6 1 36 36 ABC-type uncharacterized transport system N pfam09822 250 380 162 51 455 570 Ga0063436_10732 Domain of unknown function (DUF4340) - 145 565 178 69 431 512 Ga0063436_10733 deoxyguanosinetriphosphate triphosphohydrolase, putative F 142 356 96 36 348 424 Ga0063436_10734 Helicase conserved C-terminal domain/UvrB/uvrC motif/Ultraviolet resistance protein B - pfam14238 pfam01966 pfam13286 pfam00271 pfam02151 pfam12344 50 40 75 27 183 169 Ga0063436_10735 Type III restriction enzyme, res subunit - pfam04851 79 75 25 24 107 182 Ga0063436_10736 tRNA dimethylallyltransferase J pfam01715 35 95 25 26 91 115 Ga0063436_10737 Beta-glucosidase-related glycosidases pfam00933 133 565 160 92 548 744 Ga0063436_10738 Serine/threonine protein kinase G RTK L 174 364 146 30 407 640 Ga0063436_10739 Transcriptional regulatory protein, C terminal/FHA domain - pfam00069 pfam00486 pfam00498 16 90 31 26 159 195 Ga0063436_107310 CpXC protein - pfam14353 91 370 93 34 232 430 Ga0063436_10723 Ga0063436_10724 Ga0063436_107311 PAS domain S-box - pfam00512 pfam02518 pfam13185 pfam13426 Ga0063436_107312 - Ga0063436_107313 hypothetical protein Universal stress protein UspA and related nucleotide-binding proteins Ga0063436_10751 hypothetical protein Ga0063436_10752 120 217 43 49 261 386 - 30 2 3 1 89 105 T pfam00582 13 109 11 11 80 178 - - 0 41 6 0 8 58 R pfam14602 27 139 28 0 127 140 Ga0063436_10753 Acetyltransferase (isoleucine patch superfamily) Membrane protein involved in the export of O-antigen and teichoic acid R pfam01943 119 139 61 8 271 357 Ga0063436_10754 Glycosyl transferases group 1 - pfam00534 6 189 37 6 94 172 Ga0063436_10755 hypothetical protein - - 14 32 4 0 4 23 Ga0063436_13861 hypothetical protein - - 36 20 33 28 140 167 Ga0063436_13862 Glycosyltransferases involved in cell wall biogenesis Peptidyl-prolyl cis-trans isomerase (rotamase) - cyclophilin family M pfam00535 95 196 78 41 327 482 O pfam00160 388 359 178 82 718 851 F pfam00268 149 708 70 44 514 725 Ga0063436_13865 Ribonucleotide reductase, beta subunit Nucleotidyltransferase/DNA polymerase involved in DNA repair L 16 12 6 0 16 28 Ga0063436_13866 DNA polymerase III, alpha subunit L pfam00817 pfam02811 pfam07733 pfam14579 25 80 15 18 20 74 Ga0063436_13867 Alpha/beta hydrolase family - 20 0 6 0 58 78 Ga0063436_13422 RND family efflux transporter, MFP subunit V pfam12697 pfam12700 pfam13533 82 179 46 58 274 274 Ga0063436_13421 Glutamine amidotransferase domain - pfam13522 133 186 69 14 323 419 Ga0063436_14071 Glycosyltransferases involved in cell wall biogenesis M pfam00535 57 127 51 27 154 275 Ga0063436_10772 conserved hypothetical integral membrane protein S 28 36 51 0 104 169 Ga0063436_10773 Altronate dehydratase G pfam02592 pfam04295 pfam08666 103 370 139 10 331 622 Ga0063436_10774 Trypsin-like peptidase domain - pfam13365 34 19 14 10 52 27 Ga0063436_10775 hypothetical protein - 12 12 2 1 34 30 C pfam00111 pfam01315 pfam01799 pfam02738 90 219 118 68 479 539 Ga0063436_13863 Ga0063436_13864 Ga0063436_10791 Aerobic-type carbon monoxide dehydrogenase, large subunit CoxL/CutL homologs Uncharacterized protein, possibly involved in utilization of glycolate and propanediol R pfam03928 15 29 16 2 66 119 Ga0063436_10792 Molybdopterin converting factor, small subunit H 68 121 55 29 105 139 Ga0063436_10793 Aldehyde:ferredoxin oxidoreductase C pfam02597 pfam01314 pfam02730 Ga0063436_10794 S-adenosylmethionine--tRNA ribosyltransferase-isomerase J Ga0063436_10795 L Ga0063436_10801 Holliday junction DNA helicase subunit RuvB ATPase components of ABC transporters with duplicated ATPase domains Ga0063436_10802 transcriptional attenuator, LytR family K Ga0063436_10803 helicase, putative, RecD/TraA family Ga0063436_10804 Domain of unknown function (DUF329) Ga0063436_10805 Ga0063436_10781 346 608 290 79 1139 1208 52 122 53 22 151 232 52 204 69 60 265 280 85 164 58 8 303 374 82 316 95 80 371 415 L pfam02547 pfam05491 pfam05496 pfam00005 pfam12848 pfam03816 pfam13399 pfam13538 pfam13604 pfam14490 66 260 66 24 297 456 - pfam03884 15 90 26 0 173 319 hypothetical protein - - 10 18 25 2 78 114 Ga0063436_10806 PUCC protein - pfam03209 90 783 61 20 191 169 Ga0063436_10807 L pfam03167 33 246 56 23 242 364 Ga0063436_10808 Uracil-DNA glycosylase Predicted oxidoreductases (related to aryl-alcohol dehydrogenases) C pfam00248 38 203 58 24 145 261 Ga0063436_10811 hypothetical protein - - Ga0063436_10812 hypothetical protein - Ga0063436_10813 ribosomal protein L25, Ctc-form - Ga0063436_10814 Dipeptidyl peptidase IV (DPP IV) N-terminal region/Domain of unknown function (DUF348)/G5 domain - pfam01386 pfam14693 pfam00930 pfam03990 pfam07501 Ga0063436_10815 Lipid A core - O-antigen ligase and related enzymes M Ga0063436_10816 Nucleoside-diphosphate-sugar epimerases MG Ga0063436_10817 rod shape-determining protein MreC Ga0063436_10818 Ga0063436_10819 R 35 162 43 0 181 213 109 157 49 11 234 230 281 715 264 109 1049 1180 84 237 63 10 263 326 pfam13425 51 222 31 6 187 262 pfam01370 122 397 107 36 434 653 M pfam04085 15 171 12 0 153 310 rod shape-determining protein MreD M 22 121 6 0 44 46 penicillin-binding protein 2 M pfam04093 pfam00905 pfam03717 91 252 74 14 202 333 Ga0063436_108110 Protein of unknown function (DUF2723) - pfam11028 9 189 66 8 138 Ga0063436_108111 septum site-determining protein MinC D pfam03775 269 491 186 80 838 294 801 Ga0063436_108112 septum site-determining protein MinD D pfam01656 340 868 270 178 1137 1545 Ga0063436_108113 cell division topological specificity factor MinE D pfam03776 172 497 148 52 688 754 Ga0063436_108114 Bacterial cell division membrane protein D pfam01098 42 298 74 32 267 402 Ga0063436_108115 J pfam00749 40 164 42 18 218 345 Ga0063436_108117 glutamyl-tRNA synthetase, bacterial family Adenosylmethionine-8-amino-7-oxononanoate aminotransferase H pfam00202 81 547 97 50 583 748 Ga0063436_108118 thiol peroxidase (atypical 2-Cys peroxiredoxin) (EC 1.11.1.5) O 153 236 123 40 486 551 Ga0063436_108119 Glycosyltransferase M pfam08534 pfam00534 pfam13439 40 83 20 6 84 128 Ga0063436_108120 Predicted membrane protein S pfam04138 22 39 12 14 86 98 Ga0063436_108121 Lipid A core - O-antigen ligase and related enzymes M pfam04932 34 427 33 14 274 577 Ga0063436_108122 D 883 208 124 768 950 D pfam06723 pfam01171 pfam11734 154 Ga0063436_108123 rod shape-determining protein MreB tRNA(Ile)-lysidine synthetase, N-terminal domain/tRNA(Ile)lysidine synthetase, C-terminal domain 16 210 24 6 178 380 Ga0063436_108124 hypothetical protein - - 2 9 8 4 27 70 Ga0063436_10821 - 445 98 39 257 527 O pfam13365 pfam00226 pfam01556 81 Ga0063436_10822 Trypsin-like peptidase domain DnaJ-class molecular chaperone with C-terminal Zn finger domain 44 199 29 10 201 254 Ga0063436_10823 Acetyltransferase (GNAT) family - pfam00583 30 359 14 12 160 342 Ga0063436_10824 Uncharacterized protein conserved in bacteria S pfam06107 1 65 8 6 63 116 Ga0063436_10825 HAD-superfamily hydrolase, subfamily IIB R pfam08282 7 57 24 14 84 110 Ga0063436_10826 Glucose/sorbosone dehydrogenases G pfam07995 66 262 53 29 273 378 Ga0063436_10827 - 361 58 15 491 838 L pfam00521 pfam03989 119 Ga0063436_10828 hypothetical protein Type IIA topoisomerase (DNA gyrase/topo II, topoisomerase IV), A subunit 381 1284 266 52 925 1134 Ga0063436_10829 Asp/Glu/Hydantoin racemase - pfam01177 12 50 7 8 60 123 Ga0063436_10831 hypothetical protein - - 14 38 12 0 97 133 Ga0063436_10832 ATPases involved in chromosome partitioning D pfam01656 64 277 74 60 329 322 Ga0063436_10833 ribosomal protein L34, bacterial type J pfam00468 48 301 53 13 368 522 Ga0063436_10834 ribonuclease P protein component, eubacterial J pfam00825 76 484 119 34 369 711 Ga0063436_10835 putative membrane protein insertion efficiency factor S pfam01809 38 65 16 7 47 114 Ga0063436_10836 S pfam13360 136 459 199 89 544 662 Ga0063436_10837 FOG: WD40-like repeat membrane protein insertase, YidC/Oxa1 family, C-terminal domain - pfam02096 232 314 141 91 508 418 Ga0063436_10838 Predicted RNA-binding protein R pfam01424 178 311 140 12 429 659 Ga0063436_10839 chromosome segregation ATPase D 47 257 45 40 166 261 Ga0063436_108310 ParB/RepB/Spo0J family partition protein K pfam01656 pfam02195 pfam08535 2 162 37 14 162 301 Ga0063436_108311 Amidohydrolase family - pfam07969 64 231 73 22 356 477 Ga0063436_108312 Amidohydrolase family - 22 140 17 2 151 189 Ga0063436_108313 selenocysteine-specific elongation factor SelB J pfam07969 pfam00009 pfam03144 pfam09106 pfam09107 Ga0063436_108314 hypothetical protein - - Ga0063436_108315 M Ga0063436_108316 Glycosyltransferase Glycosyl transferase 4-like domain/Glycosyl transferases group 1 Ga0063436_10841 Indoleamine 2,3-dioxygenase Ga0063436_10842 38 249 49 35 233 382 134 278 135 16 540 586 10 24 10 0 80 110 - pfam00534 pfam13579 pfam13692 23 36 30 0 80 114 - pfam01231 113 550 108 18 519 754 H pfam01329 34 57 19 4 134 220 Ga0063436_10843 pterin-4-alpha-carbinolamine dehydratase (EC 4.2.1.96) DhnA-type fructose-1,6-bisphosphate aldolase and related enzymes G pfam01791 281 437 149 68 572 725 Ga0063436_10844 histidinol phosphate phosphatase, HisJ family ER - 75 468 94 28 528 664 Ga0063436_10851 hypothetical protein - 61 56 27 22 53 114 Ga0063436_10852 Aspartate ammonia-lyase E pfam00206 pfam10415 Ga0063436_10853 Ferredoxin C Ga0063436_10854 Superoxide dismutase P Ga0063436_10855 transcriptional regulator, ArsR family K Ga0063436_10856 copper-(or silver)-translocating P-type ATPase/heavy metal(Cd/Co/Hg/Pb/Zn)-translocating P-type ATPase P 112 216 77 36 340 425 pfam00037 pfam00081 pfam02777 591 1236 533 253 2052 2648 999 1777 681 352 2671 3144 pfam01022 pfam00122 pfam00403 pfam00702 83 74 26 6 288 380 115 572 116 121 1269 1591 Ga0063436_10857 cation diffusion facilitator family transporter P pfam01545 90 196 60 29 606 526 Ga0063436_10858 G pfam13347 184 381 166 58 1010 1117 Ga0063436_10859 Na+/melibiose symporter and related transporters Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) QIR pfam13561 14 68 34 0 99 114 Ga0063436_10861 Predicted endonuclease containing a URI domain L pfam01541 0 78 6 0 54 79 Ga0063436_10862 Zn-dependent proteases R 24 168 36 22 109 149 Ga0063436_10863 Peptidase MA superfamily/Immune inhibitor A peptidase M6 - pfam02163 pfam05547 pfam13485 225 478 137 28 373 482 Ga0063436_10864 hypothetical protein - - 23 172 53 22 178 232 Ga0063436_10865 YtxH-like protein - pfam12732 22 92 4 2 56 123 Ga0063436_10866 hypothetical protein - 13 109 20 4 97 148 Ga0063436_10871 WD40-like Beta Propeller Repeat/Prolyl oligopeptidase family - pfam00326 pfam07676 57 60 41 6 132 222 Ga0063436_10872 WD40-like Beta Propeller Repeat - pfam07676 6 0 26 3 88 94 Ga0063436_10873 Asparaginase E pfam01112 34 163 77 30 253 241 Ga0063436_10874 probable F420-dependent oxidoreductase, Rv3520c family C pfam00296 14 46 53 4 161 160 Ga0063436_10875 Oligoendopeptidase F E pfam01432 155 157 65 60 249 335 Ga0063436_10876 hypothetical protein - - 4 31 14 0 55 49 Ga0063436_10877 Trimethylamine:corrinoid methyltransferase H pfam06253 42 126 58 0 156 216 Ga0063436_14101 dihydroxy-acid dehydratase EG pfam00920 206 200 140 36 473 475 Ga0063436_14094 DinB superfamily - pfam12867 73 64 75 6 264 303 Ga0063436_14093 Molybdopterin-guanine dinucleotide biosynthesis protein A H pfam12804 40 44 43 14 115 163 Ga0063436_14092 2'-5' RNA ligase J pfam13563 15 109 18 2 83 171 Ga0063436_14091 Glycine/D-amino acid oxidases (deaminating) E pfam01266 71 238 90 26 288 354 Ga0063436_10881 Dolichyl-phosphate-mannose-protein mannosyltransferase - pfam13231 12 199 32 12 77 104 Ga0063436_10882 Zn-dependent dipeptidase, microsomal dipeptidase homolog E pfam01244 66 165 80 0 199 213 Ga0063436_10883 2-amino-3-ketobutyrate coenzyme A ligase (EC 2.3.1.29) H 65 187 63 32 136 175 Ga0063436_10884 L-threonine 3-dehydrogenase (EC 1.1.1.103) ER pfam00155 pfam00107 pfam08240 84 134 53 24 148 181 Ga0063436_10891 PHP domain - pfam02811 4 23 17 12 65 119 Ga0063436_10892 Predicted methylated DNA-protein cysteine methyltransferase L 2 29 6 0 33 77 Ga0063436_10893 DNA polymerase IV (family X) L pfam01035 pfam02811 pfam14520 pfam14716 pfam14791 182 621 139 67 609 928 Ga0063436_10894 ribosome silencing factor RsfS/YbeB/iojap S pfam02410 43 154 106 22 326 340 Ga0063436_10895 nucleoside diphosphate kinase (EC 2.7.4.6) F pfam00334 141 281 69 46 289 383 Ga0063436_11671 FtsX-like permease family - pfam02687 10 47 6 16 48 70 Ga0063436_11672 Ion transport protein - pfam00520 48 108 71 6 253 312 Ga0063436_11673 cytidine deaminase (EC 3.5.4.5) F pfam00383 71 39 28 4 97 194 Ga0063436_11674 EM pfam00701 81 318 112 57 508 638 Ga0063436_11675 Dihydrodipicolinate synthase/N-acetylneuraminate lyase Uncharacterized membrane protein (homolog of Drosophila rhomboid) R pfam01694 68 128 42 16 99 148 Ga0063436_11676 Geranylgeranyl pyrophosphate synthase H pfam00348 24 319 47 10 184 425 Ga0063436_11677 isopentenyl-diphosphate delta-isomerase (EC 5.3.3.2) C pfam01070 74 263 126 24 267 507 Ga0063436_11678 IQ pfam00550 225 333 184 77 671 712 R pfam00994 pfam13180 pfam13365 46 254 70 4 179 281 Ga0063436_116710 acyl carrier protein Predicted nucleotide-utilizing enzyme related to molybdopterin-biosynthesis enzyme MoeA Trypsin-like serine proteases, typically periplasmic, contain Cterminal PDZ domain 458 1308 149 100 316 466 Ga0063436_116711 TIGR00252 family protein L 86 141 56 30 153 145 Ga0063436_116712 C-terminal peptidase (prc) M pfam02021 pfam03572 pfam13180 96 131 36 0 109 198 Ga0063436_116713 SsrA-binding protein O pfam01668 30 89 6 0 42 68 Ga0063436_116715 dimethyladenosine transferase (EC 2.1.1.-) J 40 43 20 14 73 129 Ga0063436_116716 Domain of unknown function (DUF348)/3D domain/G5 domain - pfam00398 pfam03990 pfam06725 pfam07501 56 246 50 0 209 228 Ga0063436_116718 hypothetical protein - - 18 10 18 0 57 38 Ga0063436_116719 Alpha/beta hydrolase family - 22 143 93 48 203 210 Ga0063436_10921 RNA polymerase sigma factor, sigma-70 family K pfam12697 pfam00140 pfam04539 pfam04542 pfam04545 232 343 165 86 448 714 Ga0063436_11679 O Ga0063436_10922 NADH dehydrogenase subunit A (EC 1.6.5.3) C pfam00507 64 214 53 15 202 191 Ga0063436_10923 NADH dehydrogenase subunit B (EC 1.6.5.3) C pfam01058 101 466 94 24 284 449 Ga0063436_10924 NADH dehydrogenase subunit C (EC 1.6.5.3) C pfam00329 222 189 77 46 244 360 Ga0063436_10925 NADH dehydrogenase subunit D (EC 1.6.5.3) C pfam00346 253 802 198 52 565 713 Ga0063436_10926 NADH dehydrogenase subunit E (EC 1.6.5.3) C 56 302 67 18 180 279 Ga0063436_10927 NADH dehydrogenase subunit F (EC 1.6.5.3) C 385 1385 321 97 985 1460 Ga0063436_10928 NADH-quinone oxidoreductase, chain G C pfam01257 pfam01512 pfam10531 pfam10589 pfam00384 pfam01568 pfam04879 pfam10588 pfam13510 305 1248 213 111 960 1402 Ga0063436_10929 NADH dehydrogenase subunit H (EC 1.6.5.3) C pfam00146 136 555 131 80 476 672 Ga0063436_109210 NADH dehydrogenase subunit I (EC 1.6.5.3) C pfam12838 131 301 64 63 300 442 Ga0063436_109211 NADH dehydrogenase subunit J (EC 1.6.5.3) C pfam00499 52 171 9 3 65 151 Ga0063436_109212 NADH dehydrogenase subunit K (EC 1.6.5.3) C 16 185 12 6 62 88 Ga0063436_109213 NADH dehydrogenase subunit L (EC 1.6.5.3) CP 209 887 211 82 757 941 Ga0063436_109214 NADH dehydrogenase subunit M (EC 1.6.5.3) C pfam00420 pfam00361 pfam00662 pfam00361 pfam01059 246 652 132 105 663 724 Ga0063436_109215 NADH dehydrogenase subunit N (EC 1.6.5.3) C 205 798 310 70 788 948 Ga0063436_109216 shikimate dehydrogenase (EC 1.1.1.25) E pfam00361 pfam01488 pfam08501 111 280 109 28 364 406 Ga0063436_109217 ABC-type sugar transport system, periplasmic component G pfam13407 1746 2420 1257 638 7943 7614 Ga0063436_109218 ABC-type sugar transport system, ATPase component G pfam00005 132 114 95 56 450 459 Ga0063436_109219 ribose ABC transporter membrane protein Predicted spermidine synthase with an N-terminal membrane domain G pfam02653 31 71 45 26 113 156 R pfam01564 22 36 36 0 79 130 - - 8 2 4 19 19 15 Ga0063436_10931 hypothetical protein branched chain amino acid aminotransferase apoenzyme (EC 2.6.1.42) EH pfam01063 228 371 154 67 761 829 Ga0063436_10932 Hsp70 protein - pfam00012 238 738 126 118 527 821 Ga0063436_10941 Glycosyltransferases involved in cell wall biogenesis M pfam00535 32 128 76 41 266 393 Ga0063436_10942 NAD-dependent aldehyde dehydrogenases C pfam00171 45 184 75 22 271 534 Ga0063436_10943 cold-shock DNA-binding protein family K pfam00313 99 124 103 32 390 360 Ga0063436_10944 - 345 115 16 275 446 38 290 56 24 195 314 Ga0063436_10951 transcription elongation factor GreA K 223 262 79 40 203 323 Ga0063436_10952 lysyl-tRNA synthetase, class II (EC 6.1.1.6) J 90 226 68 8 307 441 Ga0063436_10953 Oligoendopeptidase F E 108 297 76 14 315 509 Ga0063436_10954 Predicted GTPase, probable translation factor J pfam05572 pfam00534 pfam03808 pfam01272 pfam03449 pfam00152 pfam01336 pfam01432 pfam08439 pfam01926 pfam06071 74 Ga0063436_10945 Pregnancy-associated plasma protein-A bacterial polymer biosynthesis proteins, WecB/TagA/CpsF family 91 242 64 16 257 336 Ga0063436_10956 hypothetical protein - - 18 11 8 13 30 67 Ga0063436_10957 hypothetical protein - - 7 16 4 0 0 7 Ga0063436_11344 hypothetical protein - - 28 200 47 12 130 171 Ga0063436_11345 Glycosyltransferase M 41 131 65 0 145 281 Ga0063436_11346 M 58 183 76 8 144 305 Ga0063436_11347 Glycosyltransferase exopolysaccharide biosynthesis polyprenyl glycosylphosphotransferase M pfam00534 pfam00534 pfam13439 pfam02397 pfam13727 74 180 75 34 277 348 Ga0063436_11348 Glycosyl transferase family 2 - pfam00535 42 135 21 21 82 126 Ga0063436_10961 Adenylosuccinate synthetase - pfam00709 36 54 9 2 64 58 Ga0063436_10962 Fumarylacetoacetate (FAA) hydrolase family protein R pfam01557 86 219 83 12 243 377 Ga0063436_10964 hypothetical protein - - 53 48 33 28 118 147 Ga0063436_10965 Uncharacterized protein conserved in bacteria S 87 116 42 12 147 191 Ga0063436_10966 DAK2 domain fusion protein YloV R pfam03780 pfam02734 pfam13684 116 Ga0063436_10967 hypothetical protein - Ga0063436_10968 ATP-dependent DNA helicase RecG (EC 3.6.1.-) Ga0063436_10969 Phosphopantetheine adenylyltransferase (EC 2.7.7.3) Ga0063436_109610 Ga0063436_109611 Ga0063436_109220 Ga0063436_109221 M 426 116 15 428 563 61 124 53 26 220 271 LK pfam00270 pfam00271 117 636 139 44 503 863 H pfam01467 68 112 78 22 210 414 hypothetical protein - - 76 242 118 29 415 504 ribosomal protein L32 J pfam01783 48 228 73 65 444 509 Ga0063436_109612 Acyl transferase domain - pfam00698 26 40 9 37 66 76 Ga0063436_109613 (acyl-carrier-protein) S-malonyltransferase I 17 167 19 8 177 246 Ga0063436_109614 condensin subunit Smc D pfam00698 pfam02463 pfam06470 187 Ga0063436_10971 Domain of unknown function (DUF309) - Ga0063436_10972 ribonucleoside-diphosphate reductase class II (EC 1.17.4.-) Ga0063436_10973 transcriptional regulator NrdR Ga0063436_10974 429 69 60 534 769 17 66 26 30 104 119 F pfam03745 pfam00317 pfam02867 pfam12637 995 3111 911 563 6006 7026 K pfam03477 93 272 116 151 468 643 hypothetical protein - - 16 146 25 20 223 287 Ga0063436_13081 SufE protein probably involved in Fe-S center assembly R pfam02657 59 120 35 29 101 171 Ga0063436_13082 hypothetical protein - 9 74 9 6 52 91 Ga0063436_13083 Glycosyltransferase M pfam00534 pfam13439 43 75 26 26 100 156 Ga0063436_13084 ribokinase ATPase family associated with various cellular activities (AAA)/Clp amino terminal domain C-terminal, D2-small domain, of ClpB protein/AAA domain (Cdc48 subfamily) G pfam00294 pfam00004 pfam02861 pfam07724 pfam10431 34 230 43 30 186 240 72 239 92 18 184 334 72 105 43 4 183 197 T 168 18 6 151 159 TK pfam00481 pfam00072 pfam00486 80 Ga0063436_13088 Serine/threonine protein phosphatase Response regulators consisting of a CheY-like receiver domain and a winged-helix DNA-binding domain 43 130 59 20 193 248 Ga0063436_13089 acetylornithine deacetylase (EC 3.5.1.16) E pfam01546 14 41 28 19 46 81 Ga0063436_10991 Uncharacterized protein conserved in bacteria S pfam02646 105 315 136 21 450 484 Ga0063436_10992 hypothetical protein - - 84 179 85 46 361 489 Ga0063436_10993 diaminopimelate epimerase (EC 5.1.1.7) E pfam01678 0 58 22 10 78 90 Ga0063436_10994 hypothetical protein - - 0 0 0 0 0 6 Ga0063436_10995 urocanate hydratase (EC 4.2.1.49) E pfam01175 98 228 49 2 311 406 Ga0063436_10996 histidine ammonia-lyase (EC 4.3.1.3) E pfam00221 32 166 39 12 274 318 Ga0063436_10997 imidazolonepropionase (EC 3.5.2.7) Q pfam13147 27 140 20 0 129 233 Ga0063436_11001 - pfam01408 46 407 30 26 128 264 Ga0063436_11002 Oxidoreductase family, NAD-binding Rossmann fold Predicted hydrolases or acyltransferases (alpha/beta hydrolase superfamily) R pfam12697 14 303 70 33 147 174 Ga0063436_11003 hypothetical protein - - 82 319 54 12 196 262 Ga0063436_11011 Formiminotransferase-cyclodeaminase - pfam04961 0 22 2 0 17 78 Ga0063436_11012 Acyl CoA:acetate/3-ketoacid CoA transferase, beta subunit I pfam01144 14 6 4 0 38 90 Ga0063436_11013 Acyl CoA:acetate/3-ketoacid CoA transferase, alpha subunit I pfam01144 20 59 38 10 69 159 Ga0063436_11014 hypothetical protein - - 24 109 36 0 105 151 Ga0063436_11021 Bacitracin resistance protein BacA - pfam02673 38 28 6 10 41 50 Ga0063436_11022 tRNA-guanine transglycosylase (EC 2.4.2.29) J pfam01702 30 50 22 0 102 82 Ga0063436_11031 RNAse Z (EC 3.1.26.11) R 31 75 27 0 116 134 Ga0063436_11032 UDP-glucose dehydrogenase M 241 739 151 44 681 1005 Ga0063436_11033 Predicted Zn-dependent peptidases R 124 247 119 48 337 669 Ga0063436_11034 Predicted Zn-dependent peptidases R pfam12706 pfam00984 pfam03720 pfam03721 pfam00675 pfam05193 pfam00675 pfam05193 102 379 110 30 483 697 Ga0063436_11035 - pfam04389 60 384 89 32 427 563 OU pfam01343 43 162 64 18 275 314 Ga0063436_11037 Peptidase family M28 signal peptide peptidase A. Serine peptidase. MEROPS family S49 4-aminobutyrate aminotransferase and related aminotransferases E pfam00202 139 474 129 31 367 552 Ga0063436_11041 hypothetical protein - 55 174 55 38 136 251 Ga0063436_11042 GTP-binding protein HflX R pfam01926 pfam13167 50 373 77 31 248 328 Ga0063436_11043 tRNA:m(5)U-54 methyltransferase J 87 197 53 44 246 339 Ga0063436_11044 RNA polymerase, sigma 54 subunit, RpoN/SigL K pfam01134 pfam00309 pfam04552 pfam04963 283 352 187 60 724 977 Ga0063436_11045 hypothetical protein - - 20 36 37 22 172 154 Ga0063436_11051 - - 22 36 21 1 76 86 Ga0063436_11052 hypothetical protein 6-pyruvoyl-tetrahydropterin synthase related domain; membrane protein - 136 282 129 27 398 721 Ga0063436_11053 succinyl-CoA synthetase, alpha subunit C 116 312 153 30 455 621 Ga0063436_11054 succinyl-CoA synthetase, beta subunit C pfam10131 pfam00549 pfam02629 pfam00549 pfam08442 168 325 85 10 291 383 Ga0063436_13085 Ga0063436_13086 Ga0063436_13087 Ga0063436_11036 - Ga0063436_11055 NHL repeat O pfam01436 144 446 94 98 500 530 Ga0063436_11061 - pfam08211 pfam00132 pfam12804 18 72 8 8 88 73 Ga0063436_11062 Cytidine and deoxycytidylate deaminase zinc-binding region UDP-N-acetylglucosamine diphosphorylase/glucosamine-1phosphate N-acetyltransferase Ga0063436_11064 chorismate synthase (EC 4.2.3.5) E Ga0063436_11065 shikimate dehydrogenase Ga0063436_11066 3-phosphoshikimate 1-carboxyvinyltransferase (EC 2.5.1.19) Ga0063436_11067 119 640 150 62 480 809 99 327 96 28 397 588 E pfam01264 pfam01488 pfam08501 33 51 29 12 134 201 E pfam00275 24 124 29 30 146 205 Prephenate dehydrogenase - pfam02153 20 10 0 0 30 58 Ga0063436_11068 Prephenate dehydrogenase - pfam02153 7 140 42 22 148 135 Ga0063436_11069 - pfam00793 7 39 19 0 74 86 - pfam00793 38 213 58 38 327 432 Ga0063436_110611 DAHP synthetase I family 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase (EC 2.5.1.54) 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase (EC 2.5.1.54) E pfam00793 106 264 87 63 425 540 Ga0063436_11071 hypothetical protein - 12 81 15 0 71 116 Ga0063436_11072 alanine racemase M pfam00842 pfam01168 41 243 44 2 155 274 Ga0063436_11073 Uncharacterized protein conserved in bacteria S pfam06155 41 98 27 10 181 427 Ga0063436_11074 hypothetical protein - - 18 69 20 0 77 152 Ga0063436_11075 Zn-dependent hydrolases, including glyoxylases R pfam00753 42 54 47 26 192 242 Ga0063436_11076 tRNA (Guanine37-N(1)-) methyltransferase (EC 2.1.1.31) J pfam01746 36 68 13 0 116 94 Ga0063436_11077 MAF protein D 4 111 31 4 122 203 Ga0063436_11078 Cell division protein FtsI/penicillin-binding protein 2 M pfam02545 pfam00905 pfam03717 pfam05223 86 386 116 75 407 580 Ga0063436_11079 LysM domain - pfam01476 24 71 11 2 81 128 Ga0063436_110710 hypothetical protein - - 109 204 233 73 629 596 Ga0063436_110711 Dephospho-CoA kinase - 10 0 6 0 26 22 Ga0063436_11081 IQ 129 129 72 70 280 409 Ga0063436_11082 Acyl-CoA synthetases (AMP-forming)/AMP-acid ligases II 4-amino-4-deoxy-L-arabinose transferase and related glycosyltransferases of PMT family pfam01121 pfam00501 pfam13193 M pfam13231 27 49 16 10 121 153 Ga0063436_11083 Bacterial membrane protein YfhO - pfam09586 82 308 69 22 220 223 Ga0063436_11084 hypothetical protein - - 4 15 13 1 51 66 Ga0063436_14411 Uncharacterized protein conserved in bacteria S pfam02381 27 306 31 20 247 323 Ga0063436_11092 16S rRNA (cytosine(1402)-N(4))-methyltransferase M pfam01795 129 655 187 47 1009 1128 Ga0063436_11093 hypothetical protein - 138 334 118 64 457 710 Ga0063436_11094 Cell division protein FtsI/penicillin-binding protein 2 M 214 498 178 185 1124 1673 74 551 86 27 739 1210 101 416 72 10 369 559 26 108 22 4 98 130 Ga0063436_110610 M M pfam00905 pfam03717 pfam01225 pfam02875 pfam08245 pfam00953 pfam10555 - - Ga0063436_11098 hypothetical protein UDP-N-acetylmuramoylalanine--D-glutamate ligase (EC 6.3.2.9) M pfam08245 108 404 114 38 539 840 Ga0063436_13941 Iron-containing alcohol dehydrogenase - pfam00465 132 210 77 10 209 278 Ga0063436_12331 Iron-containing alcohol dehydrogenase - 0 12 2 0 21 30 Ga0063436_12332 Zn-dependent alcohol dehydrogenases, class III C pfam00465 pfam00107 pfam08240 50 215 42 53 263 219 Ga0063436_12333 Glycine/D-amino acid oxidases (deaminating) E pfam01266 151 377 126 22 281 352 Ga0063436_12334 Trimethylamine:corrinoid methyltransferase H 86 488 106 35 298 365 Ga0063436_12335 C 184 328 130 36 288 448 Ga0063436_12336 Aldehyde:ferredoxin oxidoreductase L-alanine-DL-glutamate epimerase and related enzymes of enolase superfamily pfam06253 pfam01314 pfam02730 36 164 50 27 191 337 Ga0063436_11102 Dipeptidyl aminopeptidases/acylaminoacyl-peptidases E pfam01188 pfam00326 pfam07676 70 149 51 12 277 359 Ga0063436_11103 Uncharacterized protein, putative amidase R pfam02633 14 76 6 2 80 169 Ga0063436_11104 L-aminopeptidase/D-esterase EQ pfam03576 180 385 164 50 655 813 Ga0063436_11121 hypothetical protein - - 0 0 0 0 0 2 Ga0063436_11122 chorismate mutase (EC 5.4.99.5) E 17 52 24 7 93 102 Ga0063436_11123 nucleotide sugar dehydrogenase M pfam07736 pfam00984 pfam03720 pfam03721 42 248 48 22 199 252 Ga0063436_11124 Nucleoside-diphosphate-sugar epimerases MG pfam01370 38 124 37 23 174 256 Ga0063436_11095 Ga0063436_11096 Ga0063436_11097 UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase (EC 6.3.2.10) Phospho-N-acetylmuramoyl-pentapeptide-transferase (EC 2.7.8.13) M MR Ga0063436_11125 Glycosyl transferases group 1 - pfam13692 12 88 11 37 68 126 Ga0063436_11131 Predicted sugar kinase G pfam01513 51 159 77 22 206 348 Ga0063436_11132 hypothetical protein - - 30 58 9 0 14 89 Ga0063436_11133 Predicted RNA-binding protein of the translin family J pfam01997 86 109 44 26 225 441 Ga0063436_11134 Protein of unknown function (DUF3467) - pfam11950 153 235 120 17 369 621 Ga0063436_11135 single stranded DNA-binding protein (ssb) L 591 1052 788 160 2742 3463 Ga0063436_11136 E 60 157 81 8 301 594 E pfam00528 42 83 52 2 70 191 Ga0063436_11138 spermidine/putrescine ABC transporter ATP-binding subunit ABC-type spermidine/putrescine transport system, permease component II ABC-type spermidine/putrescine transport system, permease component I pfam00436 pfam00005 pfam08402 E pfam00528 14 0 21 10 83 146 Ga0063436_11139 Spermidine/putrescine-binding periplasmic protein E 108 196 183 36 398 628 Ga0063436_111310 K 28 29 20 22 89 143 Ga0063436_111311 Transcriptional regulators Phenylpropionate dioxygenase and related ring-hydroxylating dioxygenases, large terminal subunit PR pfam13343 pfam01037 pfam13404 pfam00355 pfam00848 49 231 71 19 504 440 Ga0063436_11141 hypothetical protein - - 35 122 31 6 106 205 Ga0063436_11142 undecaprenyl diphosphate synthase I pfam01255 103 210 87 22 314 463 Ga0063436_11143 ribosome recycling factor J pfam01765 159 330 179 139 442 521 Ga0063436_11144 uridylate kinase (EC 2.7.4.22) F 216 806 146 115 811 932 Ga0063436_11145 translation elongation factor Ts (EF-Ts) - pfam00696 pfam00627 pfam00889 195 542 216 48 934 932 Ga0063436_11146 SSU ribosomal protein S2P J pfam00318 466 1162 381 124 1378 1357 Ga0063436_11147 Fucose permease G pfam07690 90 149 57 8 163 252 Ga0063436_11148 hypothetical protein - - 28 92 18 6 47 90 Ga0063436_11149 diphosphomevalonate decarboxylase (EC 4.1.1.33) I - 44 202 14 6 152 217 Ga0063436_111410 Iron-containing alcohol dehydrogenase - pfam00465 14 50 2 0 37 117 Ga0063436_11151 hypothetical protein - - 131 268 180 46 393 555 Ga0063436_11152 hypothetical protein - - 48 160 33 20 188 291 Ga0063436_11153 hypothetical protein - - 24 11 13 2 31 54 Ga0063436_11154 Pentapeptide repeats (8 copies) - pfam00805 31 7 7 4 46 87 Ga0063436_11155 - - 42 61 9 0 114 165 Ga0063436_11156 hypothetical protein Membrane protein TerC, possibly involved in tellurium resistance P pfam03741 118 146 68 18 362 435 Ga0063436_11157 FOG: LysM repeat M pfam01476 360 461 259 110 1369 1755 Ga0063436_11158 hypothetical protein - 27 45 30 12 77 60 Ga0063436_11159 DNA gyrase, B subunit L pfam00204 pfam00986 pfam01751 pfam02518 383 869 218 64 837 1008 Ga0063436_111510 hypothetical protein - - 96 320 60 12 295 434 Ga0063436_111511 M pfam00275 15 242 42 40 210 278 Ga0063436_111512 UDP-N-acetylglucosamine enolpyruvyl transferase EPSP synthase (3-phosphoshikimate 1carboxyvinyltransferase) - pfam00275 26 85 25 0 121 178 Ga0063436_111513 hypothetical protein - - 181 253 93 12 546 778 Ga0063436_111514 EDD domain protein, DegV family S pfam02645 478 664 347 162 1607 1843 Ga0063436_11161 CheW-like domain - pfam01584 16 32 6 2 77 114 Ga0063436_11162 Methyl-accepting chemotaxis protein (MCP) signalling domain - pfam00015 116 382 80 41 587 915 Ga0063436_11163 dTDP-glucose 4,6-dehydratase (EC 4.2.1.46) M pfam01370 99 272 145 88 573 690 Ga0063436_11164 Glucose-1-phosphate thymidylyltransferase (EC 2.7.7.24) M pfam00483 92 184 69 55 287 436 Ga0063436_11165 Nucleoside-diphosphate-sugar epimerases MG pfam01370 93 121 56 24 279 346 Ga0063436_11166 Glycosyltransferase M pfam00534 92 217 58 54 263 361 Ga0063436_11167 phosphate transport system regulatory protein PhoU P pfam01895 175 608 210 156 882 1048 Ga0063436_11168 SET domain - 240 286 182 119 882 1021 Ga0063436_11169 DNA polymerase III, beta subunit (EC 2.7.7.7) L pfam00856 pfam00712 pfam02767 pfam02768 244 519 264 177 943 1165 Ga0063436_111610 Sulfite oxidase and related enzymes R pfam00174 237 586 102 108 430 439 Ga0063436_111611 Glycosyltransferases involved in cell wall biogenesis M pfam00535 32 137 16 20 60 135 Ga0063436_111612 Enterochelin esterase and related enzymes P pfam00756 81 129 50 45 184 215 Ga0063436_111613 SurA N-terminal domain - pfam13624 60 251 40 66 204 344 Ga0063436_111614 PPIC-type PPIASE domain - pfam13616 101 255 65 21 297 421 Ga0063436_11137 Ga0063436_111615 hypothetical protein - - 93 163 65 31 286 407 Ga0063436_111616 16S rRNA (cytidine(1402)-2'-O)-methyltransferase R 22 114 16 6 109 183 Ga0063436_111617 bifunctional phosphoglucose/phosphomannose isomerase G pfam00590 pfam01380 pfam10432 46 143 42 9 201 316 Ga0063436_111618 Pyruvate phosphate dikinase, PEP/pyruvate binding domain - pfam01326 130 291 72 30 330 542 Ga0063436_111619 hypothetical protein - - 4 148 14 4 93 157 Ga0063436_111620 L 80 72 49 18 116 143 73 175 78 22 259 318 V pfam01555 pfam00005 pfam00664 pfam00005 pfam00664 73 300 81 22 237 397 Ga0063436_11183 DNA modification methylase ABC-type multidrug transport system, ATPase and permease components ABC-type multidrug transport system, ATPase and permease components 6-pyruvoyl-tetrahydropterin synthase related domain; membrane protein - pfam10131 12 64 17 2 59 123 Ga0063436_11184 hypothetical protein - - 16 172 47 8 125 284 Ga0063436_11185 Uncharacterized enzyme involved in pigment biosynthesis Q pfam04227 17 210 41 24 131 224 Ga0063436_11191 - 62 0 0 20 84 GM pfam00005 pfam01061 14 Ga0063436_11192 hypothetical protein ABC-type polysaccharide/polyol phosphate transport system, ATPase component 38 48 40 4 155 377 Ga0063436_11193 FecR family protein - pfam04773 66 111 28 2 225 423 Ga0063436_11201 proton-translocating NADH-quinone oxidoreductase, chain M - pfam00361 83 427 98 60 438 527 Ga0063436_11202 NADH dehydrogenase subunit N (EC 1.6.5.3) C pfam00361 99 209 57 60 367 500 Ga0063436_11203 Putative F0F1-ATPase subunit (ATPase_gene1) - pfam09527 234 711 199 148 700 725 Ga0063436_11204 F0F1-type ATP synthase, subunit a C pfam00119 726 1402 505 226 2349 2330 Ga0063436_11205 ATP synthase F0 subcomplex C subunit - 437 485 309 124 1443 1232 Ga0063436_11206 ATP synthase, F0 subunit b C 1205 1512 824 437 3381 3161 Ga0063436_11207 UDP-N-acetylmuramoylalanyl-D-glutamate--2,6diaminopimelate ligase (EC 6.3.2.13) M 41 108 31 4 185 245 Ga0063436_11208 Undecaprenyl-phosphate glucose phosphotransferase M 118 285 120 54 481 502 Ga0063436_112010 methionine adenosyltransferase (EC 2.5.1.6) H pfam00137 pfam00213 pfam00430 pfam01225 pfam02875 pfam08245 pfam02397 pfam13727 pfam00438 pfam02772 pfam02773 312 631 193 143 1102 1210 Ga0063436_112011 Predicted pyrophosphatase R pfam03819 30 31 31 18 168 139 Ga0063436_112012 NUDIX domain - pfam00293 30 119 27 20 81 137 Ga0063436_112013 Predicted phosphoesterases, related to the Icc protein R pfam12850 19 9 8 6 64 90 Ga0063436_112014 Predicted Rossmann fold nucleotide-binding protein R 106 291 84 24 333 379 Ga0063436_112015 prolyl-tRNA synthetase (EC 6.1.1.15) J pfam03641 pfam00587 pfam03129 pfam09180 144 295 130 43 483 606 Ga0063436_112016 J 186 125 83 422 495 528 1163 286 237 1144 1393 Ga0063436_112018 polyribonucleotide nucleotidyltransferase - pfam00312 pfam01138 pfam03725 pfam00013 pfam00575 pfam01138 pfam03725 124 Ga0063436_112017 SSU ribosomal protein S15P 3' exoribonuclease family, domain 1/3' exoribonuclease family, domain 2 531 1264 343 274 1599 1966 Ga0063436_112019 hypothetical protein - - 94 182 31 17 149 156 Ga0063436_112020 Methyltransferase domain - pfam13659 137 506 95 62 638 732 Ga0063436_112021 Aldehyde dehydrogenase family - pfam00171 45 115 16 30 123 206 Ga0063436_11211 ribosomal protein L2, bacterial/organellar - pfam03947 137 800 195 88 1026 1273 Ga0063436_11212 ribosomal protein S19, bacterial/organelle J pfam00203 76 313 124 70 563 768 Ga0063436_11213 ribosomal protein L22, bacterial type J 143 316 104 62 458 578 Ga0063436_11214 SSU ribosomal protein S3P J pfam00237 pfam00189 pfam07650 271 726 187 98 1204 1578 Ga0063436_11215 LSU ribosomal protein L16P J pfam00252 254 447 164 137 1047 929 Ga0063436_11216 LSU ribosomal protein L29P J pfam00831 127 203 77 69 508 437 Ga0063436_11217 SSU ribosomal protein S17P J pfam00366 83 245 59 52 475 567 Ga0063436_11218 LSU ribosomal protein L14P J pfam00238 239 583 191 180 845 913 Ga0063436_11219 ribosomal protein L24, bacterial/organelle J 115 231 61 47 535 486 Ga0063436_112110 Ribosomal protein L5 J pfam00467 pfam00281 pfam00673 211 309 147 59 730 929 Ga0063436_112111 SSU ribosomal protein S14P J pfam00253 101 312 108 40 497 659 Ga0063436_112112 SSU ribosomal protein S8P J pfam00410 138 443 129 85 711 993 Ga0063436_112113 LSU ribosomal protein L6P J pfam00347 114 866 125 99 925 1065 Ga0063436_112114 LSU ribosomal protein L18P J pfam00861 155 677 141 136 868 1153 Ga0063436_11181 Ga0063436_11182 V - Ga0063436_112115 SSU ribosomal protein S5P J pfam00333 pfam03719 319 890 299 Ga0063436_112116 LSU ribosomal protein L30P J pfam00327 68 428 Ga0063436_112117 ribosomal protein L15, bacterial/organelle J pfam00828 93 567 Ga0063436_112118 protein translocase subunit secY/sec61 alpha U pfam00344 291 Ga0063436_112119 Adenylate kinase (EC 2.7.4.3) F pfam00406 Ga0063436_112120 methionine aminopeptidase, type I J pfam00557 Ga0063436_112121 30S ribosomal protein S13 J Ga0063436_112122 30S ribosomal protein S11 J Ga0063436_112123 SSU ribosomal protein S4P J Ga0063436_112124 DNA-directed RNA polymerase, alpha subunit, bacterial and chloroplast-type Ga0063436_112125 Ga0063436_112126 116 1378 1677 62 73 272 496 84 102 531 828 1195 229 93 1315 1433 216 726 240 113 1017 1142 318 880 325 122 1302 1744 pfam00416 271 591 169 153 887 1269 197 525 183 82 829 1014 324 946 303 136 1183 1631 K pfam00411 pfam00163 pfam01479 pfam01000 pfam01193 pfam03118 561 1217 412 173 1803 2279 LSU ribosomal protein L17P J pfam01196 275 461 197 86 924 1062 tRNA pseudouridine(38-40) synthase J pfam01416 353 643 263 89 1053 1117 Ga0063436_112127 LSU ribosomal protein L13P J pfam00572 169 590 165 93 625 926 Ga0063436_112128 SSU ribosomal protein S9P J pfam00380 151 415 104 62 335 687 Ga0063436_112129 Tetrapyrrole (Corrin/Porphyrin) Methylases - pfam00590 8 14 6 4 85 66 Ga0063436_11221 hypothetical protein - - 16 104 12 16 43 74 Ga0063436_11222 hypothetical protein - - 24 68 13 0 91 122 Ga0063436_11223 D-tyrosyl-tRNA(Tyr) deacylase J pfam02580 6 59 20 20 87 177 Ga0063436_11224 tryptophanyl-tRNA synthetase (EC 6.1.1.2) J pfam00579 106 239 117 69 358 476 Ga0063436_11225 Rdx family - pfam10262 131 341 116 51 483 665 Ga0063436_11226 Enoyl-CoA hydratase/carnithine racemase I pfam00378 30 327 62 56 169 306 Ga0063436_11227 hypothetical protein - - 188 362 152 51 625 811 Ga0063436_11228 dephospho-CoA kinase H pfam01121 12 50 13 0 44 140 Ga0063436_11231 Isocitrate/isopropylmalate dehydrogenase - pfam00180 12 4 4 6 34 36 Ga0063436_11232 3-isopropylmalate dehydratase, small subunit (EC 4.2.1.33) homoaconitate hydratase family protein/3-isopropylmalate dehydratase, large subunit E pfam00694 0 0 6 0 27 14 E pfam00330 4 18 8 14 63 40 P pfam13343 2169 3497 1650 990 9061 10946 R pfam00528 68 38 75 11 192 182 E 18 20 5 134 109 E pfam00528 pfam00005 pfam08402 15 Ga0063436_11244 ABC-type Fe3+ transport system, periplasmic component ABC-type uncharacterized transport system, permease component ABC-type spermidine/putrescine transport system, permease component II ABC-type spermidine/putrescine transport systems, ATPase components 81 229 83 24 289 415 Ga0063436_11245 hypothetical protein - - 54 172 53 34 166 205 Ga0063436_11246 Glycine/D-amino acid oxidases (deaminating) E pfam01266 122 194 146 29 333 481 Ga0063436_11247 GMP synthase - Glutamine amidotransferase domain F pfam00117 35 104 48 43 257 325 Ga0063436_11248 Type I phosphodiesterase / nucleotide pyrophosphatase - pfam01663 69 179 47 22 188 250 Ga0063436_11249 C pfam03009 37 210 64 36 307 408 Ga0063436_112410 Glycerophosphoryl diester phosphodiesterase Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) IQR pfam00106 158 351 87 67 254 331 Ga0063436_112411 Predicted phosphoribosyltransferases R pfam00156 95 256 54 64 273 322 Ga0063436_112412 SSU ribosomal protein S21P - pfam01165 383 727 248 110 992 1145 Ga0063436_112413 bacterial translation initiation factor 1 (bIF-1) J 116 117 57 31 270 313 Ga0063436_112414 tRNA-i(6)A37 thiotransferase enzyme MiaB J pfam01176 pfam01938 pfam04055 163 292 131 20 315 389 Ga0063436_11251 negative transcriptional regulator, PaiB family K pfam04299 81 52 53 10 115 150 Ga0063436_11252 transaldolase (EC 2.2.1.2) G pfam00923 49 139 40 32 192 260 Ga0063436_11253 NAD-dependent aldehyde dehydrogenases C 211 378 194 128 779 940 Ga0063436_11254 J 168 341 88 24 370 519 82 288 83 31 157 336 Ga0063436_11262 aspartyl-tRNA synthetase (EC 6.1.1.12) ABC-type multidrug transport system, ATPase and permease components ABC-type multidrug transport system, ATPase and permease components pfam00171 pfam00152 pfam01336 pfam02938 pfam00005 pfam00664 pfam00005 pfam00664 70 120 62 6 129 188 Ga0063436_11271 DinB superfamily - 41 41 19 4 74 54 Ga0063436_11272 folate-binding protein YgfZ E pfam12867 pfam01571 pfam08669 32 190 32 10 169 257 Ga0063436_11273 hypothetical protein - - 46 279 57 44 443 497 Ga0063436_11274 hypothetical protein - - 134 198 74 70 335 595 Ga0063436_11233 Ga0063436_11241 Ga0063436_11242 Ga0063436_11243 Ga0063436_11261 V V E pfam00224 pfam02887 pfam01546 pfam07687 TK pfam00486 50 242 60 33 417 582 J pfam01649 pfam00512 pfam00672 pfam02518 pfam08448 pfam13185 209 258 183 50 402 650 149 478 184 38 800 873 62 301 65 8 199 322 - pfam02779 pfam02780 48 46 44 14 210 290 - pfam00456 81 199 58 84 333 400 Uncharacterized protein involved in methicillin resistance V pfam02388 34 87 19 8 116 192 Ga0063436_11292 Uncharacterized protein involved in methicillin resistance V pfam02388 30 152 49 16 137 183 Ga0063436_11293 hypothetical protein - - 7 82 15 0 58 147 Ga0063436_11301 H pfam06253 pfam00528 pfam12911 35 123 18 2 106 203 Ga0063436_11302 Trimethylamine:corrinoid methyltransferase ABC-type dipeptide/oligopeptide/nickel transport systems, permease components 132 198 88 67 535 678 Ga0063436_11311 hypothetical protein - 77 73 20 20 124 157 Ga0063436_11312 proton-translocating NADH-quinone oxidoreductase, chain M C 176 693 125 38 817 906 Ga0063436_11313 NADH dehydrogenase subunit L (EC 1.6.5.3) CP pfam00361 pfam01059 pfam00361 pfam00662 pfam06455 224 787 202 121 812 1104 Ga0063436_11314 NADH dehydrogenase subunit K (EC 1.6.5.3) C pfam00420 33 55 40 50 196 175 Ga0063436_11315 NADH dehydrogenase subunit J (EC 1.6.5.3) C pfam00499 106 212 48 35 293 375 Ga0063436_11316 NADH:ubiquinone oxidoreductase subunit 1 (chain H) C pfam00146 188 539 165 83 788 791 Ga0063436_11317 C pfam00507 68 308 76 36 358 423 Ga0063436_11318 NADH dehydrogenase subunit A (EC 1.6.5.3) 2-polyprenyl-3-methyl-5-hydroxy-6-metoxy-1,4-benzoquinol methylase H 20 18 10 1 61 78 Ga0063436_11319 Glycosyltransferase M pfam13847 pfam00534 pfam13579 40 133 42 19 196 282 Ga0063436_113110 6-pyruvoyl-tetrahydropterin synthase H pfam01242 18 39 16 9 53 98 Ga0063436_113111 S pfam03992 28 0 5 7 80 78 Ga0063436_113112 Uncharacterized conserved protein Threonine dehydrogenase and related Zn-dependent dehydrogenases ER pfam08240 16 57 32 2 132 231 Ga0063436_113113 Pyrimidine reductase, riboflavin biosynthesis H pfam01872 20 177 14 1 159 277 Ga0063436_113114 hypothetical protein - - 42 127 31 1 83 147 Ga0063436_113115 hypothetical protein - - 11 29 2 3 12 11 Ga0063436_11321 - pfam04224 pfam05496 pfam12002 44 52 16 4 56 94 28 184 45 22 167 274 Ga0063436_11323 Protein of unknown function, DUF417 ATPase related to the helicase subunit of the Holliday junction resolvase haloacid dehalogenase superfamily, subfamily IA, variant 3 with third motif having DD or ED/haloacid dehalogenase superfamily, subfamily IA, variant 1 with third motif having Dx(3-4)D or Dx(3-4)E 16 183 54 44 202 285 Ga0063436_11324 Membrane proteins related to metalloendopeptidases M 44 251 55 22 210 414 Ga0063436_11325 transcription termination factor Rho K pfam00535 pfam13419 pfam01476 pfam01551 pfam00006 pfam07497 pfam07498 341 861 269 124 780 1307 Ga0063436_11351 - - 14 66 4 0 24 44 Ga0063436_11352 hypothetical protein ABC-type antimicrobial peptide transport system, ATPase component V pfam00005 56 321 16 4 60 137 Ga0063436_11353 Predicted ornithine cyclodeaminase, mu-crystallin homolog E 68 103 36 36 135 234 Ga0063436_11361 arginyl-tRNA synthetase (EC 6.1.1.19) J pfam02423 pfam00750 pfam03485 pfam05746 24 142 51 21 210 361 Ga0063436_11362 deoxycytidine triphosphate deaminase F pfam00692 41 164 105 10 225 314 Ga0063436_11363 WD40-like Beta Propeller Repeat - pfam07676 238 817 139 76 622 697 Ga0063436_11364 Dipeptidyl peptidase IV (DPP IV) N-terminal region - pfam00930 297 819 127 40 500 607 Ga0063436_11365 hypothetical protein - - 18 81 27 2 134 178 Ga0063436_11371 hypothetical protein - 11 95 4 20 68 64 Ga0063436_11372 Predicted metal-dependent phosphoesterases (PHP family) R pfam02811 pfam13263 18 117 27 12 107 148 Ga0063436_11374 hypothetical protein - 6 79 22 0 79 89 Ga0063436_11375 Superfamily I DNA and RNA helicases L pfam00580 pfam13361 48 386 101 62 386 456 Ga0063436_11275 G Ga0063436_11277 pyruvate kinase Acetylornithine deacetylase/Succinyl-diaminopimelate desuccinylase and related deacylases Response regulators consisting of a CheY-like receiver domain and a winged-helix DNA-binding domain Ga0063436_11278 SSU ribosomal protein S20P Ga0063436_11279 T Ga0063436_11281 Signal transduction histidine kinase NADPH-dependent glutamate synthase beta chain and related oxidoreductases Transketolase, pyrimidine binding domain/Transketolase, Cterminal domain Ga0063436_11282 Transketolase, thiamine diphosphate binding domain Ga0063436_11291 Ga0063436_11276 Ga0063436_112710 Ga0063436_11322 RE EP L R 130 176 112 34 447 487 43 25 30 20 68 157 Ga0063436_11376 UDP-galactose 4-epimerase (EC 5.1.3.2) M pfam01370 pfam13950 29 22 13 Ga0063436_11377 Arabinose efflux permease G pfam07690 64 187 35 Ga0063436_11378 hypothetical protein - - 20 6 15 Ga0063436_11379 chaperonin GroL O pfam00118 9039 16632 4589 Ga0063436_113710 Co-chaperonin GroES (HSP10) O 2506 4377 Ga0063436_113711 threonyl-tRNA synthetase (EC 6.1.1.3) J pfam00166 pfam00587 pfam03129 pfam07973 140 Ga0063436_113714 endoribonuclease L-PSP J pfam01042 117 Ga0063436_113715 F pfam00293 Ga0063436_113716 ADP-ribose pyrophosphatase 3'-nucleotidase (EC 3.1.3.6)/5'-nucleotidase (EC 3.1.3.5)/exopolyphosphatase (EC 3.6.1.11) R pfam01975 Ga0063436_113717 hypothetical protein - - Ga0063436_113718 hypothetical protein - - 115 Ga0063436_113719 short chain enoyl-CoA hydratase (EC 4.2.1.17) I pfam00378 142 Ga0063436_113720 Short-chain alcohol dehydrogenase of unknown specificity R pfam00106 Ga0063436_113721 - Ga0063436_113723 conserved hypothetical protein Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide acyltransferase (E2) component, and related enzymes Pyruvate/2-oxoglutarate dehydrogenase complex, dehydrogenase (E1) component, eukaryotic type, beta subunit C pfam07110 pfam00198 pfam00364 pfam02817 pfam02779 pfam02780 Ga0063436_113724 pyruvate dehydrogenase E1 component, alpha subunit C pfam00676 Ga0063436_113725 Lipoate-protein ligase A H pfam03099 6 0 16 0 31 32 Ga0063436_113726 Excinuclease ABC subunit A L pfam00005 329 825 162 76 472 670 Ga0063436_113727 LysM domain - pfam01476 115 219 119 26 205 295 Ga0063436_15032 HEAT repeats - 2 88 6 10 76 40 Ga0063436_11381 ATP-dependent Zn proteases O 1345 2993 543 204 2021 2248 Ga0063436_11382 peptide chain release factor 2 J pfam13646 pfam00004 pfam01434 pfam00472 pfam03462 75 211 94 10 251 341 Ga0063436_11383 - 37 11 8 73 56 - pfam00448 pfam00448 pfam02881 40 Ga0063436_11384 SRP54-type protein, GTPase domain SRP54-type protein, GTPase domain/SRP54-type protein, helical bundle domain 44 128 28 26 109 197 Ga0063436_11385 Subtilase family - pfam00082 18 203 41 5 271 544 Ga0063436_11391 hydroxymethylglutaryl-CoA synthase, putative I pfam08541 105 503 15 8 273 428 Ga0063436_11392 Acetyl-CoA acetyltransferase I 51 316 14 20 148 327 Ga0063436_11393 Predicted nucleic-acid-binding protein containing a Zn-ribbon R pfam00108 pfam01796 pfam12172 74 404 18 0 246 330 Ga0063436_11394 uracil-DNA glycosylase, family 4 L 26 196 37 16 275 430 Ga0063436_11395 dTDP-D-glucose 4,6-dehydratase M pfam03167 pfam01370 pfam13950 50 340 63 0 212 307 Ga0063436_11396 ribosomal-protein-alanine acetyltransferase R pfam00583 26 43 12 4 112 188 Ga0063436_11397 tRNA threonylcarbamoyl adenosine modification protein YeaZ O pfam00814 26 145 24 24 171 392 Ga0063436_11398 tRNA threonylcarbamoyl adenosine modification protein YjeE R pfam02367 4 76 48 0 136 140 Ga0063436_11399 Predicted phosphoesterase R pfam12850 24 111 30 8 122 228 Ga0063436_113910 Domain of unknown function (DUF4349) - pfam14257 24 107 18 0 75 110 Ga0063436_113911 - pfam14257 10 46 14 0 50 70 Ga0063436_113912 Domain of unknown function (DUF4349) cyclic pyranopterin monophosphate synthase subunit MoaC (EC 4.1.99.18) H 16 62 17 24 152 145 Ga0063436_113913 Molybdopterin converting factor, large subunit H pfam01967 pfam02391 pfam02597 52 45 70 12 139 244 Ga0063436_113914 Threonine synthase E pfam00291 0 111 18 0 89 135 Ga0063436_113915 hypothetical protein - - 91 260 55 32 206 354 Ga0063436_113916 - - 68 202 65 13 201 266 Ga0063436_11401 hypothetical protein Uncharacterized protein SCO1/SenC/PrrC, involved in biogenesis of respiratory and photosynthetic systems R pfam02630 48 223 19 6 68 127 Ga0063436_11402 methylthioribose-1-phosphate isomerase (EC 5.3.1.23) J pfam01008 113 281 75 21 199 263 Ga0063436_11403 - pfam00293 22 80 28 11 57 80 Ga0063436_11404 NUDIX domain purine nucleoside phosphorylase I, inosine and guanosinespecific F pfam01048 68 199 47 2 153 235 Ga0063436_11405 hypothetical protein - - Ga0063436_11406 Acetyltransferase (GNAT) family - pfam00583 Ga0063436_11407 hypothetical protein - Ga0063436_11408 FtsK/SpoIIIE family/Ftsk gamma domain - Ga0063436_113722 C 16 90 85 6 47 198 18 107 68 3608 21333 26338 1208 1085 5839 6155 108 73 46 438 448 149 56 50 224 289 31 113 32 7 131 124 30 187 50 25 139 203 187 468 132 56 567 513 462 99 42 310 422 424 140 61 377 397 76 333 44 29 292 373 74 252 52 28 208 271 233 718 136 57 672 810 204 317 135 24 484 498 200 359 115 86 437 520 6 124 10 0 45 115 92 278 91 24 257 406 - 208 485 174 50 659 910 pfam01580 193 608 190 34 644 851 pfam09397 R Ga0063436_114010 Large extracellular alpha-helical protein Methionine synthase I (cobalamin-dependent), methyltransferase domain E pfam00207 pfam01835 pfam07678 pfam07703 pfam13205 pfam02219 pfam02574 Ga0063436_114011 hypothetical protein - - Ga0063436_11421 - Ga0063436_11422 Adenosine/AMP deaminase phosphate ABC transporter ATP-binding protein, PhoT family (TC 3.A.1.7.1) Ga0063436_11423 ABC-type tungstate transport system, periplasmic component Ga0063436_13302 ABC-type tungstate transport system, permease component Ga0063436_13301 Ga0063436_11431 Ga0063436_11409 114 545 143 74 660 890 85 205 72 24 337 350 10 46 6 2 10 10 pfam00962 0 22 3 4 21 33 P pfam00005 12 50 12 0 57 61 H 32 19 24 2 68 32 H pfam00528 pfam01476 pfam12849 35 66 51 34 135 237 hypothetical protein - - 3 0 8 10 48 29 Protein-disulfide isomerase O pfam13462 55 90 29 37 184 221 Ga0063436_11432 Beta-lactamase class C and other penicillin binding proteins V 109 119 51 53 201 161 Ga0063436_14222 Fe-S oxidoreductase C pfam00144 pfam02754 pfam13183 136 467 125 22 425 664 Ga0063436_14221 hypothetical protein ABC-type dipeptide/oligopeptide/nickel transport systems, permease components ABC-type dipeptide/oligopeptide/nickel transport systems, permease components - 6 43 7 1 67 122 EP pfam00528 pfam12911 59 278 79 34 437 559 EP pfam00528 141 492 136 89 909 1083 - pfam00496 3790 6760 3255 1091 21380 22337 V 48 208 35 15 173 250 EP pfam02016 pfam00528 pfam12911 26 169 66 46 301 452 Ga0063436_12214 Bacterial extracellular solute-binding proteins, family 5 Middle Uncharacterized proteins, homologs of microcin C7 resistance protein MccF ABC-type dipeptide/oligopeptide/nickel transport systems, permease components ABC-type dipeptide/oligopeptide/nickel transport systems, permease components EP pfam00528 156 439 159 72 756 996 Ga0063436_12215 Bacterial extracellular solute-binding proteins, family 5 Middle - pfam00496 1479 2275 1167 458 5065 5216 Ga0063436_12216 Bacterial extracellular solute-binding proteins, family 5 Middle - pfam00496 1583 2486 1059 552 6256 6085 Ga0063436_12217 hypothetical protein - - 14 29 14 5 60 145 Ga0063436_12218 E 409 65 17 309 485 288 1151 400 189 1705 1928 E pfam00496 pfam00005 pfam08352 pfam00005 pfam08352 123 Ga0063436_11446 ABC-type dipeptide transport system, periplasmic component oligopeptide/dipeptide ABC transporter, ATP-binding protein, C-terminal domain oligopeptide/dipeptide ABC transporter, ATP-binding protein, C-terminal domain 424 832 295 159 1550 2036 Ga0063436_11451 Predicted SAM-dependent methyltransferases R pfam10672 104 258 71 13 290 304 Ga0063436_11452 7-keto-8-aminopelargonate synthetase and related enzymes H 121 542 140 52 423 679 Ga0063436_11453 L 50 223 66 8 174 243 Ga0063436_11454 DNA ligase, NAD-dependent Predicted dithiol-disulfide isomerase involved in polyketide biosynthesis pfam00155 pfam00533 pfam01653 pfam03119 pfam03120 pfam12826 pfam14520 Q pfam01323 30 165 49 20 262 417 Ga0063436_11455 hypothetical protein - - 31 119 29 7 164 242 Ga0063436_11456 Putative threonine efflux protein E 5 153 10 7 105 178 Ga0063436_11457 Glycosyltransferase M pfam01810 pfam00534 pfam13579 pfam00111 pfam01315 pfam01799 pfam02738 126 230 47 36 231 301 253 517 175 62 587 831 72 198 44 24 185 284 44 159 67 28 244 308 Ga0063436_14521 Ga0063436_14522 Ga0063436_14523 Ga0063436_12212 Ga0063436_12213 Ga0063436_11445 EP Ga0063436_11462 xanthine dehydrogenase, molybdenum binding subunit apoprotein (EC 1.17.1.4) Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) Ga0063436_11463 Argininosuccinate lyase C-terminal/Lyase - pfam00106 pfam00206 pfam14698 Ga0063436_11471 hypothetical protein - - 22 34 13 0 93 53 Ga0063436_11472 Membrane dipeptidase (Peptidase family M19) - pfam01244 78 125 49 22 108 180 Ga0063436_11473 - 320 70 28 282 590 34 117 22 0 106 142 Ga0063436_11475 asparagine synthase (glutamine-hydrolyzing) - pfam00534 pfam13579 pfam00733 pfam13537 58 Ga0063436_11474 hypothetical protein Glycosyl transferases group 1/Glycosyl transferase 4-like domain 6 37 24 14 88 200 Ga0063436_11476 Asparagine synthase - pfam00733 14 100 27 4 148 187 Ga0063436_11477 hypothetical protein - - 54 79 38 3 127 191 Ga0063436_11478 hypothetical protein - - 33 28 36 9 115 169 Ga0063436_11479 Glycosyltransferase M pfam00534 26 159 22 24 160 176 Ga0063436_11461 C IQR - Ga0063436_114710 hypothetical protein - - 6 8 11 0 36 27 Ga0063436_11481 R pfam01588 0 0 2 2 18 26 Ga0063436_11482 export-related chaperone protein CsaA Methionine synthase I (cobalamin-dependent), methyltransferase domain E pfam02574 66 282 58 58 290 317 Ga0063436_11491 hypothetical protein - - 13 63 4 0 23 82 Ga0063436_11492 heme ABC exporter, ATP-binding protein CcmA V 10 112 16 6 36 87 Ga0063436_11501 bacterial translation initiation factor 3 (bIF-3) J pfam00005 pfam00707 pfam05198 513 1303 448 193 1973 2424 Ga0063436_11502 LSU ribosomal protein L35P J pfam01632 183 351 149 42 545 785 Ga0063436_11503 LSU ribosomal protein L20P J 141 599 97 98 543 813 Ga0063436_11504 rRNA methylases J pfam00453 pfam00588 pfam08032 35 170 39 14 143 240 Ga0063436_11505 SecD/SecF GG Motif - pfam07549 172 503 157 104 537 743 Ga0063436_11506 MMPL family - pfam03176 53 78 62 15 201 253 Ga0063436_11507 protein translocase subunit secF U pfam02355 194 439 187 134 900 773 Ga0063436_11508 hypothetical protein - - 0 0 0 0 20 12 Ga0063436_11509 Dolichyl-phosphate-mannose-protein mannosyltransferase - pfam13231 15 30 4 2 30 68 Ga0063436_11511 hypothetical protein - - 0 4 0 0 15 27 Ga0063436_11512 hypothetical protein - - 14 11 14 0 46 21 Ga0063436_11513 hypothetical protein - 12 2 10 10 31 43 Ga0063436_11514 E 64 246 111 36 302 451 Ga0063436_11515 Glycine cleavage system T protein (aminomethyltransferase) haloacid dehalogenase superfamily, subfamily IA, variant 1 with third motif having Dx(3-4)D or Dx(3-4)E pfam01266 pfam01571 pfam08669 R pfam13419 64 156 62 0 174 291 Ga0063436_11516 Phosphate/sulphate permeases P pfam01384 2 54 13 2 122 146 Ga0063436_11517 Phosphate transport regulator (distant homolog of PhoU) P pfam01865 14 37 24 10 86 117 Ga0063436_11518 2-aminoethylphosphonate--pyruvate transaminase E pfam00266 48 152 48 24 213 183 Ga0063436_11519 R 204 142 57 378 435 - pfam13419 pfam00557 pfam01321 69 Ga0063436_11521 phosphonoacetaldehyde hydrolase (EC 3.11.1.1) Creatinase/Prolidase N-terminal domain/Metallopeptidase family M24 36 112 21 8 73 166 Ga0063436_11522 Platelet-activating factor acetylhydrolase, isoform II - pfam03403 38 99 28 40 186 253 Ga0063436_11523 Predicted metal-dependent hydrolase with the TIM-barrel fold R 138 566 178 62 994 1251 Ga0063436_11524 R 14 59 44 4 105 104 Ga0063436_11525 Predicted permeases Asp-tRNAAsn/Glu-tRNAGln amidotransferase A subunit and related amidases pfam07969 pfam00892 pfam13536 58 181 39 18 258 313 Ga0063436_11526 glutamate formiminotransferase E pfam01425 pfam02971 pfam04961 pfam07837 108 281 120 34 285 535 Ga0063436_11527 hypothetical protein - - 22 67 12 0 55 55 Ga0063436_11541 hypothetical protein - - 7 38 12 16 20 17 Ga0063436_11542 Kynureninase (EC 3.7.1.3) E 28 92 42 63 232 333 Ga0063436_11543 Bacterial PH domain/Cyclic nucleotide-binding domain - pfam00266 pfam00027 pfam03703 87 277 61 18 353 465 Ga0063436_11544 N-formylmethionyl-tRNA deformylase J pfam01327 17 50 28 4 59 124 Ga0063436_11545 Predicted membrane protein S pfam06210 57 64 33 3 120 171 Ga0063436_11546 PAP2 superfamily - pfam01569 7 20 4 3 29 51 Ga0063436_11547 hypothetical protein - - 24 73 9 14 57 62 Ga0063436_11548 - 123 17 0 41 46 R pfam14340 pfam00005 pfam12848 40 Ga0063436_11551 Domain of unknown function (DUF4395) ATPase components of ABC transporters with duplicated ATPase domains 172 343 189 56 487 739 Ga0063436_11552 Glycosyltransferases, probably involved in cell wall biogenesis M pfam13641 27 138 44 23 266 360 Ga0063436_11553 Cellulase M and related proteins (peptidase) G pfam05343 43 121 40 0 183 243 Ga0063436_11554 Cellulase M and related proteins (peptidase) G pfam05343 14 84 26 16 99 190 Ga0063436_11555 Cellulase M and related proteins (peptidase) G pfam05343 115 112 49 24 184 350 Ga0063436_11556 Glycosyl transferase family 2 - pfam00535 105 289 47 22 269 431 Ga0063436_11557 Phosphoheptose isomerase G pfam13580 125 178 91 22 323 444 Ga0063436_11558 MG pfam01370 8 46 36 12 155 145 Ga0063436_11559 Nucleoside-diphosphate-sugar epimerases D-alpha,beta-D-heptose 1,7-bisphosphate phosphatase (EC 3.1.3.-) E pfam13242 38 142 23 16 138 253 Ga0063436_115510 Nucleoside-diphosphate-sugar epimerases MG pfam01370 42 109 51 68 168 293 Ga0063436_115511 hypothetical protein UDP-N-acetylmuramyl pentapeptide phosphotransferase/UDP-N-acetylglucosamine-1-phosphate - - 131 219 90 43 440 439 M pfam00953 137 590 104 34 525 814 Ga0063436_115512 J transferase Ga0063436_115513 Glycosyltransferase M Ga0063436_115514 Nucleoside-diphosphate-sugar epimerases MG Ga0063436_115515 Glycosyltransferase Ga0063436_115516 Ga0063436_11561 pfam00534 pfam13579 83 232 61 39 267 293 85 100 53 28 230 234 M pfam01370 pfam00534 pfam13579 12 158 48 18 138 185 hypothetical protein - - 32 40 38 0 53 62 hypothetical protein - - 3 44 9 8 35 33 Ga0063436_11562 phosphoribosyl-ATP pyrophosphatase (EC 3.6.1.31) E pfam01503 14 45 6 10 51 101 Ga0063436_11563 Predicted secreted hydrolase R pfam07143 99 335 10 2 64 74 Ga0063436_11564 - 40 143 6 4 36 46 M pfam02687 pfam02687 pfam12704 135 669 18 0 100 121 Ga0063436_11566 FtsX-like permease family ABC-type transport system, involved in lipoprotein release, permease component ABC-type antimicrobial peptide transport system, ATPase component V pfam00005 44 246 17 26 15 60 Ga0063436_11567 phosphoribosyl-AMP cyclohydrolase (EC 3.5.4.19) E pfam01502 2 5 3 0 27 67 Ga0063436_11568 E pfam00977 2 10 6 10 45 66 Ga0063436_11569 imidazoleglycerol phosphate synthase, cyclase subunit 1-(5-phosphoribosyl)-5-[(5phosphoribosylamino)methylideneamino] imidazole-4carboxamide isomerase (EC 5.3.1.16) E pfam00117 pfam00977 0 44 8 2 67 76 Ga0063436_115610 imidazoleglycerol-phosphate dehydratase (EC 4.2.1.19) E pfam00475 0 103 20 12 69 151 Ga0063436_115611 histidinol-phosphate aminotransferase E pfam00155 0 6 8 0 32 49 Ga0063436_13281 hypothetical protein - - 4 35 12 0 62 40 Ga0063436_13282 hypothetical protein - - 6 34 8 0 16 14 Ga0063436_13283 hypothetical protein - - 0 20 4 0 16 4 Ga0063436_13284 hypothetical protein - - 20 35 12 5 70 104 Ga0063436_13285 Dolichyl-phosphate-mannose-protein mannosyltransferase - pfam13231 28 190 24 17 74 230 Ga0063436_13286 Glycosyltransferase M pfam00534 16 40 18 0 60 129 Ga0063436_11571 Glycosyl transferases group 1 - pfam00534 0 40 4 0 6 13 Ga0063436_11572 Glycosyl transferases group 1 - pfam00534 0 2 0 0 4 16 Ga0063436_11573 Glycosyl transferase 4-like domain - 0 34 26 33 26 54 Ga0063436_11574 Glycosyltransferase M 42 203 20 3 144 253 Ga0063436_11575 nucleotide sugar dehydrogenase M pfam13579 pfam00534 pfam13579 pfam00984 pfam03720 pfam03721 109 429 102 14 473 689 Ga0063436_11576 Nucleoside-diphosphate-sugar epimerases GM pfam01370 142 478 118 26 398 471 Ga0063436_11577 Predicted metal-binding, possibly nucleic acid-binding protein R pfam02620 80 115 26 29 158 220 Ga0063436_11578 - pfam13231 44 180 40 3 168 267 Ga0063436_11579 Dolichyl-phosphate-mannose-protein mannosyltransferase 4-amino-4-deoxy-L-arabinose transferase and related glycosyltransferases of PMT family M 12 104 13 14 126 272 Ga0063436_115710 methionyl-tRNA synthetase (EC 6.1.1.10) J pfam13231 pfam08264 pfam09334 121 373 132 50 599 555 Ga0063436_115711 Dolichyl-phosphate-mannose-protein mannosyltransferase - 26 100 27 4 159 228 Ga0063436_115712 glycerol kinase C pfam13231 pfam00370 pfam01408 pfam02782 pfam02894 95 264 73 30 293 468 Ga0063436_115713 hypothetical protein - - 39 19 11 12 63 128 Ga0063436_115714 YdjC-like protein - pfam04794 62 99 66 36 251 253 Ga0063436_11581 hypothetical protein - - 625 597 539 195 2690 3797 Ga0063436_11582 Small-conductance mechanosensitive channel M pfam00924 435 463 294 212 2057 2820 Ga0063436_11583 hypothetical protein - - 63 190 90 25 317 465 Ga0063436_11584 Protein of unknown function (DUF2939) - pfam11159 22 38 41 30 28 75 Ga0063436_11585 hypothetical protein - - 3 48 23 43 59 119 Ga0063436_11586 hypothetical protein - - 50 74 67 0 164 239 Ga0063436_11591 hypothetical protein - 0 0 0 0 8 0 Ga0063436_11592 choline dehydrogenase E pfam00732 pfam05199 90 399 83 72 435 649 Ga0063436_11593 Predicted membrane protein S pfam01794 15 70 7 0 28 60 Ga0063436_11594 Oxidoreductase molybdopterin binding domain - pfam00174 71 317 48 2 119 202 Ga0063436_11601 Dehydratase large subunit L-alanine-DL-glutamate epimerase and related enzymes of enolase superfamily L-alanine-DL-glutamate epimerase and related enzymes of enolase superfamily - pfam02286 pfam02746 pfam13378 pfam02746 pfam13378 0 0 4 6 8 37 215 164 108 60 341 545 127 111 90 24 372 444 Ga0063436_11565 Ga0063436_11602 Ga0063436_11603 MR MR Ga0063436_11604 Nucleoside-diphosphate-sugar epimerases MG pfam01370 143 136 96 39 260 387 Ga0063436_11605 D-mannonate dehydratase G pfam03786 127 241 76 64 402 494 Ga0063436_11606 - 18 8 24 14 62 63 48 64 34 30 88 233 86 129 78 36 263 361 EP pfam01188 pfam02746 pfam00005 pfam08352 pfam00005 pfam08352 102 214 69 64 240 409 EP pfam00528 57 200 89 32 394 453 Ga0063436_116011 hypothetical protein L-alanine-DL-glutamate epimerase and related enzymes of enolase superfamily oligopeptide/dipeptide ABC transporter, ATP-binding protein, C-terminal domain oligopeptide/dipeptide ABC transporter, ATP-binding protein, C-terminal domain ABC-type dipeptide/oligopeptide/nickel transport systems, permease components ABC-type dipeptide/oligopeptide/nickel transport systems, permease components EP pfam00528 206 235 133 55 680 722 Ga0063436_116012 ABC-type dipeptide transport system, periplasmic component E pfam00496 8909 13080 7759 3833 37246 32817 Ga0063436_116013 Glucose/sorbosone dehydrogenases G pfam07995 35 30 52 22 101 171 Ga0063436_116014 Demethylmenaquinone methyltransferase H pfam03737 92 91 57 20 349 403 Ga0063436_116015 R 140 60 18 227 300 MR pfam02633 pfam01188 pfam13378 45 Ga0063436_116016 Uncharacterized protein, putative amidase L-alanine-DL-glutamate epimerase and related enzymes of enolase superfamily 48 32 35 38 178 173 Ga0063436_116017 Glutamate-1-semialdehyde aminotransferase H pfam00202 20 57 44 12 127 200 Ga0063436_116018 Predicted amidohydrolase Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) R pfam13594 50 122 42 0 128 176 IQR pfam13561 54 95 52 2 216 288 - 302 120 55 517 520 G pfam13377 pfam01547 pfam10518 259 Ga0063436_116021 Periplasmic binding protein-like domain carbohydrate ABC transporter, Nacetylglucosamine/diacetylchitobiose-binding protein 4749 6161 3451 1592 17587 17407 Ga0063436_116022 ABC-type sugar transport systems, permease components G pfam00528 173 296 103 57 728 869 Ga0063436_116023 ABC-type sugar transport system, permease component G pfam00528 113 144 61 30 422 390 Ga0063436_116024 Domain of unknown function (DUF4432) - pfam14486 104 270 82 41 212 314 Ga0063436_116025 hypothetical protein - - 32 35 26 8 77 122 Ga0063436_116026 MG 112 41 14 251 272 MR pfam01370 pfam02746 pfam13378 55 Ga0063436_116027 Nucleoside-diphosphate-sugar epimerases L-alanine-DL-glutamate epimerase and related enzymes of enolase superfamily 56 104 70 18 255 383 Ga0063436_116028 3-carboxymuconate cyclase G pfam10282 41 110 81 46 249 430 Ga0063436_116029 D-mannonate dehydratase G pfam03786 89 101 55 29 206 313 Ga0063436_116030 2-deoxy-D-gluconate 3-dehydrogenase IQR pfam00106 23 19 28 18 34 97 Ga0063436_116031 5-deoxyglucuronate isomerase (EC 5.3.1.-) G pfam04962 74 72 20 10 67 150 Ga0063436_116032 Glycosyl Hydrolase Family 88 - 14 45 32 0 56 64 Ga0063436_11611 signal recognition particle subunit FFH/SRP54 (srp54) U pfam07470 pfam00448 pfam02881 pfam02978 167 329 94 25 335 505 Ga0063436_11612 ribosomal protein S16 J pfam00886 156 832 163 49 799 799 Ga0063436_11613 RNA-binding protein (KH domain) R 81 272 46 35 350 510 Ga0063436_11614 16S rRNA processing protein RimM J pfam13083 pfam01782 pfam05239 179 516 164 66 629 711 Ga0063436_11615 DNA protecting protein DprA LU 147 703 131 68 555 773 Ga0063436_11616 DNA topoisomerase I, bacterial - 198 503 107 88 510 651 Ga0063436_11621 transcription elongation factor GreA K pfam02481 pfam01131 pfam01751 pfam01272 pfam03449 227 597 209 60 611 864 Ga0063436_11622 - pfam01131 pfam01396 124 711 189 87 744 1155 Ga0063436_11623 hypothetical protein DNA topoisomerase/Topoisomerase DNA binding C4 zinc finger 166 492 146 47 613 567 Ga0063436_11631 Acetamidase/Formamidase family - 49 287 82 36 281 415 Ga0063436_12068 dihydrolipoamide dehydrogenase C pfam03069 pfam00070 pfam02852 pfam07992 234 525 159 129 611 749 Ga0063436_12067 L,D-transpeptidase catalytic domain - pfam03734 64 216 59 58 224 261 Ga0063436_12066 Cytochrome c-type biogenesis protein CcmE O pfam03100 34 275 32 16 72 180 Ga0063436_12065 O pfam01578 274 777 77 46 237 465 Ga0063436_12064 Cytochrome c biogenesis factor Uncharacterized protein involved in biosynthesis of c-type cytochromes O pfam03918 57 187 16 12 95 158 Ga0063436_12063 Thiol-disulfide isomerase and thioredoxins OC pfam00578 20 157 21 8 44 76 Ga0063436_12062 hypothetical protein - - 18 76 10 0 22 66 Ga0063436_12061 Cytochrome C oxidase, cbb3-type, subunit III - 0 66 6 0 8 30 Ga0063436_11641 Di- and tricarboxylate transporters P 74 256 64 72 170 262 Ga0063436_11642 Predicted hydrolase of the metallo-beta-lactamase superfamily R pfam13442 pfam02080 pfam03600 pfam00753 pfam07521 416 929 189 71 735 1021 Ga0063436_11607 Ga0063436_11608 Ga0063436_11609 Ga0063436_116010 Ga0063436_116019 Ga0063436_116020 MR E - Ga0063436_11643 hypothetical protein - - Ga0063436_11651 CBS domain - Ga0063436_11652 EH Ga0063436_11653 acetolactate synthase, large subunit (EC 2.2.1.6) Thiamine pyrophosphate enzyme, C-terminal TPP binding domain pfam00571 pfam00205 pfam02775 pfam02776 64 220 46 4 212 369 8 100 31 4 100 82 196 347 105 62 495 548 Ga0063436_11654 acetolactate synthase, small subunit (EC 2.2.1.6) E Ga0063436_11655 ketol-acid reductoisomerase (EC 1.1.1.86) EH Ga0063436_11656 2-isopropylmalate synthase (EC 2.3.3.13) E pfam02775 pfam10369 pfam13710 pfam01450 pfam07991 pfam00682 pfam08502 47 101 9 0 96 112 67 145 36 29 150 182 528 1341 408 176 1916 1956 Ga0063436_11657 hypothetical protein - - 230 1094 285 91 1047 1491 74 375 106 36 363 Ga0063436_11658 Aconitase family (aconitate hydratase) - 441 pfam00330 47 256 58 15 337 389 Ga0063436_11659 3-isopropylmalate dehydratase, small subunit (EC 4.2.1.33) E 51 42 55 19 176 184 E pfam00694 pfam00682 pfam08502 Ga0063436_116510 2-isopropylmalate synthase (EC 2.3.3.13) Ga0063436_116511 D-alanine-D-alanine ligase and related ATP-grasp enzymes 122 310 117 66 404 546 M pfam07478 102 215 84 29 266 384 Ga0063436_116512 Ga0063436_116513 Uncharacterized conserved protein S pfam04073 42 241 44 8 172 377 Isocitrate/isopropylmalate dehydrogenase - pfam00180 26 62 31 0 60 115 Ga0063436_13612 Isocitrate/isopropylmalate dehydrogenase - pfam00180 70 145 24 29 236 331 Ga0063436_13613 Prephenate dehydratase E pfam00800 22 93 22 51 101 191 Ga0063436_11661 hypothetical protein - - 47 110 36 6 154 202 Ga0063436_11662 Predicted transcriptional regulator K pfam13280 Ga0063436_11663 hypothetical protein - - Ga0063436_11664 Peroxiredoxin O Ga0063436_11681 hypothetical protein - Ga0063436_11682 ATP-dependent Lon protease, bacterial type O Ga0063436_11683 Lon protease (S16) C-terminal proteolytic domain - Ga0063436_11684 transcription-repair coupling factor (mfd) Ga0063436_11685 Ga0063436_11686 - 75 205 86 18 353 502 260 534 188 31 705 1321 pfam00578 83 240 76 34 249 275 pfam00004 pfam02190 8 76 10 0 48 99 388 1030 295 126 987 1296 71 326 107 37 322 618 LK pfam05362 pfam00270 pfam00271 pfam02559 pfam03461 188 485 94 56 320 628 aminoacyl-tRNA hydrolase J pfam01195 14 77 12 38 60 117 hypothetical protein - - 40 105 15 0 23 47 Ga0063436_11691 PAP2 superfamily - pfam01569 22 40 12 0 92 133 Ga0063436_11692 Chromate transport protein ChrA P pfam02417 18 138 29 0 56 109 Ga0063436_11693 Chromate transport protein ChrA P pfam02417 14 16 11 0 25 91 Ga0063436_11701 parallel beta-helix repeat-containing protein - pfam13229 70 102 52 4 165 253 Ga0063436_11702 Aldo/keto reductase family - 34 192 58 18 224 339 Ga0063436_11703 amidohydrolase R pfam00248 pfam01546 pfam07687 151 576 184 62 509 756 Ga0063436_11704 Glycine/D-amino acid oxidases (deaminating) E 33 158 33 22 231 258 Ga0063436_11705 Tfp pilus assembly protein PilF NU pfam01266 pfam07719 pfam13231 pfam13414 79 338 57 18 320 404 Ga0063436_11706 Glycosyl transferase family 2 - pfam00535 6 28 10 0 23 39 Ga0063436_11711 hypothetical protein - - 12 12 3 2 10 4 Ga0063436_11712 hypothetical protein - - 0 0 0 0 0 8 Ga0063436_11713 hypothetical protein - - 34 184 33 52 35 20 Ga0063436_11721 Exonuclease III L 46 68 38 14 89 269 Ga0063436_14451 Glycosyltransferase M pfam03372 pfam00534 pfam13579 11 5 7 13 56 77 Ga0063436_14452 hypothetical protein - - 5 34 6 0 39 39 Ga0063436_11731 Glycosyl transferases group 1 - pfam00534 12 19 21 4 30 37 Ga0063436_11732 hypothetical protein - - 20 111 44 0 195 391 Ga0063436_11741 E pfam00496 3855 5419 2799 1226 13009 12430 EP 223 101 24 472 398 46 95 32 22 171 178 Ga0063436_11744 polyphosphate kinase 1 P pfam00528 pfam00528 pfam12911 pfam02503 pfam13089 pfam13090 140 Ga0063436_11743 ABC-type dipeptide transport system, periplasmic component ABC-type dipeptide/oligopeptide/nickel transport systems, permease components ABC-type dipeptide/oligopeptide/nickel transport systems, permease components 158 294 189 100 568 761 Ga0063436_11745 Helix-turn-helix domain - pfam13518 2 2 0 0 13 54 Ga0063436_11742 EP Ga0063436_11761 hypothetical protein - - 6 53 28 0 107 79 Ga0063436_11762 Lipid A core - O-antigen ligase and related enzymes M 16 40 20 0 69 121 Ga0063436_11771 site-2 protease. Metallo peptidase. MEROPS family M50B M pfam13425 pfam02163 pfam13180 196 508 121 48 699 701 Ga0063436_11772 HEAT repeats - pfam13646 119 232 66 34 184 335 Ga0063436_11773 FecR family protein - pfam04773 73 60 44 10 132 194 Ga0063436_10031 hypothetical protein - - 9 4 14 0 26 28 Ga0063436_10032 Dolichyl-phosphate-mannose-protein mannosyltransferase - pfam13231 48 184 34 14 145 199 Ga0063436_11781 Domain of unknown function (DUF1974) oligopeptide/dipeptide ABC transporter, ATP-binding protein, C-terminal domain - 32 120 46 16 238 483 6 52 12 6 81 142 EP pfam09317 pfam00005 pfam08352 pfam00005 pfam00528 pfam08352 pfam12911 14 38 20 6 106 128 EP pfam00528 20 78 20 0 104 203 Ga0063436_11782 EP Ga0063436_11784 oligopeptide/dipeptide ABC transporter, ATP-binding protein, C-terminal domain ABC-type dipeptide/oligopeptide/nickel transport systems, permease components Ga0063436_11785 Bacterial extracellular solute-binding proteins, family 5 Middle - pfam00496 106 216 101 8 467 636 Ga0063436_11791 BadF/BadG/BcrA/BcrD ATPase family - pfam01869 20 81 21 20 138 162 Ga0063436_11792 Nucleoside-diphosphate-sugar epimerases MG pfam01370 60 103 22 12 156 194 Ga0063436_11801 Amidase - pfam01425 8 11 6 1 56 31 Ga0063436_11802 Amidase - pfam01425 35 75 17 2 49 67 Ga0063436_11803 C pfam03009 106 134 62 42 167 222 Ga0063436_11804 Glycerophosphoryl diester phosphodiesterase Phenylpropionate dioxygenase and related ring-hydroxylating dioxygenases, large terminal subunit PR 76 170 58 8 173 277 Ga0063436_11805 3-hydroxyacyl-CoA dehydrogenase I pfam00355 pfam00725 pfam02737 pfam13279 94 234 88 72 320 591 Ga0063436_11811 Putative transcriptional regulators (Ypuh-like) - pfam04079 116 156 69 34 314 522 Ga0063436_11812 mttA/Hcf106 family - pfam02416 121 389 104 41 627 670 Ga0063436_11813 Glycosyl transferases group 1 - pfam13524 56 146 70 40 299 410 Ga0063436_11814 Predicted dehydrogenases and related proteins R pfam01408 52 140 30 40 326 403 Ga0063436_11815 Uncharacterized protein conserved in bacteria S pfam06283 56 131 59 4 204 267 Ga0063436_11816 Cytidylyltransferase family - pfam01148 12 4 34 8 64 132 Ga0063436_11817 Major Facilitator Superfamily - pfam07690 32 62 34 8 117 191 Ga0063436_11818 Sugar kinases, ribokinase family G pfam00294 14 125 7 0 67 124 Ga0063436_11819 pfkB family carbohydrate kinase - pfam00294 0 31 17 0 105 126 Ga0063436_11821 Acyl-coenzyme A synthetases/AMP-(fatty) acid ligases I 71 221 26 30 150 160 Ga0063436_11831 Uncharacterized conserved protein S 82 84 44 10 174 233 Ga0063436_11832 DNA polymerase I (EC 2.7.7.7) L pfam00501 pfam07171 pfam07364 pfam00476 pfam01367 pfam01612 pfam02739 58 407 82 54 440 746 Ga0063436_11833 hypothetical protein - 20 195 44 20 219 397 Ga0063436_11851 ATP-dependent metalloprotease FtsH O pfam00004 pfam01434 1926 4107 739 324 2839 3591 Ga0063436_11852 protein translocase, SecG subunit U pfam03840 82 251 43 33 260 208 Ga0063436_11853 ABC-type dipeptide transport system, periplasmic component E pfam00496 102 362 130 14 457 444 Ga0063436_11854 Transglycosylase SLT domain - pfam01464 75 16 41 38 143 177 Ga0063436_11861 hypothetical protein - - 0 26 2 6 18 15 Ga0063436_11862 - 161 60 22 170 264 32 249 70 14 346 393 Ga0063436_11864 putative NAD(P)H quinone oxidoreductase, PIG3 family CR pfam01555 pfam03446 pfam14833 pfam00107 pfam08240 40 Ga0063436_11863 DNA methylase 3-hydroxyisobutyrate dehydrogenase and related betahydroxyacid dehydrogenases 41 102 38 6 126 255 Ga0063436_11865 Dolichyl-phosphate-mannose-protein mannosyltransferase S pfam13231 60 58 32 46 172 221 Ga0063436_11866 Secreted and surface protein containing fasciclin-like repeats M pfam02469 171 269 131 38 524 588 Ga0063436_11867 Putative lysophospholipase - pfam12146 4 24 9 6 41 25 Ga0063436_13851 hypothetical protein - - 13 0 10 0 6 7 Ga0063436_13852 propionyl-CoA carboxylase carboxyltransferase subunit I pfam01039 214 661 220 57 691 1081 Ga0063436_13853 GDSL-like Lipase/Acylhydrolase family - 86 112 52 4 244 290 Ga0063436_13854 Acetyl/propionyl-CoA carboxylase, alpha subunit I pfam13472 pfam00289 pfam02785 pfam02786 88 230 68 48 277 420 Ga0063436_13855 Biotin-requiring enzyme - pfam00364 50 50 39 6 132 262 Ga0063436_11783 I Ga0063436_13856 Predicted Zn-dependent protease R 90 416 84 46 261 553 M pfam04298 pfam03572 pfam13180 Ga0063436_13857 C-terminal peptidase (prc) Ga0063436_13858 Periplasmic protease 144 583 117 60 240 434 M pfam03572 245 753 130 43 424 Ga0063436_13859 699 hypothetical protein - 8 26 6 0 32 48 Ga0063436_138510 DNA mismatch repair protein MutL L pfam01119 pfam08676 pfam13589 28 356 42 28 354 449 Ga0063436_138511 Alcohol dehydrogenase GroES-like domain - pfam08240 8 147 10 0 106 142 Ga0063436_13871 hypothetical protein - - 6 29 4 0 15 50 Ga0063436_13872 ADP-ribose pyrophosphatase Metal-dependent hydrolases of the beta-lactamase superfamily III F pfam00293 26 94 33 16 163 256 R pfam12706 40 147 58 11 189 292 E 148 46 24 129 245 - pfam02073 pfam01411 pfam07973 82 Ga0063436_13875 Leucyl aminopeptidase (aminopeptidase T) Threonyl and Alanyl tRNA synthetase second additional domain/tRNA synthetases class II (A) 40 189 25 6 147 205 Ga0063436_13876 LysM domain - pfam01476 122 903 96 16 239 365 Ga0063436_13877 hypothetical protein - - 70 489 53 21 209 221 Ga0063436_13878 methylmalonyl-CoA epimerase (EC 5.1.99.1) E pfam13669 12 81 31 33 107 184 Ga0063436_13879 methylmalonyl-CoA mutase (EC 5.4.99.2) I pfam01642 55 233 49 26 261 367 Ga0063436_138710 hypothetical protein - - 245 556 101 98 511 638 Ga0063436_138711 hypothetical protein - - 58 263 33 30 196 289 Ga0063436_13361 hypothetical protein - 2 54 11 0 34 43 Ga0063436_13362 K+ transport systems, NAD-binding component P 32 79 38 8 163 344 Ga0063436_13363 K+ transport systems, NAD-binding component P 28 221 22 1 128 301 Ga0063436_13364 ATPase, P-type (transporting), HAD superfamily, subfamily IC P pfam02080 pfam02254 pfam02080 pfam02254 pfam00122 pfam00689 pfam00690 pfam00702 Ga0063436_11881 hypothetical protein - Ga0063436_11882 peptidase T Ga0063436_11883 Gamma-glutamyltransferase Ga0063436_11884 Ga0063436_13873 Ga0063436_13874 103 611 125 55 655 914 15 8 12 0 38 69 E pfam01546 pfam07687 75 316 128 20 264 283 E pfam01019 42 201 51 12 171 281 hypothetical protein - 16 10 18 0 55 86 Ga0063436_11891 iron-sulfur cluster-binding protein C pfam02589 pfam13183 82 631 51 40 174 353 Ga0063436_11892 Cysteine-rich domain - pfam02754 40 382 25 20 82 177 Ga0063436_11893 Highly conserved protein containing a thioredoxin domain O pfam03190 90 311 72 10 342 522 Ga0063436_11894 Cystathionine beta-lyases/cystathionine gamma-synthases E pfam01053 46 225 50 14 281 304 Ga0063436_11901 phosphoserine aminotransferase apoenzyme (EC 2.6.1.52) HE pfam00266 16 147 34 31 200 274 Ga0063436_11902 Uncharacterized proteins, LmbE homologs S 82 388 109 6 417 599 Ga0063436_12021 Predicted transmembrane sensor domain T 562 1133 595 227 1925 2787 Ga0063436_12022 phenylalanyl-tRNA synthetase, alpha subunit (EC 6.1.1.20) J 47 64 36 40 160 234 Ga0063436_12023 phenylalanyl-tRNA synthetase beta subunit (EC 6.1.1.20) J pfam02585 pfam00211 pfam05226 pfam01409 pfam02912 pfam01588 pfam03147 pfam03483 pfam03484 97 520 66 38 498 630 Ga0063436_12024 Selenocysteine lyase E pfam00266 119 521 95 54 283 407 Ga0063436_12025 Mg-chelatase subunit ChlD H pfam13519 39 84 16 0 98 110 Ga0063436_12026 Mg-chelatase subunit ChlI H pfam01078 186 507 123 72 410 420 Ga0063436_12027 S1 RNA binding domain - 539 1861 539 262 2794 2743 Ga0063436_12028 ATPases with chaperone activity, ATP-binding subunit O 2119 5929 1195 598 4651 6690 Ga0063436_12029 DNA repair protein RadA O pfam00575 pfam00004 pfam02861 pfam07724 pfam10431 pfam13481 pfam13541 39 388 93 27 464 735 Ga0063436_120210 Helix-turn-helix domain - pfam13413 21 287 50 29 191 395 Ga0063436_120211 Aspartate/tyrosine/aromatic aminotransferase E 48 303 33 0 225 410 Ga0063436_120212 acetyl-CoA acetyltransferases I 34 182 40 10 155 326 Ga0063436_120213 thioredoxin O pfam00155 pfam00108 pfam02803 pfam00085 pfam14561 212 1104 245 88 796 1066 Ga0063436_120214 Uncharacterised ACR (DUF711) - 26 190 41 8 127 244 Ga0063436_120215 radical SAM enzyme/protein acetyltransferase, ELP3 family KB 114 298 66 26 383 604 pfam05167 pfam00583 pfam04055 Ga0063436_120216 Methyltransferase domain - pfam13489 33 225 59 0 127 299 Ga0063436_11911 - 0 0 0 11 14 44 207 32 14 207 354 Ga0063436_11913 Membrane proteins related to metalloendopeptidases M pfam01546 pfam07687 pfam01476 pfam01551 0 Ga0063436_11912 hypothetical protein Acetylornithine deacetylase/Succinyl-diaminopimelate desuccinylase and related deacylases 104 239 60 10 465 773 Ga0063436_11931 AmmeMemoRadiSam system protein B - pfam01875 1 5 4 0 28 21 Ga0063436_11932 ROK family - pfam00480 31 108 23 36 140 226 Ga0063436_11951 L-rhamnose isomerase, Streptomyces subtype M 113 201 101 9 371 465 Ga0063436_11952 rhamnulose-1-phosphate aldolase/alcohol dehydrogenase S 84 290 108 20 546 686 Ga0063436_11953 Sugar (pentulose and hexulose) kinases G pfam00106 pfam00596 pfam00370 pfam02782 42 229 70 22 202 356 Ga0063436_11954 Polysaccharide deacetylase - pfam01522 14 64 21 8 38 94 Ga0063436_11955 hypothetical protein - - 31 84 55 30 63 162 Ga0063436_11956 SMP-30/Gluconolaconase/LRE-like region - pfam08450 13 43 27 16 75 85 Ga0063436_11961 hypothetical protein - - 1 0 13 4 32 31 Ga0063436_11962 hypothetical protein - - 27 66 29 6 129 144 Ga0063436_11963 hypothetical protein - - 16 0 4 6 61 44 Ga0063436_11964 Glycosyl transferases group 1 - 30 24 23 0 73 90 Ga0063436_11971 pyrimidine-nucleoside phosphorylase F pfam00534 pfam00591 pfam02885 69 168 83 36 307 325 Ga0063436_11972 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100) IRQ pfam00106 61 143 37 20 210 264 Ga0063436_11973 Uncharacterized protein conserved in bacteria S pfam03780 33 145 27 34 128 211 Ga0063436_11974 NusB antitermination factor K pfam01029 58 130 33 0 191 211 Ga0063436_11975 hypothetical protein - - 33 130 19 16 143 129 Ga0063436_11976 Twin arginine targeting (Tat) protein translocase TatC U pfam00902 27 108 14 38 149 144 Ga0063436_11977 Zn-dependent protease with chaperone function O pfam01435 68 148 62 12 243 329 Ga0063436_11978 Short-chain dehydrogenases of various substrate specificities R pfam00106 42 52 27 28 148 130 Ga0063436_11979 FAD/FMN-containing dehydrogenases C pfam01565 34 89 30 16 214 194 Ga0063436_119710 Dienelactone hydrolase and related enzymes Q pfam01738 100 241 46 45 154 282 Ga0063436_119711 transcriptional regulator, HxlR family K 54 46 25 14 45 85 Ga0063436_119712 - 124 208 49 10 85 155 Ga0063436_119713 MacB-like periplasmic core domain/FtsX-like permease family ABC-type antimicrobial peptide transport system, ATPase component pfam01638 pfam02687 pfam12704 88 261 46 45 151 255 Ga0063436_119714 WD40-like Beta Propeller Repeat/Bacterial Ig-like domain - pfam00005 pfam07676 pfam13205 94 272 103 38 375 412 Ga0063436_119715 TIGR03943 family protein S pfam09323 91 198 89 12 332 367 Ga0063436_119716 Predicted permeases R pfam03773 41 145 43 20 146 204 Ga0063436_11981 Uncharacterized membrane protein, putative virulence factor R pfam03023 111 194 58 6 164 238 Ga0063436_11982 Peptidase_C39 like family - pfam13529 45 317 47 16 217 354 Ga0063436_11983 hypothetical protein - - 201 390 130 40 442 530 Ga0063436_11984 - pfam12847 4 45 8 0 60 74 Ga0063436_11985 Methyltransferase domain Sphingosine kinase and enzymes related to eukaryotic diacylglycerol kinase IR pfam00781 25 87 42 18 99 128 Ga0063436_11986 Nucleoside-diphosphate-sugar epimerases MG pfam01370 133 453 137 46 321 413 Ga0063436_11987 DNA-3-methyladenine glycosylase (3mg) L pfam02245 23 98 21 20 116 205 Ga0063436_11988 Enoyl-[acyl-carrier-protein] reductase [NADH] (EC 1.3.1.9) I pfam13561 39 134 59 33 166 262 Ga0063436_11991 - - 19 95 26 8 106 98 Ga0063436_11992 hypothetical protein Predicted oxidoreductases (related to aryl-alcohol dehydrogenases) C pfam00248 128 259 110 64 715 796 Ga0063436_11993 hypothetical protein - - 30 70 24 36 120 181 Ga0063436_11994 Predicted amidohydrolase R pfam00795 55 135 68 12 236 280 Ga0063436_11995 HhH-GPD superfamily base excision DNA repair protein L pfam00730 8 31 16 2 66 85 Ga0063436_11996 Uncharacterized conserved protein S pfam02698 41 80 27 0 110 212 Ga0063436_11997 hypothetical protein - - 3 6 5 0 70 88 Ga0063436_11998 hypothetical protein - - 48 57 28 0 192 440 Ga0063436_11999 conserved hypothetical protein S pfam03706 38 167 44 6 238 274 Ga0063436_119910 Protoporphyrinogen oxidase H pfam01593 28 111 32 12 154 231 Ga0063436_12001 alanyl-tRNA synthetase (EC 6.1.1.7) - pfam01411 72 282 93 28 220 314 Ga0063436_12002 tyrosyl-tRNA synthetase (EC 6.1.1.1) J pfam00579 24 220 84 28 279 350 E V pfam01479 pfam02770 pfam02771 pfam08028 pfam00994 pfam03453 pfam03454 pfam12727 pfam00994 pfam03453 pfam03454 Ga0063436_12003 Acyl-CoA dehydrogenases I 15 141 42 6 159 324 Ga0063436_12004 molybdopterin molybdochelatase (EC 2.10.1.1) H 60 215 100 4 307 488 Ga0063436_12005 molybdopterin molybdochelatase (EC 2.10.1.1) H Ga0063436_12006 - Ga0063436_12008 Molydopterin dinucleotide binding domain Molybdopterin oxidoreductase Fe4S4 domain/Molybdopterin oxidoreductase 2Fe-2S iron-sulfur cluster binding domain/4Fe-4S dicluster domain Ga0063436_12009 NAD-dependent formate dehydrogenase flavoprotein subunit C Ga0063436_120010 Uncharacterized conserved protein S pfam01568 pfam00384 pfam04879 pfam12838 pfam13510 pfam01257 pfam01512 pfam10531 pfam10589 pfam02588 pfam10035 46 197 30 36 193 227 50 40 26 0 54 89 212 515 133 58 488 679 37 173 111 59 226 266 192 505 176 59 585 784 Ga0063436_12011 hypothetical protein - - 26 63 56 10 105 182 27 47 32 10 73 142 Ga0063436_12012 NAD(P)-binding Rossmann-like domain - pfam13450 14 Ga0063436_12013 Predicted NAD/FAD-binding protein R 57 Ga0063436_12014 amidohydrolase R pfam13450 pfam01546 pfam07687 46 10 32 71 107 112 101 3 290 373 136 265 121 48 471 470 Ga0063436_12015 hypothetical protein - - Ga0063436_12016 hypothetical protein - - 2 29 15 0 58 68 37 100 36 14 98 186 Ga0063436_12017 CBS domain - pfam00571 32 157 34 8 199 174 Ga0063436_12031 Chlor_Arch_YYY domain S Ga0063436_12032 Uncharacterized membrane protein (DUF2298) - pfam10060 95 220 69 20 217 282 pfam10060 28 145 42 12 116 167 Ga0063436_12041 hypothetical protein Ga0063436_12042 integral membrane protein TIGR01906 - - 10 115 9 6 42 81 - 66 210 59 5 164 235 99 309 95 58 632 751 G pfam07314 pfam01676 pfam10143 pfam00005 pfam08402 Ga0063436_12043 G Ga0063436_12044 phosphoglycerate mutase (EC 5.4.2.1) carbohydrate ABC transporter ATP-binding protein, CUT1 family (TC 3.A.1.1.-) 671 1323 818 269 4611 5564 Ga0063436_12045 Ga0063436_12046 Phosphatidylglycerophosphate synthase I pfam01066 122 198 112 21 297 332 hypothetical protein - - 19 20 8 10 39 78 Ga0063436_12047 Prolipoprotein diacylglyceryl transferase - 64 107 29 12 120 131 Ga0063436_12048 PHP domain/Divergent AAA domain - pfam01790 pfam02811 pfam04326 260 Ga0063436_12049 hypothetical protein - Ga0063436_120410 Carbonic anhydrase P Ga0063436_120411 Tetratricopeptide repeat/TPR repeat Ga0063436_12051 Ga0063436_12007 - 489 254 51 661 985 30 78 23 8 150 122 221 755 306 151 969 1396 - pfam00194 pfam01476 pfam13414 pfam14559 80 179 61 10 293 397 Glycosyltransferases involved in cell wall biogenesis Glycerol uptake facilitator and related permeases (Major Intrinsic Protein Family) M pfam00535 34 178 12 24 160 321 G pfam00230 84 287 73 10 269 500 Protein-tyrosine-phosphatase Uncharacterized ABC-type transport system, periplasmic component/surface lipoprotein T pfam01451 22 80 30 28 116 160 R pfam02608 3050 5035 3075 1140 10877 10676 R pfam00005 333 518 303 176 1511 1527 Ga0063436_12073 nucleoside ABC transporter ATP-binding protein Branched-chain amino acid transport system / permease component - pfam02653 41 62 31 34 138 192 Ga0063436_12081 GDSL-like Lipase/Acylhydrolase family - pfam13472 26 202 42 8 197 226 Ga0063436_12082 hypothetical protein - - 123 446 117 120 593 838 Ga0063436_12083 hypothetical protein - - 37 88 16 26 158 176 Ga0063436_12084 metal dependent phosphohydrolase R - 26 153 35 13 167 274 Ga0063436_12085 FOG: LysM repeat M 64 137 54 25 160 312 Ga0063436_12086 Lactate dehydrogenase and related dehydrogenases CRH 15 146 65 0 189 398 Ga0063436_12087 Glycine cleavage system T protein (aminomethyltransferase) E pfam01476 pfam00389 pfam02826 pfam01266 pfam01571 pfam08669 30 365 149 8 584 718 Ga0063436_12088 Trimethylamine:corrinoid methyltransferase H pfam06253 239 406 244 53 1089 1326 Ga0063436_12089 FCD domain - pfam07729 37 110 23 0 148 189 Ga0063436_12091 hypothetical protein - - 49 142 73 2 203 151 Ga0063436_12092 Adenine/guanine phosphoribosyltransferases and related F pfam00156 34 103 32 2 118 195 Ga0063436_12054 Ga0063436_12055 Ga0063436_12071 Ga0063436_12072 PRPP-binding proteins Ga0063436_12093 H Ga0063436_12094 Methylase involved in ubiquinone/menaquinone biosynthesis aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B (EC 6.3.5.-) pfam08241 pfam02637 pfam02934 20 135 24 2 146 226 Ga0063436_12095 Aspartate/tyrosine/aromatic aminotransferase E 33 235 75 45 293 495 70 228 73 16 175 316 E pfam00155 pfam01262 pfam05222 Ga0063436_12096 L-alanine dehydrogenase (EC 1.4.1.1) Ga0063436_12097 Ga0063436_12103 hypothetical protein Nucleoside-diphosphate-sugar pyrophosphorylase involved in lipopolysaccharide biosynthesis/translation initiation factor 2B, gamma/epsilon subunits (eIF-2Bgamma/eIF-2Bepsilon) aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit A (EC 6.3.5.-) ABC-type Fe3+-hydroxamate transport system, periplasmic component ABC-type cobalamin/Fe3+-siderophores transport systems, ATPase components ABC-type Fe3+-siderophore transport system, permease component 160 356 96 28 418 561 - - 6 22 24 16 85 96 MJ pfam00132 pfam00483 142 268 109 50 391 405 J pfam01425 19 216 61 0 205 437 P pfam01497 133 164 19 94 74 157 PH pfam00005 39 221 19 21 54 84 Ga0063436_12111 Helix-hairpin-helix motif P pfam01032 97 384 5 100 123 186 - pfam12836 21 91 8 10 97 187 Ga0063436_12112 Ga0063436_12113 Helix-hairpin-helix motif L pfam12836 105 385 123 41 355 541 Peptidase_C39 like family - pfam13529 55 129 52 0 140 213 Ga0063436_12114 hypothetical protein - - 90 1056 150 46 302 387 Ga0063436_12115 cysteine desulfurase family protein, VC1184 subfamily E 71 157 111 76 357 540 Ga0063436_12117 succinate dehydrogenase subunit B (EC 1.3.5.1) C 236 577 132 91 415 622 Ga0063436_12118 succinate dehydrogenase subunit A (EC 1.3.5.1) C pfam00266 pfam13085 pfam13183 pfam00890 pfam02910 349 1181 176 95 660 1154 Ga0063436_12119 Succinate dehydrogenase, hydrophobic anchor subunit C pfam01127 218 869 118 56 517 759 Ga0063436_121110 isocitrate dehydrogenase (NADP) (EC 1.1.1.42) C pfam00180 592 1191 189 72 817 1165 Ga0063436_121111 Protein-disulfide isomerase O pfam13462 226 537 96 84 376 446 Ga0063436_121112 Acetyltransferases R pfam00583 64 79 19 19 85 208 Ga0063436_121113 citrate synthase (EC 2.3.3.1) C pfam00285 161 457 146 43 607 774 Ga0063436_121116 Uncharacterized protein conserved in bacteria R pfam02631 20 89 33 21 193 254 Ga0063436_14702 protein RecA L pfam00154 244 840 288 145 982 1278 Ga0063436_121117 hypothetical protein - 34 42 8 6 60 86 Ga0063436_121118 ribonuclease Y R pfam00013 pfam01966 pfam12072 193 390 155 65 631 672 Ga0063436_12121 Rhomboid family - pfam01694 4 155 13 8 112 188 Ga0063436_12122 guanylate kinase (EC 2.7.4.8) F pfam00625 44 81 51 26 251 222 Ga0063436_12123 putative regulatory protein, FmdB family - pfam09723 56 78 64 28 127 187 Ga0063436_12124 thymidylate kinase (EC 2.7.4.9) F pfam02223 43 257 24 2 142 262 Ga0063436_12125 Uncharacterized protein conserved in bacteria S pfam01424 132 571 102 74 308 521 Ga0063436_12126 hypothetical protein - - 91 328 47 58 221 289 Ga0063436_12127 pyridoxal phosphate synthase yaaE subunit H pfam01174 66 452 56 35 217 417 Ga0063436_12128 pyridoxal phosphate synthase yaaD subunit H 611 1820 307 158 1116 1636 Ga0063436_12129 glutamate dehydrogenase (NAD/NADP) (EC 1.4.1.3) E pfam01680 pfam00208 pfam02812 132 397 174 28 569 754 Ga0063436_121210 Cytochrome c553 C 859 2409 318 231 1269 1545 Ga0063436_121211 Cytochrome b subunit of the bc complex C pfam00034 pfam00032 pfam13631 1935 Ga0063436_121212 hypothetical protein - Ga0063436_12131 translation elongation factor 2 (EF-2/EF-G) Ga0063436_12132 Ga0063436_12133 Ga0063436_12098 Ga0063436_12099 Ga0063436_12101 Ga0063436_12102 J 4620 694 397 2724 3066 109 170 45 10 190 124 J pfam00009 pfam03144 pfam03764 pfam14492 1197 3090 966 448 4216 5256 ribosomal protein S7, bacterial/organelle J pfam00177 357 744 155 63 942 1141 SSU ribosomal protein S12P J 408 887 234 99 1350 1658 Ga0063436_12134 DNA-directed RNA polymerase, beta subunit/140 kD subunit K pfam00164 pfam00562 pfam04560 pfam04561 pfam04563 pfam04565 pfam10385 3007 6120 1728 773 6988 8594 Ga0063436_12135 Predicted nucleoside-diphosphate-sugar epimerases MG 30 132 24 12 77 166 Ga0063436_12136 3-oxoacyl-[acyl-carrier-protein] synthase II (EC 2.3.1.41) IQ pfam13460 pfam00109 pfam02801 49 411 86 96 321 450 Ga0063436_12137 3-oxoacyl-(acyl-carrier-protein) synthase III I pfam08541 63 193 58 40 265 337 pfam08545 Ga0063436_12138 RNA polymerase sigma factor, sigma-70 family - pfam04539 pfam04545 225 391 185 48 476 813 Ga0063436_12139 Sigma-70 region 2 - pfam04542 138 217 69 11 249 369 Ga0063436_12141 Uncharacterized protein conserved in bacteria S pfam06108 14 35 13 4 15 69 Ga0063436_12142 luciferase family oxidoreductase, group 1 C pfam00296 40 145 29 16 127 229 Ga0063436_12143 hypothetical protein - - 64 70 29 0 72 82 Ga0063436_12151 hypothetical protein - - 0 28 1 0 11 51 Ga0063436_12152 Peptidase family M23 - pfam01551 16 157 37 6 96 217 Ga0063436_12153 Predicted phosphohydrolases R pfam00149 50 241 25 28 128 209 Ga0063436_12154 cystathionine gamma-synthase (EC 2.5.1.48) E pfam01053 76 239 72 6 241 282 Ga0063436_12155 Integrase core domain ABC-type multidrug transport system, ATPase and permease components - pfam00665 pfam00005 pfam00664 55 198 45 0 137 187 42 213 53 22 159 174 hypothetical protein ABC-type multidrug transport system, ATPase and permease components - pfam00005 pfam00664 14 0 10 0 41 28 30 86 38 6 104 126 hypothetical protein C-terminal, D2-small domain, of ClpB protein/AAA domain (Cdc48 subfamily) - pfam07724 pfam10431 143 375 162 32 210 474 337 897 246 116 1149 1885 D pfam01098 pfam03033 pfam04101 161 176 104 34 588 780 Ga0063436_12182 Bacterial cell division membrane protein UDP-N-acetylglucosamine:LPS N-acetylglucosamine transferase 84 322 105 36 580 947 Ga0063436_12183 Colicin V production protein - 30 83 25 8 160 232 Ga0063436_12184 UDP-N-acetylmuramate--L-alanine ligase (EC 6.3.2.8) M 114 273 102 10 464 725 Ga0063436_12185 UDP-N-acetylmuramate dehydrogenase (EC 1.1.1.158) M 64 337 52 32 341 539 Ga0063436_12186 D-alanine--D-alanine ligase (EC 6.3.2.4) M pfam02674 pfam01225 pfam02875 pfam08245 pfam01565 pfam02873 pfam01820 pfam07478 Ga0063436_12187 Cell division septal protein M Ga0063436_12188 cell division protein FtsA D Ga0063436_12189 cell division protein FtsZ D Ga0063436_12261 Holliday junction endonuclease RuvC (EC 3.1.22.4) L Ga0063436_12262 Holliday junction DNA helicase subunit RuvA L Ga0063436_12263 pyrroline-5-carboxylate reductase (EC 1.5.1.2) E Ga0063436_12264 RNA polymerase sigma factor, sigma-70 family K Ga0063436_12265 proton translocating ATP synthase, F1 alpha subunit C Ga0063436_12266 ATP synthase, F1 gamma subunit C Ga0063436_12267 ATP synthase F1 subcomplex beta subunit Ga0063436_12268 ATP synthase, F1 epsilon subunit (delta in mitochondria) Ga0063436_12269 Ga0063436_12161 Ga0063436_12162 Ga0063436_12163 Ga0063436_12164 Ga0063436_12171 Ga0063436_12181 V V - M 80 310 72 35 494 803 pfam08478 pfam02491 pfam14450 pfam00091 pfam12327 158 355 163 74 941 1355 422 844 321 137 1647 1999 887 2274 810 426 4554 4771 pfam02075 pfam01330 pfam07499 pfam14520 pfam03807 pfam14748 pfam04539 pfam04542 pfam04545 pfam00006 pfam00306 pfam02874 159 443 81 25 399 689 74 236 60 26 286 313 43 197 67 10 305 505 1211 6251 149 160 1950 2370 1119 2247 587 246 2854 3332 213 720 208 100 832 1074 C pfam00231 pfam00006 pfam00306 pfam02874 713 2219 547 349 2334 2671 C pfam02823 238 542 152 91 497 660 rod shape-determining protein MreB D 70 592 126 78 511 653 Ga0063436_122610 AT-rich DNA-binding protein R 73 195 79 7 230 393 Ga0063436_122611 Obg family GTPase CgtA R pfam06723 pfam02629 pfam06971 pfam01018 pfam01926 pfam09269 119 277 143 26 432 627 Ga0063436_122613 dTDP-4-dehydrorhamnose reductase M 50 51 32 46 114 183 Ga0063436_122614 ribonuclease III, bacterial K 46 78 33 55 191 251 Ga0063436_122615 3-oxoacyl-[acyl-carrier-protein] synthase II (EC 2.3.1.41) IQ 131 194 76 46 371 540 Ga0063436_122616 FOG: LysM repeat M 41 66 59 23 226 357 Ga0063436_122617 Dinucleotide-utilizing enzymes involved in molybdopterin and thiamine biosynthesis family 2 H pfam04321 pfam00035 pfam14622 pfam00109 pfam02801 pfam01476 pfam03734 pfam00581 pfam00899 pfam05237 437 1003 131 91 688 854 Ga0063436_122618 Rhodanese-related sulfurtransferase P pfam00581 440 714 174 39 669 716 Ga0063436_14441 Molybdopterin converting factor, small subunit H pfam02597 70 145 6 8 52 51 Ga0063436_12191 hypothetical protein Predicted metal-dependent protease of the PAD1/JAB1 superfamily - - 6 28 6 0 16 11 R pfam14464 65 95 27 0 76 62 Ga0063436_12192 Ga0063436_12193 cysteine synthase (EC 2.5.1.47) E pfam00291 194 403 47 0 216 Ga0063436_12194 Molybdopterin converting factor, small subunit H pfam02597 5 100 12 14 78 79 Ga0063436_12195 Predicted acetamidase/formamidase C pfam03069 236 682 428 91 1515 2396 Ga0063436_12196 hypothetical protein - - 4 52 25 9 71 93 Ga0063436_12201 - - 4 12 6 0 62 55 Ga0063436_12202 hypothetical protein Repeat domain in Vibrio, Colwellia, Bradyrhizobium and Shewanella - 351 480 255 102 1248 1762 Ga0063436_12203 Glycine cleavage system T protein (aminomethyltransferase) E pfam13517 pfam01571 pfam08669 77 155 83 28 474 495 Ga0063436_12204 NAD(P)-binding Rossmann-like domain - pfam13450 21 74 25 15 133 161 Ga0063436_12205 hypothetical protein - - 79 128 82 9 267 394 Ga0063436_12206 Phytoene dehydrogenase and related proteins Q pfam13450 41 196 57 50 377 498 Ga0063436_12207 Rieske [2Fe-2S] domain - pfam00355 20 192 34 0 90 160 Ga0063436_12221 - 7 18 13 1 13 43 139 315 124 29 340 479 EP pfam00005 pfam08352 pfam00528 pfam12911 117 194 85 46 242 333 Ga0063436_12224 hypothetical protein ATPase components of various ABC-type transport systems, contain duplicated ATPase ABC-type dipeptide/oligopeptide/nickel transport systems, permease components ABC-type dipeptide/oligopeptide/nickel transport systems, permease components EP pfam00528 497 703 254 143 803 1114 Ga0063436_12225 ABC-type dipeptide transport system, periplasmic component E pfam00496 6393 10163 3688 1766 14868 14884 Ga0063436_12226 putative efflux protein, MATE family V pfam01554 64 174 38 14 117 143 Ga0063436_12227 - 105 10 0 80 97 102 330 90 12 256 374 V pfam01966 pfam00005 pfam00664 pfam00005 pfam00664 11 Ga0063436_12229 HD domain ABC-type multidrug transport system, ATPase and permease components ABC-type multidrug transport system, ATPase and permease components 63 316 78 56 212 280 Ga0063436_122210 Sarcosine oxidase, gamma subunit family - pfam04268 26 29 26 17 73 90 Ga0063436_12231 Iron-sulfur cluster assembly accessory protein S pfam01521 974 1074 500 273 1710 2169 Ga0063436_12232 Predicted transcriptional regulator K pfam01022 224 385 121 59 373 540 Ga0063436_12233 FeS assembly protein IscX S pfam04384 108 185 54 46 352 314 Ga0063436_12234 NADH-quinone oxidoreductase, B subunit C 190 514 127 44 638 855 Ga0063436_12235 C 494 1015 260 194 1316 1678 Ga0063436_12236 NADH:ubiquinone oxidoreductase 49 kD subunit 7 Formate hydrogenlyase subunit 6/NADH:ubiquinone oxidoreductase 23 kD subunit (chain I) pfam01058 pfam00329 pfam00346 191 278 112 75 526 669 Ga0063436_12237 ATPases involved in chromosome partitioning D pfam12838 pfam10609 pfam13614 187 438 177 83 715 784 Ga0063436_12238 Uncharacterized homolog of PSP1 S pfam04468 330 463 213 88 767 883 Ga0063436_12251 Selenocysteine lyase E pfam00266 86 237 58 46 284 352 Ga0063436_12252 R pfam04909 6 64 21 6 105 138 Ga0063436_12253 Predicted metal-dependent hydrolase of the TIM-barrel fold Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) IQR pfam13561 23 46 26 20 101 112 Ga0063436_12254 hypothetical protein - 91 119 34 33 92 259 Ga0063436_12255 Uncharacterized conserved protein S pfam07171 pfam07364 79 14 53 2 106 228 Ga0063436_12271 - pfam13407 2356 2251 5443 4152 4100 2159 Ga0063436_12272 Periplasmic binding protein domain Uncharacterized protein containing SIS (Sugar ISomerase) phosphosugar binding domain R pfam13580 61 26 59 52 46 36 Ga0063436_12281 Protein of unknown function (DUF3090) - pfam11290 119 331 34 13 68 175 Ga0063436_12282 Fructose-2,6-bisphosphatase G pfam00300 58 660 28 63 154 219 Ga0063436_12312 Cupredoxin-like domain - pfam13473 540 751 67 73 189 245 Ga0063436_12311 putative C-S lyase E 70 167 59 28 197 322 Ga0063436_12291 Predicted dehydrogenases and related proteins R 140 148 186 58 411 658 Ga0063436_12292 Sugar phosphate isomerases/epimerases G pfam00155 pfam01408 pfam02894 pfam01261 pfam07582 Ga0063436_12293 Trehalose utilization protein G Ga0063436_12294 Predicted dehydrogenases and related proteins Ga0063436_12295 Predicted dehydrogenases and related proteins Ga0063436_12296 Ga0063436_12297 Ga0063436_12222 Ga0063436_12223 Ga0063436_12228 R V C 275 189 344 158 48 528 823 112 463 128 58 515 651 R pfam06283 pfam01408 pfam02894 216 655 292 79 954 1226 R pfam01408 77 268 76 50 302 525 Sugar phosphate isomerases/epimerases G pfam01261 127 286 117 55 441 465 FAD dependent oxidoreductase - pfam01266 30 94 22 14 68 199 Ga0063436_12301 orotate phosphoribosyltransferase (EC 2.4.2.10) F 16 73 18 10 147 135 Ga0063436_12302 aspartate carbamoyltransferase (EC 2.1.3.2) F pfam00215 pfam00185 pfam02729 44 92 40 2 242 235 Ga0063436_12303 Dihydroorotase and related cyclic amidohydrolases F pfam13147 42 53 28 12 193 235 Ga0063436_12304 Helicase conserved C-terminal domain - pfam13625 Ga0063436_12305 hypothetical protein - - 16 52 11 10 74 103 522 1533 467 242 2550 3892 Ga0063436_12321 Restriction endonuclease V pfam01844 26 33 30 10 88 152 Ga0063436_12322 hypothetical protein Ga0063436_12323 hypothetical protein - - 18 14 12 0 63 134 - - 34 67 22 16 80 Ga0063436_12324 237 ribosomal subunit interface protein J pfam02482 28 45 40 12 104 203 Ga0063436_12325 Predicted amidophosphoribosyltransferases R - 46 120 28 9 187 225 Ga0063436_12326 Flp/Fap pilin component - pfam04964 59 143 81 89 263 253 Ga0063436_12327 UDP-N-acetylglucosamine 2-epimerase M pfam02350 60 161 41 6 144 295 Ga0063436_12328 hypothetical protein - - 38 18 4 0 25 29 Ga0063436_12329 23S rRNA m(2)A-2503 methyltransferase (EC 2.1.1.192) R 42 316 87 34 292 314 Ga0063436_123210 Predicted sugar phosphatases of the HAD superfamily G pfam04055 pfam13242 pfam13344 71 253 54 20 113 201 Ga0063436_123211 Protease subunit of ATP-dependent Clp proteases OU 573 1373 308 142 825 1000 Ga0063436_123212 trigger factor O 399 623 154 87 765 857 Ga0063436_123214 translation elongation factor P (EF-P) J pfam00574 pfam00254 pfam05697 pfam05698 pfam01132 pfam08207 pfam09285 66 203 108 58 273 381 Ga0063436_123215 hypothetical protein - 22 151 37 21 196 301 Ga0063436_123216 exonuclease, DNA polymerase III, epsilon subunit family KL pfam00270 pfam00929 pfam06733 pfam13307 Ga0063436_123217 E Ga0063436_123218 L-threonine aldolase (EC 4.1.2.5) ribosomal large subunit pseudouridine synthase D (EC 5.4.99.-) Ga0063436_123219 lipoprotein signal peptidase MU Ga0063436_123220 Isoleucyl-tRNA synthetase (EC 6.1.1.5) Ga0063436_123221 Ga0063436_15001 DNA modification methylase Threonyl and Alanyl tRNA synthetase second additional domain Branched-chain amino acid transport system / permease component Branched-chain amino acid transport system / permease component Ga0063436_12371 hypothetical protein Ga0063436_12372 102 451 165 87 439 715 pfam01212 pfam00849 pfam01479 88 224 81 70 218 288 83 267 76 11 186 266 62 308 56 18 120 190 J pfam01252 pfam00133 pfam08264 154 549 155 62 648 840 L pfam01555 14 58 29 16 161 198 - pfam07973 17 43 16 0 62 115 - pfam02653 0 0 2 0 0 0 - pfam02653 0 0 3 0 2 5 - - 8 7 0 0 0 0 hypothetical protein - - 2 15 1 4 0 0 Ga0063436_12373 hypothetical protein - - 52 38 9 0 0 0 Ga0063436_12374 hypothetical protein - - 17 22 5 0 0 0 Ga0063436_12375 hypothetical protein - - 79 82 4 14 0 0 Ga0063436_12376 hypothetical protein - - Ga0063436_12377 VWA-like domain (DUF2201) - pfam09967 Ga0063436_12381 hypothetical protein - Ga0063436_12382 Fasciclin domain - Ga0063436_12391 SIS domain Ga0063436_12392 Ga0063436_12401 Ga0063436_123222 Ga0063436_12361 J 8 1 0 0 0 0 23 144 6 18 0 0 - 8 0 0 0 0 0 pfam02469 0 0 0 2 0 0 - pfam01380 12 0 10 40 40 36 N-acetylglucosamine-6-phosphate deacetylase G pfam13147 20 38 22 0 14 16 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_12402 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_12403 hypothetical protein - - 0 0 0 0 8 2 Ga0063436_12404 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_12405 hypothetical protein - - 0 0 0 0 6 0 Ga0063436_12406 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_12408 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_12409 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_124010 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_124011 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_124012 Methyltransferase domain - pfam13578 0 0 0 0 0 0 Ga0063436_124013 NAD dependent epimerase/dehydratase family - pfam01370 0 0 0 0 10 0 Ga0063436_12411 hypothetical protein - - 0 2 7 0 5 19 Ga0063436_12412 Uncharacterized protein conserved in bacteria S pfam05163 16 77 22 0 51 73 Ga0063436_12413 Uncharacterized conserved protein S pfam08818 48 30 43 26 93 96 Ga0063436_12414 hypothetical protein - - 12 30 15 4 46 64 Ga0063436_12415 Methyltransferase domain - pfam13489 0 0 14 10 19 56 Ga0063436_12416 uncharacterized flavoprotein, PP_4765 family - pfam03486 2 49 26 16 64 58 Ga0063436_12421 Predicted nucleoside-diphosphate-sugar epimerases MG pfam05368 22 142 18 8 43 33 Ga0063436_12422 hypothetical protein - - 0 0 4 0 10 22 Ga0063436_12423 Uncharacterised ACR, YkgG family COG1556 - pfam02589 0 22 14 0 0 12 Ga0063436_12424 K pfam00440 2 33 24 0 49 71 Ga0063436_12425 transcriptional regulator, TetR family Permeases of the drug/metabolite transporter (DMT) superfamily GER pfam00892 32 71 54 42 227 262 Ga0063436_12426 hypothetical protein - - 1 52 24 0 101 73 Ga0063436_12427 hypothetical protein - - 101 184 80 45 398 418 Ga0063436_12428 - pfam12804 93 86 80 24 288 428 - 28 91 24 30 133 94 V pfam00763 pfam02687 pfam12704 906 3177 150 93 480 783 Ga0063436_12433 MobA-like NTP transferase domain Tetrahydrofolate dehydrogenase/cyclohydrolase, catalytic domain ABC-type antimicrobial peptide transport system, permease component ABC-type antimicrobial peptide transport system, ATPase component V pfam00005 733 1872 77 83 395 574 Ga0063436_13981 RND family efflux transporter, MFP subunit V pfam12700 1548 4788 202 192 805 1157 Ga0063436_12441 hypothetical protein - - 0 0 3 0 10 17 Ga0063436_12442 C 84 23 6 126 138 V pfam00881 pfam00005 pfam00664 14 Ga0063436_12443 Nitroreductase ABC-type multidrug transport system, ATPase and permease components 117 331 66 65 357 616 Ga0063436_12444 AmmeMemoRadiSam system protein B - pfam01875 33 125 16 10 66 82 Ga0063436_12451 Thiamin pyrophosphokinase, catalytic domain - pfam04263 0 26 0 12 0 0 Ga0063436_12453 cation diffusion facilitator family transporter P pfam01545 Ga0063436_12461 hypothetical protein - - Ga0063436_12462 Bacterial extracellular solute-binding protein - Ga0063436_12471 argininosuccinate synthase (EC 6.3.4.5) Ga0063436_12472 Ga0063436_12482 hypothetical protein Branched-chain amino acid transport system / permease component Branched-chain amino acid transport system / permease component Ga0063436_12491 ABC-type multidrug transport system, ATPase component Ga0063436_12492 Ga0063436_12431 Ga0063436_12432 0 0 0 2 0 0 64 55 35 24 184 342 pfam01547 386 577 465 131 2425 2522 E pfam00764 204 310 110 28 642 767 - - 5 27 16 0 68 77 - pfam02653 18 30 8 10 87 133 - pfam02653 64 19 51 6 190 229 V pfam00005 0 4 7 24 52 71 ABC-type multidrug transport system, ATPase component V pfam00005 14 27 9 0 76 79 Ga0063436_12493 HlyD family secretion protein - pfam13437 28 14 20 0 71 128 Ga0063436_12501 - - 569 868 588 162 2284 2437 EP 368 140 79 615 759 62 132 75 7 416 316 E pfam00528 pfam00528 pfam12911 pfam01546 pfam07687 235 Ga0063436_12504 hypothetical protein ABC-type dipeptide/oligopeptide/nickel transport systems, permease components ABC-type dipeptide/oligopeptide/nickel transport systems, permease components Acetylornithine deacetylase/Succinyl-diaminopimelate desuccinylase and related deacylases 87 356 142 60 582 652 Ga0063436_12505 Putative translation initiation inhibitor, yjgF family J pfam01042 145 252 85 23 414 581 Ga0063436_12506 Threonine dehydratase E 51 264 70 40 285 387 Ga0063436_12507 Predicted dehydrogenases and related proteins R 117 224 112 30 631 767 Ga0063436_12508 carbamoyl-phosphate synthase, small subunit EF 110 237 57 52 280 353 Ga0063436_12509 EF 127 369 126 46 461 636 Ga0063436_125010 carbamoyl-phosphate synthase large subunit Pyruvate dehydrogenase complex, dehydrogenase (E1) component C pfam00291 pfam01408 pfam02894 pfam00117 pfam00988 pfam00289 pfam02142 pfam02786 pfam02787 pfam00456 pfam02779 144 318 155 62 731 810 Ga0063436_12511 Uncharacterized protein conserved in bacteria S pfam06224 45 15 16 0 88 146 Ga0063436_12512 EamA-like transporter family - pfam00892 0 12 6 0 1 14 Ga0063436_12521 Glycosyltransferase M pfam00534 10 68 14 8 56 117 Ga0063436_12531 hypothetical protein - 26 162 41 20 115 154 Ga0063436_12532 Glycosyltransferase M pfam00534 pfam13579 8 115 26 14 124 116 Ga0063436_12533 hypothetical protein - - 10 35 7 0 28 93 Ga0063436_12541 Complex I intermediate-associated protein 30 (CIA30) - pfam08547 46 200 33 24 99 134 Ga0063436_12481 Ga0063436_12502 Ga0063436_12503 EP Ga0063436_12542 Dolichyl-phosphate-mannose-protein mannosyltransferase - pfam13231 16 116 16 2 73 138 Ga0063436_12543 phytoene desaturase Q pfam01593 34 277 43 9 183 232 Ga0063436_14202 Glycosyltransferases involved in cell wall biogenesis M pfam00535 6 97 13 0 97 172 Ga0063436_14201 Glycerol-3-phosphate acyltransferase - pfam02660 0 0 4 15 11 20 Ga0063436_13965 Pyridoxal-phosphate dependent enzyme - pfam00291 168 234 83 47 355 306 Ga0063436_13964 D-aminopeptidase E pfam04951 94 217 47 21 181 248 Ga0063436_13963 Cysteine synthase E 238 539 226 44 510 674 Ga0063436_13962 O 412 775 230 159 735 935 Ga0063436_12551 Molecular chaperone, HSP90 family 2-polyprenyl-6-methoxyphenol hydroxylase and related FADdependent oxidoreductases pfam00291 pfam00183 pfam13589 HC pfam01494 99 329 128 36 301 418 Ga0063436_12561 Predicted membrane protein S 10 50 19 26 63 95 Ga0063436_12562 Mg chelatase-related protein O pfam09945 pfam01078 pfam13335 pfam13541 54 167 34 14 178 329 Ga0063436_12571 hypothetical protein - - 12 8 10 0 20 38 Ga0063436_12572 V pfam13354 50 131 51 21 133 117 E pfam13458 688 888 569 190 2792 3006 Ga0063436_12582 Beta-lactamase class A ABC-type branched-chain amino acid transport systems, periplasmic component ABC-type branched-chain amino acid transport systems, periplasmic component E pfam13458 1007 1221 762 194 3599 4593 Ga0063436_12583 hypothetical protein - - 68 133 46 24 296 342 Ga0063436_12591 hypothetical protein - - 17 18 20 24 86 104 Ga0063436_12592 Glycosyl transferase family 2 - pfam00535 22 88 28 18 60 77 Ga0063436_12593 Cysteine-rich domain - pfam02754 36 147 18 14 133 255 Ga0063436_12594 hypothetical protein - - 6 6 12 4 45 78 Ga0063436_12595 hypothetical protein - - 4 20 4 0 10 18 Ga0063436_12601 Radical SAM superfamily - 96 130 46 24 233 357 Ga0063436_12611 adenosylhomocysteinase (EC 3.3.1.1) H pfam04055 pfam00670 pfam05221 890 1317 521 366 2199 1886 Ga0063436_14302 Sugar kinases, ribokinase family G pfam00294 80 106 68 83 247 325 Ga0063436_14303 hypothetical protein - 26 46 17 1 92 53 Ga0063436_12621 peptide chain release factor 3 J 60 93 43 16 258 304 Ga0063436_12622 J 67 22 41 26 86 123 Ga0063436_12623 TlyA family rRNA methyltransferase/putative hemolysin 4-hydroxybenzoate polyprenyltransferase and related prenyltransferases pfam00009 pfam03144 pfam01479 pfam01728 H pfam01040 24 65 40 0 90 59 Ga0063436_12624 hypothetical protein - - 10 10 0 0 13 24 Ga0063436_12631 Xaa-Pro aminopeptidase E pfam00557 132 222 107 22 317 530 Ga0063436_12632 O-sialoglycoprotein endopeptidase (EC 3.4.24.57) O pfam00814 78 180 63 16 311 458 Ga0063436_12641 hypothetical protein - - 0 25 15 0 24 29 Ga0063436_12642 Methyltransferase domain - pfam13659 24 80 38 11 100 159 Ga0063436_12643 Hydroxypyruvate isomerase G pfam01261 26 96 13 24 91 79 Ga0063436_12644 R pfam00106 14 90 28 6 70 120 Ga0063436_12645 Short-chain alcohol dehydrogenase of unknown specificity Predicted branched-chain amino acid permease (azaleucine resistance) E pfam03591 30 82 22 8 68 86 Ga0063436_12651 6,7-dimethyl-8-ribityllumazine synthase - pfam00885 45 23 12 16 73 77 Ga0063436_12652 G 156 45 18 187 162 44 329 41 26 204 262 Ga0063436_12654 aspartate kinase (EC 2.7.2.4) E 83 136 56 24 187 244 Ga0063436_12655 E 160 134 72 22 210 401 Ga0063436_12661 aspartate semialdehyde dehydrogenase (EC 1.2.1.11) Branched-chain amino acid transport system / permease component pfam13580 pfam06037 pfam08239 pfam00696 pfam13840 pfam01118 pfam02774 65 Ga0063436_12653 phosphoheptose isomerase (EC 5.3.1.-) Bacterial protein of unknown function (DUF922)/Bacterial SH3 domain - pfam02653 208 448 150 62 553 659 Ga0063436_12662 Periplasmic binding protein - pfam13458 3257 4399 2646 1151 10235 9493 Ga0063436_12663 Periplasmic binding protein Xanthine and CO dehydrogenases maturation factor, XdhC/CoxF family - 1939 2668 1408 547 5474 4592 107 209 98 31 373 469 C 1824 3050 1520 433 6551 8312 531 259 80 1002 1392 C pfam01799 pfam00941 pfam03450 312 Ga0063436_12674 carbon-monoxide dehydrogenase, large subunit Aerobic-type carbon monoxide dehydrogenase, small subunit CoxS/CutS homologs Aerobic-type carbon monoxide dehydrogenase, middle subunit CoxM/CutM homologs pfam13458 pfam02625 pfam13478 pfam01315 pfam02738 690 1021 517 144 2194 2793 Ga0063436_13994 drug resistance transporter, EmrB/QacA subfamily - pfam07690 88 327 84 33 438 573 Ga0063436_12581 Ga0063436_12671 Ga0063436_12672 Ga0063436_12673 - O C Ga0063436_14085 Methyl-accepting chemotaxis protein (MCP) signalling domain - pfam00015 336 511 105 75 613 Ga0063436_14084 uncharacterized peroxidase-related enzyme - - 16 65 8 0 72 87 Ga0063436_14083 Carboxymuconolactone decarboxylase family - pfam02627 46 123 46 25 175 323 Ga0063436_14082 Predicted oxidoreductase R pfam00248 132 431 107 91 581 734 Ga0063436_14081 MORN repeat - pfam02493 19 226 98 15 236 352 Ga0063436_12681 hypothetical protein - - 56 18 33 24 88 109 Ga0063436_12682 F pfam14681 78 237 81 14 360 425 Ga0063436_12683 uracil phosphoribosyltransferase (EC 2.4.2.9) UDP-N-acetylmuramyl pentapeptide phosphotransferase/UDP-N-acetylglucosamine-1-phosphate transferase M pfam00953 29 174 41 14 168 317 Ga0063436_12684 hypothetical protein - - 28 151 40 0 116 165 Ga0063436_12685 Triosephosphate isomerase - pfam00121 47 138 20 14 104 128 Ga0063436_12686 Triosephosphate isomerase - pfam00121 9 0 16 20 48 95 Ga0063436_12687 Phosphoglycerate kinase - 12 50 20 18 64 61 Ga0063436_12691 enolase (EC 4.2.1.11) G pfam00162 pfam00113 pfam03952 110 289 100 46 449 450 Ga0063436_12701 D-alanyl-D-alanine carboxypeptidase - pfam02557 0 0 2 11 0 0 Ga0063436_12702 hypothetical protein - - 0 15 0 3 0 0 Ga0063436_12711 - pfam01547 1680 3099 1128 623 6137 5862 E pfam00005 pfam00005 pfam12399 154 271 153 34 485 655 143 254 124 55 401 517 Ga0063436_12714 Bacterial extracellular solute-binding protein amino acid/amide ABC transporter ATP-binding protein 2, HAAT family (TC 3.A.1.4.-) amino acid/amide ABC transporter ATP-binding protein 1, HAAT family (TC 3.A.1.4.-) ABC-type branched-chain amino acid transport system, permease component 86 507 157 47 814 1081 Ga0063436_12721 homoserine dehydrogenase (EC 1.1.1.3) E 16 86 10 0 112 224 Ga0063436_12722 Site-specific recombinase XerD L pfam02653 pfam00742 pfam03447 pfam00589 pfam02899 40 80 30 6 142 252 Ga0063436_12723 transcriptional attenuator, LytR family - pfam03816 64 127 44 0 160 163 Ga0063436_12724 Domain of unknown function (DUF1083) - pfam06452 24 92 18 0 122 227 Ga0063436_12731 1,4-Dihydroxy-2-naphthoyl-CoA synthase (EC 4.1.3.36) H 67 209 44 20 180 170 Ga0063436_12732 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1carboxylic-acid synthase H pfam00378 pfam00205 pfam02775 pfam02776 93 155 21 26 183 168 42 89 9 8 135 174 98 520 74 21 369 432 Ga0063436_12712 Ga0063436_12713 Ga0063436_12733 E E 565 R Ga0063436_12734 Predicted flavin-nucleotide-binding protein isochorismate synthase (EC 5.4.4.2)/transcriptional regulator, TetR family HQ pfam12900 pfam00425 pfam00440 Ga0063436_12735 Uncharacterized protein conserved in bacteria S pfam12681 25 82 12 24 46 106 Ga0063436_12736 hypothetical protein - 25 0 6 0 44 57 Ga0063436_12741 Deoxyribodipyrimidine photolyase L pfam00875 pfam03441 29 187 75 40 377 443 Ga0063436_12742 hypothetical protein - - 22 152 51 29 150 207 Ga0063436_12743 Nucleoside-diphosphate-sugar epimerases MG pfam01370 74 133 85 34 226 210 Ga0063436_12751 S pfam01933 54 193 41 26 133 216 - 22 13 0 91 56 62 253 89 21 201 378 Ga0063436_12762 PAS domain S-box T pfam00044 pfam00072 pfam00158 pfam00512 pfam02518 pfam13426 10 Ga0063436_12761 conserved hypothetical protein, cofD-related Glyceraldehyde 3-phosphate dehydrogenase, NAD binding domain Sigma-54 interaction domain/Response regulator receiver domain 94 237 70 29 470 394 Ga0063436_12771 methyltransferase, FkbM family - pfam05050 47 2 8 0 76 73 Ga0063436_12772 hypothetical protein - - 18 6 31 10 74 104 Ga0063436_14211 1,4-dihydroxy-2-naphthoate prenyltransferase (EC 2.5.1.74) H 31 70 32 32 145 108 Ga0063436_14212 AMP-binding enzyme C-terminal domain/AMP-binding enzyme - pfam01040 pfam00501 pfam13193 44 132 51 40 124 193 Ga0063436_14213 AMP-binding enzyme - 34 102 9 12 41 85 Ga0063436_12781 L-seryl-tRNA(Sec) selenium transferase (EC 2.9.1.1) E pfam00501 pfam03841 pfam12390 58 234 68 34 271 379 Ga0063436_12782 hypothetical protein - - 12 80 18 1 73 108 Ga0063436_12783 ABC transporter - pfam00005 18 28 20 22 49 184 Ga0063436_12791 Multi-copper polyphenol oxidoreductase laccase - pfam02578 37 62 25 4 78 87 Ga0063436_12792 pyridoxal phosphate enzyme, YggS family R pfam01168 65 114 53 6 189 231 Ga0063436_12793 Predicted integral membrane protein S pfam02325 46 22 19 2 75 125 Ga0063436_12794 Uncharacterized conserved protein S pfam02594 17 2 14 0 43 88 Ga0063436_12801 hypothetical protein - - 5 0 0 0 16 16 Ga0063436_12752 - Ga0063436_12802 ABC-type dipeptide transport system, periplasmic component E pfam00496 1355 2604 1125 664 5939 Ga0063436_14742 RmlD substrate binding domain - pfam04321 18 8 1 22 27 6257 30 Ga0063436_12811 Probable RNA and SrmB- binding site of polymerase A - pfam12627 48 92 25 44 94 132 Ga0063436_12812 Uncharacterized protein conserved in bacteria S pfam02583 74 116 37 46 229 157 Ga0063436_12813 Copper binding proteins, plastocyanin/azurin family - pfam00127 125 259 61 76 303 239 Ga0063436_12814 Copper chaperone P 135 242 126 31 550 431 Ga0063436_12815 copper-(or silver)-translocating P-type ATPase P pfam00403 pfam00122 pfam00403 pfam00702 468 1332 246 190 1825 1463 Ga0063436_12816 L,D-transpeptidase catalytic domain - pfam03734 155 452 88 74 783 823 Ga0063436_12831 Aminotransferase class-III - pfam00202 82 24 28 8 90 174 Ga0063436_12832 K 14 4 0 41 83 - pfam13411 pfam08487 pfam13768 18 Ga0063436_12833 Predicted transcriptional regulators Vault protein inter-alpha-trypsin domain/von Willebrand factor type A domain 69 78 27 34 172 201 Ga0063436_12834 hypothetical protein - - 50 68 39 0 163 148 Ga0063436_12835 hypothetical protein - - 4 33 0 0 11 20 Ga0063436_12836 EamA-like transporter family - pfam00892 10 4 0 0 17 16 Ga0063436_12841 dihydropyrimidinase (EC 3.5.2.2) F pfam13147 44 151 22 0 212 275 Ga0063436_12851 Aldo/keto reductase family - pfam00248 75 34 38 35 107 176 Ga0063436_12861 Predicted glycosyltransferases R pfam13641 33 96 40 8 104 161 Ga0063436_12862 hypothetical protein - - 1 20 2 0 23 22 Ga0063436_12871 - pfam01476 181 379 147 56 625 1054 Ga0063436_12872 LysM domain Repeat domain in Vibrio, Colwellia, Bradyrhizobium and Shewanella - 71 229 47 37 414 524 Ga0063436_12881 riboflavin kinase/FMN adenylyltransferase H pfam13517 pfam01687 pfam06574 36 93 23 29 76 157 Ga0063436_12882 Thymidylate synthase complementing protein - pfam02511 52 84 24 4 80 112 Ga0063436_12883 Thymidylate synthase complementing protein - 64 189 55 9 129 210 Ga0063436_12884 GTP-binding protein Era R pfam02511 pfam01926 pfam07650 165 319 84 65 343 349 Ga0063436_12885 hypothetical protein - - 81 123 18 0 74 159 Ga0063436_12891 hypothetical protein - - Ga0063436_12892 Glutamate-1-semialdehyde aminotransferase H pfam00202 Ga0063436_12894 hypothetical protein - - Ga0063436_12901 Uncharacterized low-complexity proteins S pfam00805 Ga0063436_12902 Peptidase family M23 - pfam01551 Ga0063436_12903 hypothetical protein - Ga0063436_12911 oligoendopeptidase F E Ga0063436_12912 Predicted NAD/FAD-dependent oxidoreductase R pfam01432 pfam08439 pfam01593 pfam13450 Ga0063436_12913 FAD dependent oxidoreductase - pfam01266 Ga0063436_12921 Predicted metal-dependent hydrolase with the TIM-barrel fold R Ga0063436_12922 Aldo/keto reductase family - Ga0063436_12931 Ga0063436_12934 hypothetical protein ABC-type antimicrobial peptide transport system, ATPase component ABC-type antimicrobial peptide transport system, permease component Homocysteine/selenocysteine methylase (Smethylmethionine-dependent) Ga0063436_12935 hypothetical protein Ga0063436_12941 Ga0063436_12942 70 129 33 11 117 232 786 1551 398 95 685 1181 46 107 28 28 189 252 122 342 94 85 337 442 56 205 28 12 181 262 14 40 8 0 42 30 323 615 242 117 1123 1246 108 331 81 41 522 547 2 3 0 0 4 5 pfam07969 63 163 75 14 201 383 pfam00248 33 51 18 6 35 87 - - 24 20 10 0 41 55 V 20 128 26 6 68 133 V pfam00005 pfam02687 pfam12704 50 157 31 14 153 186 E pfam02574 58 172 45 30 119 230 - - 22 25 23 0 91 102 Enoyl-CoA hydratase/isomerase family - pfam00378 71 145 22 14 105 124 Putative translation initiation inhibitor, yjgF family J pfam01042 27 33 44 16 118 126 Ga0063436_12943 Acyl-CoA dehydrogenase, N-terminal domain - pfam02771 47 72 23 8 78 109 Ga0063436_12951 hypothetical protein - - 325 629 291 37 769 1014 Ga0063436_12952 Amino acid transporters E pfam13520 196 403 168 56 3678 4659 Ga0063436_12953 glutamine synthetase, type III E pfam00120 373 577 285 147 4019 4446 Ga0063436_12954 Conserved region in glutamate synthase - pfam01645 42 174 64 8 239 412 Ga0063436_12961 putative selenium metabolism protein SsnA FR 64 260 67 16 265 340 Ga0063436_12971 heavy metal-(Cd/Co/Hg/Pb/Zn)-translocating P-type ATPase P 98 200 57 24 277 499 Ga0063436_12972 small GTP-binding protein domain T pfam01979 pfam00122 pfam00702 pfam00009 pfam03144 234 766 140 187 569 539 Ga0063436_12932 Ga0063436_12933 Ga0063436_12981 MerR HTH family regulatory protein - 56 450 14 10 34 102 R pfam13411 pfam02310 pfam02607 Ga0063436_12982 Predicted cobalamin binding protein Ga0063436_12983 Predicted transcriptional regulators 10 22 7 0 63 118 K pfam13411 20 16 8 0 49 Ga0063436_12991 49 Domain of unknown function (DUF4432) - pfam14486 48 56 33 18 90 167 Ga0063436_14951 Domain of unknown function (DUF4432) - pfam14486 42 81 38 0 116 172 Ga0063436_13001 DisA bacterial checkpoint controller nucleotide-binding - pfam02457 15 188 41 0 193 154 Ga0063436_13002 Uncharacterized protein conserved in bacteria S 152 241 150 29 490 701 Ga0063436_13003 ribosome-associated GTPase EngA R pfam07949 pfam01926 pfam14714 70 127 52 16 170 260 Ga0063436_13004 signal peptidase I, bacterial type U 14 109 25 34 122 175 Ga0063436_13011 DNA primase (EC 2.7.7.-) L 102 294 74 46 238 366 Ga0063436_13012 glycyl-tRNA synthetase beta chain (EC 6.1.1.14) J pfam00717 pfam01807 pfam08275 pfam10410 pfam13662 pfam02091 pfam02092 113 600 106 39 526 728 Ga0063436_13013 hypothetical protein - 16 56 16 7 35 108 Ga0063436_13021 FtsK/SpoIIIE family/Ftsk gamma domain - pfam01580 pfam09397 94 425 115 94 401 689 Ga0063436_13022 hypothetical protein - - 8 407 39 36 226 294 Ga0063436_13023 Dolichyl-phosphate-mannose-protein mannosyltransferase - 0 38 26 0 138 172 Ga0063436_13031 ornithine carbamoyltransferase E pfam13231 pfam00185 pfam02729 22 53 30 14 159 174 Ga0063436_13032 - - 10 0 2 0 12 3 Ga0063436_13061 hypothetical protein Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) IRQ pfam00106 0 12 33 26 22 16 Ga0063436_13062 hypothetical protein - - 20 12 22 79 72 19 Ga0063436_13071 hypothetical protein - - 0 26 0 0 9 49 Ga0063436_13072 Peptidase family M28 - pfam04389 30 156 31 9 245 390 Ga0063436_13073 - - 12 30 6 12 18 68 Ga0063436_13091 hypothetical protein Pyruvate:ferredoxin oxidoreductase and related 2oxoacid:ferredoxin oxidoreductases, beta subunit C 205 446 122 110 301 504 Ga0063436_13092 2-oxoacid:acceptor oxidoreductase, alpha subunit C 634 1330 202 192 933 1077 Ga0063436_13093 Transcriptional regulators K pfam02775 pfam01558 pfam01855 pfam00392 pfam07729 138 245 36 43 159 167 Ga0063436_13101 hypothetical protein - 34 85 14 2 73 103 Ga0063436_13102 Glycine cleavage system T protein (aminomethyltransferase) E pfam01266 pfam01571 pfam08669 152 822 128 18 391 802 Ga0063436_13103 Predicted acyl-CoA transferases/carnitine dehydratase C pfam02515 26 324 43 16 148 337 Ga0063436_13104 Amidohydrolase - pfam04909 36 80 34 0 65 208 Ga0063436_13105 Permeases of the major facilitator superfamily R pfam11700 79 216 76 24 372 594 Ga0063436_13106 hypothetical protein - - 2 78 10 6 62 138 Ga0063436_13107 hypothetical protein - 2 0 1 0 28 41 Ga0063436_13108 L 0 144 9 0 75 209 Ga0063436_14595 A/G-specific DNA-adenine glycosylase (EC 3.2.2.-) Iron-sulfur cluster binding domain of dihydroorotate dehydrogenase B pfam00633 pfam00730 pfam14815 - pfam10418 5 51 10 10 52 98 Ga0063436_14596 Dihydroorotate dehydrogenase - pfam01180 4 48 6 0 52 81 Ga0063436_13111 xanthine phosphoribosyltransferase F pfam00156 28 46 34 31 120 178 Ga0063436_13112 Transglycosylase - pfam00912 49 36 18 0 103 110 Ga0063436_13121 hypothetical protein - - 6 37 14 0 60 75 Ga0063436_13122 hypothetical protein - - 58 139 31 0 72 184 Ga0063436_13131 Sigma-70 factor, region 1 - pfam03979 303 430 129 67 558 740 Ga0063436_13132 hypothetical protein - - 0 0 0 3 0 6 Ga0063436_13141 hypothetical protein - - 0 0 7 0 11 15 Ga0063436_13142 F 338 223 30 4758 7965 54 268 54 29 368 407 C pfam00860 pfam00117 pfam00733 pfam00958 pfam02779 pfam02780 302 Ga0063436_13161 Xanthine/uracil permeases GMP synthase (glutamine-hydrolyzing), C-terminal domain or B subunit/GMP synthase (glutamine-hydrolyzing), N-terminal domain or A subunit Pyruvate/2-oxoglutarate dehydrogenase complex, dehydrogenase (E1) component, eukaryotic type, beta subunit 57 80 40 3 81 140 Ga0063436_13162 Glycosyl transferase family 2 - pfam00535 22 47 32 4 108 114 Ga0063436_13171 Amino acid kinase family - pfam00696 2 12 0 0 20 9 Ga0063436_13172 Aminotransferase class-III - pfam00202 0 44 2 0 26 30 Ga0063436_13151 F Ga0063436_13181 MutS domain V - pfam00488 pfam00488 pfam05190 Ga0063436_13182 MutS family domain IV/MutS domain V Ga0063436_13191 Uncharacterized conserved protein S pfam02586 Ga0063436_13192 Aspartate/tyrosine/aromatic aminotransferase E Ga0063436_13193 hypothetical protein Ga0063436_13194 ATP-grasp domain Ga0063436_13201 Ga0063436_13891 1 6 24 8 16 73 22 50 171 50 6 66 106 25 235 pfam00155 64 58 38 182 2 226 201 - - 58 14 - pfam13535 10 40 16 0 127 132 38 0 60 95 ATP:cob(I)alamin adenosyltransferase S pfam01923 94 - pfam00805 6 IQR pfam00106 Ga0063436_13893 Pentapeptide repeats (8 copies) Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) Threonine dehydrogenase and related Zn-dependent dehydrogenases 149 57 61 187 222 0 2 0 6 2 16 18 4 0 7 50 ER pfam08240 8 0 2 8 26 20 Ga0063436_13894 Uncharacterized low-complexity proteins Ga0063436_13895 Rhodanese-related sulfurtransferase S pfam00805 42 105 42 28 233 292 P pfam00581 16 146 25 3 154 Ga0063436_13896 259 3'-5' exonuclease - 26 60 6 14 37 65 Ga0063436_13897 Ribonuclease D J 10 65 23 6 98 201 Ga0063436_13898 Predicted exonuclease of the beta-lactamase fold involved in RNA processing J pfam01612 pfam00570 pfam01612 pfam00753 pfam07521 pfam10996 54 262 58 30 331 476 Ga0063436_13899 Pyridoxamine 5'-phosphate oxidase - pfam01243 8 35 16 32 50 122 Ga0063436_13215 Protein of unknown function (DUF2029) - pfam09594 22 79 29 0 79 112 Ga0063436_13216 Protein of unknown function (DUF2029) - pfam09594 28 104 33 18 105 169 Ga0063436_13217 hypothetical protein - - 27 88 47 24 156 224 Ga0063436_13218 Membrane protease subunits, stomatin/prohibitin homologs O pfam01145 1139 2587 445 270 1558 1869 Ga0063436_13219 response regulator receiver and ANTAR domain protein - 22 68 3 4 64 117 Ga0063436_13221 Secreted and surface protein containing fasciclin-like repeats M pfam03861 pfam02469 pfam13345 10121 24463 596 610 543 554 Ga0063436_13231 Translation-initiation factor 2 - pfam11987 126 156 71 16 250 310 Ga0063436_13232 ribosome-binding factor A J pfam02033 80 92 37 8 157 206 Ga0063436_13233 hypothetical protein - - 64 36 6 10 17 57 Ga0063436_13241 - pfam00106 8 56 9 0 38 48 Ga0063436_13242 short chain dehydrogenase Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) IQR pfam13561 53 81 57 45 147 214 Ga0063436_13251 NAD-dependent aldehyde dehydrogenases C pfam00171 60 252 66 28 268 451 Ga0063436_13252 replicative DNA helicase loader DnaI L pfam01695 69 241 88 30 351 461 Ga0063436_13253 DnaD and phage-associated domain - pfam07261 50 144 46 25 267 406 Ga0063436_13254 DnaB-like helicase C terminal domain - pfam03796 28 254 39 1 244 266 Ga0063436_13261 Cytosine deaminase and related metal-dependent hydrolases FR pfam07969 12 86 45 18 451 669 Ga0063436_13271 Methyltransferase domain - pfam13489 45 66 37 6 79 40 Ga0063436_13272 hypothetical protein - - 46 20 8 0 63 55 Ga0063436_13291 Phosphorylase superfamily - pfam01048 21 33 13 0 60 74 Ga0063436_13292 DinB superfamily 2-keto-4-pentenoate hydratase/2-oxohepta-3-ene-1,7-dioic acid hydratase (catechol pathway) - pfam12867 35 127 35 16 196 320 Q pfam01557 98 288 52 24 306 385 hypothetical protein Integral membrane protein CcmA involved in cell shape determination - - 40 30 15 22 96 171 M 28 52 18 12 31 118 V 59 114 62 28 353 450 Ga0063436_13313 Uncharacterized vancomycin resistance protein Phosphatidylserine/phosphatidylglycerophosphate/cardiolipin synthases and related enzymes pfam04519 pfam04294 pfam12229 20 62 58 18 127 139 Ga0063436_13314 Dipeptidyl aminopeptidases/acylaminoacyl-peptidases E pfam13091 pfam00326 pfam07676 114 165 114 45 394 427 Ga0063436_13315 ADP-ribose pyrophosphatase F pfam00293 31 101 33 7 106 137 Ga0063436_13321 hypothetical protein - - 0 30 2 0 35 84 Ga0063436_13322 Cellulase (glycosyl hydrolase family 5) - 38 39 53 6 147 208 Ga0063436_14812 glucose-1-phosphate adenylyltransferase G pfam00150 pfam00132 pfam00483 34 129 55 28 199 267 Ga0063436_13332 Acyl dehydratase I 20 56 15 12 82 168 Ga0063436_13333 competence protein ComEA helix-hairpin-helix repeat region Uncharacterized membrane protein, possible Na+ channel or pump carbohydrate ABC transporter membrane protein 2, CUT1 family (TC 3.A.1.1.-) L pfam01575 pfam10531 pfam12836 6 124 16 0 131 212 R pfam04474 6 65 33 20 113 252 G pfam00528 34 26 18 14 99 93 Ga0063436_13892 Ga0063436_13293 Ga0063436_13294 Ga0063436_13311 Ga0063436_13312 Ga0063436_13334 Ga0063436_13341 I 0 0 Ga0063436_13342 hypothetical protein - - 20 9 11 0 53 50 Ga0063436_13811 MacB-like periplasmic core domain - pfam12704 52 350 4 10 91 113 Ga0063436_13831 Protein related to penicillin acylase R pfam01804 107 304 91 70 382 576 Ga0063436_13832 DNA replication and repair protein RecF L 16 31 32 6 157 185 Ga0063436_13833 Uncharacterized conserved protein S pfam02463 pfam00069 pfam03781 297 613 217 147 980 1487 Ga0063436_13834 hypothetical protein - - 0 32 18 4 92 61 Ga0063436_13835 - pfam07676 112 263 91 32 326 657 Ga0063436_13836 WD40-like Beta Propeller Repeat FGGY family of carbohydrate kinases, C-terminal domain/FGGY family of carbohydrate kinases, N-terminal domain - pfam00370 pfam02782 60 78 19 10 159 192 Ga0063436_13837 FGGY family of carbohydrate kinases, N-terminal domain - pfam00370 0 0 2 0 20 10 Ga0063436_13838 Domain of unknown function (DUF4380) - pfam14315 62 261 117 50 393 519 Ga0063436_13839 D-alanine-D-alanine ligase and related ATP-grasp enzymes M pfam07478 758 1278 573 166 2165 2785 Ga0063436_138310 Dienelactone hydrolase and related enzymes Q 31 117 23 18 87 189 Ga0063436_138311 Thrombospondin type 3 repeat/LysM domain - 479 727 410 232 1840 2381 Ga0063436_138312 ComEC/Rec2-related protein R pfam01738 pfam01476 pfam02412 pfam03772 pfam13567 13 93 10 6 118 173 Ga0063436_138313 hypothetical protein - - 45 152 27 10 83 163 Ga0063436_11532 Domain of unknown function (DUF4432) - pfam14486 90 105 38 12 197 313 Ga0063436_11533 short chain dehydrogenase - pfam00106 117 224 32 50 83 176 Ga0063436_11534 NADH(P)-binding - pfam13460 5 12 1 0 6 10 Ga0063436_11172 Sugar kinases, ribokinase family G pfam00294 42 200 39 19 167 322 Ga0063436_11171 KDPG and KHG aldolase - pfam01081 36 52 8 3 106 113 Ga0063436_13371 hypothetical protein - - 94 198 94 18 202 286 Ga0063436_13372 hypothetical protein - - 48 71 16 20 44 29 Ga0063436_13373 Cupin domain - pfam07883 95 232 28 4 120 113 Ga0063436_13374 YHYH protein - pfam14240 74 180 73 41 244 267 Ga0063436_13375 Peptidase MA superfamily - 232 168 184 60 494 435 Ga0063436_13381 CTP synthase (EC 6.3.4.2) F pfam13485 pfam00117 pfam06418 146 671 253 54 906 1270 Ga0063436_13382 thioredoxin - pfam00085 397 1201 202 87 757 816 Ga0063436_13383 hypothetical protein - - 69 184 59 49 165 223 Ga0063436_13384 Putative sterol carrier protein I 120 284 87 57 213 321 Ga0063436_13385 Beta-lactamase class A V pfam02036 pfam12229 pfam13354 153 324 139 30 384 453 Ga0063436_13386 Major Facilitator Superfamily - pfam07690 0 34 16 0 36 51 Ga0063436_13387 hypothetical protein - - 0 16 0 0 0 6 Ga0063436_13411 Putative hemolysin R pfam01553 12 26 34 42 105 113 Ga0063436_13412 hypothetical protein - - 0 0 4 22 16 2 Ga0063436_138413 tryptophan synthase, alpha chain (EC 4.2.1.20) E pfam00290 18 66 7 0 293 238 Ga0063436_138412 tryptophan synthase, beta subunit E pfam00291 8 32 32 2 917 682 Ga0063436_138411 phosphoribosylanthranilate isomerase (EC 5.3.1.24) E pfam00697 0 10 2 0 226 224 Ga0063436_138410 indole-3-glycerol phosphate synthase (EC 4.1.1.48) E 2 41 6 0 492 339 Ga0063436_13849 anthranilate phosphoribosyltransferase E pfam00218 pfam00591 pfam02885 0 18 16 8 1051 765 Ga0063436_13848 anthranilate synthase, component II (EC 4.1.3.27) EH 0 6 6 0 272 225 Ga0063436_13847 anthranilate synthase, component I (EC 4.1.3.27) EH 11 44 17 2 723 697 Ga0063436_13845 Glycine cleavage system T protein (aminomethyltransferase) E pfam00117 pfam00425 pfam04715 pfam01266 pfam01571 pfam08669 Ga0063436_13844 electron transfer flavoprotein beta subunit C Ga0063436_13843 electron transfer flavoprotein alpha subunit apoprotein Ga0063436_13842 Ga0063436_13841 484 605 882 119 3172 3207 203 410 307 76 1147 1441 C pfam01012 pfam00766 pfam01012 66 114 165 26 645 581 NHL repeat - pfam01436 4 0 0 0 34 34 hypothetical protein - - 2 29 11 0 11 59 Ga0063436_14013 condensin subunit ScpA S pfam02616 26 77 62 5 300 375 Ga0063436_13431 Uncharacterized low-complexity proteins S 245 573 296 110 1200 1457 Ga0063436_13432 selenophosphate synthase (EC 2.7.9.3) E pfam00805 pfam00586 pfam02769 89 533 138 57 533 753 Ga0063436_13491 hypothetical protein - - 29 170 76 2 166 336 Ga0063436_13492 Uncharacterized protein conserved in bacteria S pfam01503 Ga0063436_13493 hypothetical protein - - Ga0063436_13511 Putative transposase - pfam04986 Ga0063436_13512 putative efflux protein, MATE family V pfam01554 Ga0063436_13513 hypothetical protein - - Ga0063436_14036 MutS domain I - Ga0063436_14035 - Ga0063436_14034 hypothetical protein Acetylornithine deacetylase/Succinyl-diaminopimelate desuccinylase and related deacylases Ga0063436_14033 hypothetical protein Ga0063436_14032 22 113 33 6 119 139 279 779 73 25 232 467 16 0 14 0 37 38 4 69 32 2 61 69 26 20 18 3 53 81 pfam01624 2 0 0 0 0 2 2 0 0 0 0 4 E pfam01546 pfam07687 204 143 94 81 356 324 - - 32 52 20 23 98 92 hypothetical protein - 0 41 16 3 58 54 Ga0063436_14031 NusA antitermination factor K pfam00575 pfam08529 pfam13184 Ga0063436_13531 - Ga0063436_13532 hypothetical protein NADPH:quinone reductase and related Zn-dependent oxidoreductases Ga0063436_13533 Ga0063436_14001 428 665 253 156 1243 1491 33 94 8 10 61 105 CR pfam08240 pfam13602 53 197 100 18 345 387 AICARFT/IMPCHase bienzyme - pfam01808 26 106 22 5 81 195 radical SAM-linked protein S pfam10105 52 62 13 20 40 101 Ga0063436_14002 LSU ribosomal protein L21P J pfam00829 156 234 128 42 389 560 Ga0063436_14003 LSU ribosomal protein L27P J pfam01016 131 373 122 38 424 554 Ga0063436_14004 ribosomal protein L31 J pfam01197 281 820 272 175 1293 1646 Ga0063436_14005 thymidine kinase (EC 2.7.1.21) F pfam00265 62 48 32 14 158 194 Ga0063436_14006 F 6 46 22 14 104 98 - pfam00156 pfam01743 pfam12627 36 80 48 18 206 398 P pfam09084 199 763 74 70 149 170 Ga0063436_13572 hypoxanthine phosphoribosyltransferase Poly A polymerase head domain/Probable RNA and SrmBbinding site of polymerase A ABC-type nitrate/sulfonate/bicarbonate transport systems, periplasmic components ABC-type nitrate/sulfonate/bicarbonate transport system, permease component P pfam00528 8 127 12 16 44 44 Ga0063436_13601 hypothetical protein - - 11 16 7 56 10 2 Ga0063436_13602 - pfam03786 38 38 30 55 17 0 Ga0063436_13603 D-mannonate dehydratase (UxuA) Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) IQR pfam00106 50 0 16 6 2 0 Ga0063436_14251 hypothetical protein - - 104 169 89 22 152 244 Ga0063436_14252 Protein of unknown function (DUF970) - pfam06153 55 166 112 40 293 429 Ga0063436_14253 Uncharacterized protein conserved in bacteria S pfam06153 86 124 71 20 251 335 Ga0063436_13624 Uncharacterized conserved protein S pfam03992 9 60 44 22 94 96 Ga0063436_13631 transcriptional regulator, IclR family K pfam01614 53 9 61 2 256 309 Ga0063436_13632 hypothetical protein - - 25 39 38 4 153 176 Ga0063436_13633 hypothetical protein - - 12 2 13 4 48 108 Ga0063436_13634 hypothetical protein - - 0 0 0 0 16 13 Ga0063436_14234 - pfam13599 19 33 28 0 81 137 Ga0063436_14235 Pentapeptide repeats (9 copies) Dehydrogenases with different specificities (related to shortchain alcohol dehydrogenases) IQR pfam00106 45 138 44 10 136 208 Ga0063436_13651 chaperone protein DnaK O pfam00012 641 950 203 270 708 998 Ga0063436_13661 hypothetical protein - - 46344 340264 32573 30354 52872 37343 Ga0063436_13662 Penicillin binding protein transpeptidase domain - pfam00905 11 40 20 23 104 178 Ga0063436_13681 ATPase involved in DNA repair L pfam13476 56 152 48 29 150 205 Ga0063436_13693 Uncharacterized proteins, LmbE homologs S pfam02585 8 64 24 0 118 82 Ga0063436_13711 ABC-type sugar transport systems, ATPase components G pfam00005 20 16 20 148 8 6 Ga0063436_13712 hypothetical protein - - 0 0 2 59 4 0 Ga0063436_13713 hypothetical protein - - 0 0 0 16 0 2 Ga0063436_13721 Predicted membrane protein S pfam09858 18 96 3 0 73 66 Ga0063436_13722 hypothetical protein - - 0 3 0 0 2 0 Ga0063436_13723 Multi-copper polyphenol oxidoreductase laccase - pfam02578 25 134 10 12 199 147 Ga0063436_13731 Putative intracellular protease/amidase R pfam01965 3 28 2 51 12 0 Ga0063436_13741 Clp amino terminal domain - pfam02861 116 75 40 38 205 343 Ga0063436_13742 Iron-containing alcohol dehydrogenase - pfam00465 0 21 8 0 40 69 Ga0063436_13751 hypothetical protein - - 41 95 45 14 233 242 Ga0063436_14007 Ga0063436_13571 Ga0063436_13752 Aldehyde dehydrogenase family - pfam00171 37 109 24 20 233 222 Ga0063436_13761 Glycosyl transferase family 2 - 8 15 0 2 0 0 Ga0063436_13762 Fumarase C C-terminus/Lyase - pfam00535 pfam00206 pfam10415 41 48 26 230 52 24 Ga0063436_13771 hypothetical protein - - 9 36 17 0 35 51 Ga0063436_13772 ABC transporter transmembrane region - pfam00664 20 28 15 20 64 67 Ga0063436_14311 hypothetical protein - 8 0 1 0 4 7 Ga0063436_14312 ABC transporter/ABC transporter transmembrane region - pfam00005 pfam00664 121 299 107 24 397 489 Ga0063436_13781 hypothetical protein - - 61 172 81 17 250 266 Ga0063436_13791 hypothetical protein - - 0 64 10 0 11 6 Ga0063436_13801 Lumazine binding domain - pfam00677 4 45 6 4 42 33 Ga0063436_13802 riboflavin biosynthesis protein RibD H pfam01872 26 68 12 20 101 123 Ga0063436_13901 hypothetical protein - - 0 0 0 0 0 2 Ga0063436_13902 hypothetical protein - - 0 0 0 29 0 1 Ga0063436_13903 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13904 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13905 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13906 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13907 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13908 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13909 hypothetical protein - - 0 0 0 0 0 2 Ga0063436_139010 hypothetical protein - - 0 0 0 0 0 2 Ga0063436_139011 DNA binding domain, excisionase family - pfam12728 0 0 0 0 0 0 Ga0063436_139012 hypothetical protein - - 0 0 0 0 2 4 Ga0063436_139013 hypothetical protein - - 0 0 0 0 0 4 Ga0063436_139014 hypothetical protein - - 0 0 0 0 0 2 Ga0063436_139015 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13911 ABC transporter transmembrane region - 0 42 6 0 26 34 Ga0063436_13931 Exodeoxyribonuclease VII large subunit (EC 3.1.11.6) L pfam00664 pfam02601 pfam13742 40 283 61 0 287 430 Ga0063436_13932 Exodeoxyribonuclease VII small subunit (EC 3.1.11.6) L pfam02609 0 84 33 36 118 183 Ga0063436_13933 Uncharacterized protein conserved in bacteria S pfam02681 43 126 54 4 172 331 Ga0063436_13934 murein biosynthesis integral membrane protein MurJ R 78 240 44 22 234 522 Ga0063436_13935 FecR family protein - pfam03023 pfam01476 pfam03160 pfam04773 928 2200 805 357 3188 4264 Ga0063436_13951 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13952 hypothetical protein - - 0 0 0 0 2 2 Ga0063436_13953 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13954 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13955 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13956 hypothetical protein - - 0 4 0 0 7 0 Ga0063436_13957 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13958 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13959 hypothetical protein - - 0 0 0 0 4 0 Ga0063436_139510 ERF superfamily - pfam04404 0 0 0 0 0 0 Ga0063436_139511 hypothetical protein - - 0 0 0 0 0 5 Ga0063436_139512 hypothetical protein - - 0 0 0 0 1 1 Ga0063436_139513 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_139514 hypothetical protein - - 0 0 0 0 0 0 Ga0063436_13971 Predicted choline kinase involved in LPS biosynthesis M pfam01633 4 28 12 2 50 77 Ga0063436_13972 Amino acid transporters E pfam13520 36 22 39 0 263 409 Ga0063436_13973 Predicted phosphoesterases, related to the Icc protein R pfam12850 14 50 11 0 45 85 Ga0063436_13974 hypothetical protein - - 0 0 2 0 14 18 Ga0063436_13975 hypothetical protein - - 4 6 9 2 12 61 Ga0063436_14111 Secreted and surface protein containing fasciclin-like repeats M pfam02469 414 553 277 129 1123 1330 Ga0063436_14112 Acyl-CoA synthetases (AMP-forming)/AMP-acid ligases II IQ pfam00501 pfam13193 202 194 128 75 557 787 Ga0063436_14161 hypothetical protein - - 127 202 48 26 287 292 Ga0063436_14162 Peptidase family M23 - pfam01551 20 27 8 13 42 31 Ga0063436_14163 CAAX protease self-immunity - pfam02517 5 72 23 0 44 94 Ga0063436_14164 - - 0 13 0 0 0 15 Ga0063436_14165 hypothetical protein Acetyltransferases, including N-acetylases of ribosomal proteins J pfam13302 8 40 21 6 24 67 Ga0063436_11841 DNA gyrase C-terminal domain, beta-propeller - pfam03989 44 162 28 10 231 386 Ga0063436_11842 hypothetical protein - - 74 355 85 63 285 451 Ga0063436_11843 hypothetical protein - - 234 325 189 79 590 739 Ga0063436_11844 hypothetical protein - - 131 554 151 52 484 598 Ga0063436_11845 Domain of unknown function (DUF1083) - pfam06452 6 20 4 10 34 56 Ga0063436_14321 PA domain - pfam02225 78 46 44 399 127 56 Ga0063436_14351 Dolichyl-phosphate-mannose-protein mannosyltransferase - pfam13231 28 20 13 16 65 84 Ga0063436_14352 16S rRNA (guanine(966)-N(2))-methyltransferase RsmD L pfam03602 65 90 41 24 83 180 Ga0063436_14353 hypothetical protein - - 12 34 39 16 49 95 Ga0063436_14381 Glutamate synthase domain 1 E pfam00310 42 274 100 18 496 792 Ga0063436_14391 Adenosine/AMP deaminase - pfam00962 0 0 0 0 0 0 Ga0063436_14392 ABC transporter - pfam00005 0 0 4 0 4 4 Ga0063436_14461 Flavin containing amine oxidoreductase - pfam01593 19 148 26 2 93 189 Ga0063436_14472 Major intrinsic protein - pfam00230 2 61 16 0 5 0 Ga0063436_14491 Ribonucleotide reductase, beta subunit F pfam00268 109 223 20 42 8 23 Ga0063436_14492 NAD dependent epimerase/dehydratase family - 0 10 1 0 5 2 Ga0063436_14533 xylulokinase (EC 2.7.1.17) G pfam01370 pfam00370 pfam02782 50 114 25 22 163 241 Ga0063436_14711 Vacuole effluxer Atg22 like - Ga0063436_14712 Methylase of chemotaxis methyl-accepting proteins Ga0063436_14713 Response regulator receiver domain Ga0063436_14731 79 174 96 38 471 603 NT pfam11700 pfam01739 pfam03705 175 358 142 73 587 676 - pfam00072 17 18 21 11 46 65 hypothetical protein - - 0 0 0 54 14 0 Ga0063436_14732 hypothetical protein - - 0 0 0 10 3 0 Ga0063436_14733 hypothetical protein - - 0 0 0 45 11 2 Ga0063436_14734 hypothetical protein - - 0 0 0 45 19 0 Ga0063436_14735 hypothetical protein - - 0 0 0 53 32 0 Ga0063436_14736 hypothetical protein - - 0 0 0 72 25 0 Ga0063436_14737 Type III restriction enzyme, res subunit - 0 0 0 50 26 0 Ga0063436_14851 Glutamate synthase domain 2 E pfam04851 pfam01645 pfam04898 42 151 38 14 343 457 Ga0063436_14852 Glutamate synthase central domain - pfam04898 0 16 4 10 24 55 Table S6: DOC uptake COGs in freshwater (and SAR11) lineages with streamlined genomes. List of COGs is modified from Poretsky et al., Environ Microbiol 12: 616-627 (2010) and Ghylin et al., ISME J 8: 2503-2516 (2014). For CL500-11-LM, the average expression level across all datasets was added between parentheses. Phylum/class Chloroflexi β-Proteobacteria β-Proteobacteria Actinobacteria Actinobacteria α-Proteobacteria α-Proteobacteria α-Proteobacteria Strain (lineage) CL500-11LM Polynucleobacter necessarius asymbioticus QLW-P1DMWA1 (PnecC) FNE-F8 bin_6_1 (PnecC) AAA027-L06 (acI-B1) AAA278-O22 (acI-A1) SCGC AAA028D10 (LD12) SCGC AAA028C07 (LD12) Pelagibacter ubique HTCC1062 (SAR11) # proteins 2153 2088 2343 1244 1193 1057 955 1354 % completeness 90% 100% 98% 98% 91% n.d. n.d. 100% 2602041904 640427129 2596583565 2505679121 2236661007 2236347069 2236661008 637000058 2 (0.13 %) 1 - 1 3 - - - 1 5 - - 2 1 3 1 3 - - 2 1 1 2 6 - - 3 1 2 - - - - 1 1 1 - - - - - - 1 - - - - - - 1 5 4 2 2 2 1 3 4 5 2 1 1 1 3 4 5 2 1 2 1 3 4 4 2 1 2 1 3 3 4 2 1 2 1 3 IMG taxon ID Amino Acids, general COG0531 Amino acid transporters COG0834 ABC-type amino acid transport/signal transduction systems, periplasmic component/domain COG1126 ABC-type polar amino acid transport system, ATPase component COG0765 ABC-type amino acid transport system, permease component COG4597 ABC-type amino acid transport system, permease component COG4215 ABC-type arginine transport system, permease component COG4160 ABC-type arginine/histidine transport system, permease component 1 (0.25 %) 1 (0.03 %) 1 (0.02 %) - Amino acids, branched chain COG0683 ABC-type branched-chain amino acid transport systems, periplasmic component COG0410 ABC-type branched-chain amino acid transport systems, ATPase component COG0559 Branched-chain amino acid ABC-type transport system, permease components COG4177 ABC-type branched-chain amino acid transport system, permease component COG0411 ABC-type branched-chain amino acid 2 (0.38 %) 1 (0.04 %) 1 (0.04 %) 1 (0.03 %) transport systems, ATPase component COG1296 Predicted branched-chain amino acid permease (azaleucine resistance) 1 (0.01 %) 2 2 1 1 2 2 2 - - 1 1 - - - - - 1 1 - - - - - 1 1 - - - - - 1 1 - - - 1 1 - - - - - - - 1 1 - - - - - 2 1 - - 1 1 1 3 1 - - 2 - - 2 1 - - 1 - - 2 1 - - 1 - - 1 1 1 - - 1 2 - - 1 1 1 - - - - 1 - 1 - - 2 1 - - - - - 1 - - - - - - - 2 - - - 3 2 2 - - - 2 - - 2 4 - - - 1 1 1 - - - - Amino acids, di- and oligopeptides COG1173 ABC-type dipeptide/oligopeptide/nickel transport systems, permease components COG0601 ABC-type dipeptide/oligopeptide/nickel transport systems, permease components COG0747 ABC-type dipeptide transport system, periplasmic component COG0444 ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component COG4166 ABC-type oligopeptide transport system, periplasmic component COG4608 ABC-type oligopeptide transport system, ATPase component 8 (0.18 %) 9 (0.4 %) 8 (6.71 %) 4 (0.14 %) 2 (0.13 %) Polyamines COG1176 ABC-type spermidine/putrescine transport system, permease component I COG3842 ABC-type spermidine/putrescine transport systems, ATPase components COG1177 ABC-type spermidine/putrescine transport system, permease component II COG0687 Spermidine/putrescine-binding periplasmic protein 1 (0.01 %) 2 (0.04 %) 2 (0.01 %) 1 (0.04 %) Lipids COG0580 Glycerol uptake facilitator and related permeases (Major Intrinsic Protein Family) COG2867 Oligoketide cyclase/lipid transport protein COG1133 ABC-type long-chain fatty acid transport system, fused permease and ATPase components 1 (0.02 %) - Nucleotides and coenzymes COG3201 Nicotinamide mononucleotide transporter COG2233 Xanthine/uracil permeases 1 (0.17 %) Carbohydrates, general COG1129 ABC-type sugar transport system, ATPase component COG1682 ABC-type polysaccharide/polyol phosphate export systems, permease COG1879 ABC-type sugar transport system, periplasmic component COG1134 ABC-type polysaccharide/polyol phosphate transport system, ATPase 1 (0.03 %) 1 (0.42 %) 1 (0.01 %) COG1653 ABC-type sugar transport system, periplasmic component COG1175 ABC-type sugar transport systems, permease components COG0395 ABC-type sugar transport system, permease component COG0738 Fucose permease COG3839 ABC-type sugar transport systems, ATPase components COG2211 Na+/melibiose symporter and related transporters COG2271 Sugar phosphate permease COG3822 ABC-type sugar transport system, auxiliary component COG1925 Phosphotransferase system, HPr-related proteins COG1762 Phosphotransferase system mannitol/fructose-specific IIA domain (Ntr-type) COG2893 Phosphotransferase system, mannose/fructose-specific component IIA 2 (1.7 %) 2 (0.07 %) 3 (0.06 %) 1 (0.01 %) 2 (0.26 %) 1 (0.06 %) - - - - 2 - - 2 - - - 2 - - 2 - - - 2 - - 2 - - - 4 - - - - - - - - - 3 - - - 1 - - - 1 3 - - - 1 - - - 1 - - - - 1 1 - - - - - 1 1 - - - - - 1 1 - - - - - - - 2 4 - - - - - 1 1 - - - 2 2 - - 2 1 - 2 2 - - 2 2 - 2 2 - - 2 2 - 5 6 - - 1 1 - 1 1 - - 1 1 - 1 1 - - 1 - 3 1 1 - - 1 - 3 1 1 - - 1 - 3 Carbohydrates, pentoses COG1172 Ribose/xylose/arabinose/galactoside ABC-type transport systems, permease components COG4214 ABC-type xylose transport system, permease component 1 (0.01 %) - Carboxylic acids COG1593 TRAP-type C4-dicarboxylate transport system, large permease component COG1638 TRAP-type C4-dicarboxylate transport system, periplasmic component COG3090 TRAP-type C4-dicarboxylate transport system, small permease component COG2358 TRAP-type uncharacterized transport system, periplasmic component COG4666 TRAP-type uncharacterized transport system, fused permease components COG4664 TRAP-type mannitol/chloroaromatic compound transport system, large permease component COG4665 TRAP-type mannitol/chloroaromatic compound transport system, small permease component COG4663 TRAP-type mannitol/chloroaromatic compound transport system, periplasmic component - COG1301 Na+/H+-dicarboxylate symporters - 2 1 - - - - - COG0471 Di- and tricarboxylate transporters 1 (0.02 %) - - - - - - - - - 1 - - - - - - - - - - - 2 - - - - - - 2 - - - - - - 2 - - - 2 - - - - - - 1 - - - 1 1 - - 1 1 1 COG1620 L-lactate permease Compatible solutes COG2113 ABC-type proline/glycine betaine transport systems, periplasmic components COG4175 ABC-type proline/glycine betaine transport system, ATPase component COG4176 ABC-type proline/glycine betaine transport system, permease component COG1174 ABC-type proline/glycine betaine transport systems, permease component COG1125 ABC-type proline/glycine betaine transport systems, ATPase components COG0591 Na+/proline symporter - Table S7: Genes annotated as carbon monoxide dehydrogenase large subunit (coxL). Analysis of the closest matches to coxL-characteristic motifs (King and Weber, Nat Rev Microbiol 5, 107-118 (2007)) and gene neighborhoods support the presence of a Type I and Type II coxL. Amino acids deviating from the conserved motif sequence are underlined and written in bold type. X in conserved motif indicates any amino acid, while X in gene neighborhood indicates gene not involved in CO oxidation. Locus_tag Both types Type I Type II gene neighborhood GGFGXK QGQHXTX GSRST CGTRIN AYXCSFR AYRGAGR Ga0063436_106011 GGFGSK HGQGHETT GSRSG CGTLPV - AVRGAGR coxSLM XXXX DEF Ga0063436_12672 GGFGGK QGQGHETT ASRST CGTRIN AYRCSFR - coxMSLF, end contig on both sides Ga0063436_10048 GGFGPK QGQGHFTS - - - PYRGAGR coxS upstream Ga0063436_10535 - - - - - - coxS Ga0063436_10781 GGFGGK - - - - - single gene contig Table S8: Genes putatively involved in methylovory. Genes were identified based on genome annotation and by Blastp analysis using the protein sequences of the genes suggested to be involved in methylovory by Pelagibacter ubique (Sun et al., PLoS One 6: e23973 (2011)). Function Annotation Locus tag THF-linked oxidation formate dehydrogenase, alpha subunit (fdhF) Ga0063436_12007 NAD-dependent formate dehydrogenase, beta subunit (fdsB) Ga0063436_12009 formate dehydrogenase, chain D (fdhD) not identified molybdopterin-guanine dinucleotide biosynthesis protein A (mobA) Ga0063436_14093 molybdopterin biosynthesis protein (moeA) Ga0063436_101417 molybdopterin biosynthesis protein (moeA) Ga0063436_12004 molybdopterin biosynthesis protein (moeA) Ga0063436_12005 formate-THF ligase (fhs) not identified methylene-THF reductase (metF) not identified bifunctional methylene-THF dehydrogenase-methenyl-THF cyclohydrolase (folD) Ga0063436_12431 glycine system cleavage T-protein (gcvT) Ga0063436_11514 glycine system cleavage T-protein (gcvT) Ga0063436_12087 glycine system cleavage T-protein (gcvT) Ga0063436_12203 glycine system cleavage T-protein (gcvT) Ga0063436_13102 glycine system cleavage T-protein (gcvT) Ga0063436_13845 Trimethylamine:corrinoid methyltransferase Ga0063436_10877 Trimethylamine:corrinoid methyltransferase Ga0063436_11301 Trimethylamine:corrinoid methyltransferase Ga0063436_12334 Trimethylamine:corrinoid methyltransferase Ga0063436_12088 Other components glycine cleavage complex glycine cleavage system H protein (gcvH) Ga0063436_10273 glycine dehydrogenase (decarboxylating) (gcvP) Ga0063436_10274 methanol oxidation iron-containing alcohol dehydrogenase (Fe-ADH) Ga0063436_12331 iron-containing alcohol dehydrogenase (Fe-ADH) Ga0063436_111410 iron-containing alcohol dehydrogenase (Fe-ADH) Ga0063436_13941 Aminomethyltransferases (glycine and betaine) Table S9. Genome STAMP analysis. Protein functions over- and underrepresented in the CL500-11-LM sequence bin (CL500) relative to other streamlined genomes of ubiquitous freshwater bacteria (SFH), the metagenomic data set generated from the summer deep water 0.22-3 µm fraction microbial community at the offshore station (MetaG), and of the genome of its most closely related fully sequenced isolate (A.t.). Effect size is the differential between the CL500-11-LM and the compared genome relative proportions (after rarefication to the smallest dataset size, which was CL500-11-LM, and q-value is calculated based on Benjamini-Hochberg false discovery rate correction of the p-values resulting from the Chi-square or Fisher’s exact test for COGs and Pfams, respectively. All functions that are significantly differentially represented in at least one of the comparisons (p < 0.05, q < 0.15 were listed. COG / pfam Number of genes (% of all proteins with COG/Pfam assignment) effect size (q-value) CL500 SFH metaG A.t. SFH metaG A.t. Amino acid transport and metabolism Posttranslational modification, protein turnover, chaperones pfam01266 - FAD dependent oxidoreductase (aminomethyltransferases) pfam00496 - ABC transporter (di/oligopeptides) pfam01546 - Peptidase family M20/M25/M40 pfam00528 - ABC transporter (oligopeptides, carbohydrates, ions) pfam07676 - WD40-like Beta Propeller Repeat pfam12911 - Oligopeptide transport permease C 194 (13.5 %) 781 (10.8 %) 155 (10.8 %) 137 (9.5 %) 2.7 (0.01) 2.7 (0.16) 4 (0.01) 47 (3.3 %) 376 (5.2 %) 67 (4.7 %) 61 (4.2 %) -1.9 (0.01) -1.4 (0.17) -1 (0.3) 13 (0.6 %) 14 (0.1 %) 6 (0.3 %) 2 (0.1 %) 0.4 (0.05) 0.3 (1) 0.5 (1) 13 (0.6 %) 4 (0 %) 6 (0.3 %) 4 (0.2 %) 0.5 (<0.01) 0.3 (1) 0.4 (1) 12 (0.5 %) 11 (0.1 %) 6 (0.3 %) 2 (0.1 %) 0.4 (0.03) 0.2 (1) 0.4 (1) 32 (1.4 %) 56 (0.5 %) 8 (0.3 %) 25 (1.1 %) 0.8 (0.01) 1 (0.12) 0.3 (1) 9 (0.4 %) 4 (0 %) 1 (0 %) 6 (0.3 %) 0.3 (0.01) 0.3 (1) 0.1 (1) 8 (0.3 %) 3 (0 %) 2 (0.1 %) 2 (0.1 %) 0.3 (0.02) 0.3 (1) 0.3 (1) Energy production and conversion pfam01315 - Aldehyde oxidase (includes cox) pfam00248 - Aldo/keto reductase family (cox) 104 (7.2 %) 655 (9 %) 112 (7.8 %) 111 (7.7 %) -1.8 (0.06) -0.6 (0.84) -0.5 (0.86) 7 (0.3 %) 0 (0 %) 0 (0 %) 5 (0.2 %) 0.3 (<0.01) 0.3 (1) 0.1 (1) 9 (0.4 %) 7 (0.1 %) 5 (0.2 %) 10 (0.4 %) 0.3 (0.08) 0.2 (1) 0 (1) Cell wall, membrane, envelope biogenesis pfam00534 - Glycosyl transferase 1 136 (9.5 %) 535 (7.4 %) 84 (5.8 %) 109 (7.6 %) 2.1 (0.02) 3.6 (<0.01) 1.9 (0.18) 35 (1.5 %) 20 (0.2 %) 5 (0.2 %) 10 (0.4 %) 1.3 (<0.01) 1.3 (<0.01) 1.1 (0.12) pfam01476 - LysM 18 (0.8 %) 4 (0 %) 1 (0 %) 10 (0.4 %) 0.7 (<0.01) 0.7 (0.05) 0.3 (1) pfam00535 - Glycosyl transferase 2 pfam13231 - Dolichyl-phosphate-mannoseprotein mannosyltransferase pfam13579 - Glycosyl transferase 4 pfam01370 - NAD dependent epimerase/dehydratase family pfam02469 - Fasciclin domain 19 (0.8 %) 30 (0.3 %) 5 (0.2 %) 13 (0.6 %) 0.5 (0.07) 0.5 (1) 0.2 (1) 14 (0.6 %) 7 (0.1 %) 1 (0 %) 11 (0.5 %) 0.5 (<0.01) 0.5 (0.54) 0.1 (1) 12 (0.5 %) 5 (0 %) 0 (0 %) 11 (0.5 %) 0.5 (<0.01) 0.5 (0.17) 0 (1) 22 (0.9 %) 29 (0.3 %) 14 (0.6 %) 9 (0.4 %) 0.7 (0.01) 0.2 (1) 0.4 (1) 6 (0.3 %) 0 (0 %) 1 (0 %) 1 (0 %) 0.3 (0.01) 0.2 (1) 0.2 (1) Signal transduction mechanisms 22 (1.5 %) 204 (2.8 %) 32 (2.2 %) 54 (3.8 %) -1.3 (0.02) -0.7 (0.47) -2.2 (<0.01) Transcription 50 (3.5 %) 265 (3.7 %) 71 (4.9 %) 84 (5.8 %) -0.2 (0.83) -1.5 (0.18) -2.4 (0.01) pfam00990 - GGDEF domain 0 (0 %) 6 (0.1 %) 1 (0 %) 15 (0.7 %) -0.1 (1) 0 (1) -0.7 (0.06) pfam13492 - GAF domain 1 (0 %) 1 (0 %) 0 (0 %) 17 (0.7 %) 0 (1) 0 (1) -0.7 (0.1) 6 (0.3 %) 37 (0.3 %) 5 (0.2 %) 42 (1.8 %) -0.1 (1) 0 (1) -1.6 (<0.01) pfam00072 - Response regulator receiver Defense mechanisms 33 (2.3 %) 98 (1.4 %) 19 (1.3 %) 29 (2 %) 0.9 (0.02) 1 (0.21) 0.3 (0.89) pfam00664 - ABC transporter 12 (0.5 %) 9 (0.1 %) 2 (0.1 %) 7 (0.3 %) 0.4 (0.01) 0.4 (1) 0.2 (1) Carbohydrate transport and metabolism pfam03401 - Tripartite tricarboxylate transporter 92 (6.4 %) 375 (5.2 %) 89 (6.2 %) 140 (9.7 %) 1.2 (0.13) 0.2 (1) -3.3 (0.01) 0 (0 %) 52 (0.5 %) 15 (0.7 %) 0 (0 %) -0.5 (0.01) -0.7 (0.06) 0 (1) 29 (2 %) 45 (0.6 %) 8 (0.6 %) 7 (0.5 %) 1.4 (<0.01) 1.5 (<0.01) 1.5 (<0.01) 64 (4.4 %) 502 (6.9 %) 64 (4.4 %) 44 (3.1 %) -2.5 (<0.01) 0 (1) 1.4 (0.16) 27 (1.9 %) 103 (1.4 %) 37 (2.6 %) 12 (0.8 %) 0.5 (0.35) -0.7 (0.47) 1 (0.06) Cell motility Coenzyme transport and metabolism Intracellular trafficking, secretion, and vesicular transport Function unknown General function prediction only Replication, recombination and repair Translation, ribosomal structure and biogenesis Lipid transport and metabolism pfam00805 - Pentapeptide repeats pfam01391 - Collagen triple helix repeat (20 copies) pfam04773 - FecR protein 76 (5.3 %) 327 (4.5 %) 77 (5.4 %) 113 (7.9 %) 0.8 (0.35) -0.1 (1) -2.6 (0.02) 169 (11.7 %) 545 (7.5 %) 158 (11 %) 177 (12.3 %) 4.2 (<0.01) 0.8 (0.81) -0.6 (0.85) 60 (4.2 %) 309 (4.3 %) 120 (8.3 %) 80 (5.6 %) -0.1 (0.94) -4.2 (<0.01) -1.4 (0.19) 116 (8.1 %) 864 (11.9 %) 133 (9.2 %) 92 (6.4 %) -3.9 (<0.01) -1.2 (0.54) 1.7 (0.18) 53 (3.7 %) 395 (5.5 %) 60 (4.2 %) 37 (2.6 %) -1.8 (0.02) -0.5 (0.84) 1.1 (0.17) 8 (0.3 %) 0 (0 %) 1 (0 %) 0 (0 %) 0.3 (<0.01) 0.3 (1) 0.3 (1) 0 (0 %) 1 (0 %) 14 (0.6 %) 0 (0 %) 0 (1) -0.6 (0.06) 0 (1) 5 (0.2 %) 0 (0 %) 0 (0 %) 1 (0 %) 0.2 (0.03) 0.2 (1) 0.2 (1) Table S10. Bowtie2 read recruitment results. Total number of metatranscriptomic reads used and the number and percentage recruited to the CL500-11-LM coding genes. Season Station Fraction Layer Daytime Total reads Matches % recruited Spring M110 0.22-3 Surface Night 65,819,740 353,019 0.54 Spring M110 0.22-3 Surface Day 95,687,714 1,052,673 1.10 Spring M110 0.22-3 Deep Day 54,796,774 261,137 0.48 Summer M110 0.22-3 ChlaMax Day 71,004,444 143,646 0.20 Summer M110 0.22-3 Deep Night 20,000,000 1,043,156 5.22 Fall M110 0.22-3 Deep Night 20,000,000 1,252,373 6.26 Table S11: STAMP analysis metatranscriptomic data. Protein functions over- and underrepresented among highly expressed (5%, 10%, and 25% highest average expression levels across all six samples) and differentially expressed genes (surface to deep) relative to the CL500-11-LM sequence bin (CL500). Effect size is the differential between the relative abundance in the expression dataset and CL500-11-LM, and q-value is calculated based on Benjamini-Hochberg false discovery rate correction of the p-values resulting from the Chi-square or Fisher’s exact test for COGs and Pfams, respectively. All functions that are significantly differentially represented in at least one of the comparisons (p < 0.05; q < 0.15) were listed. COG / pfam Number of genes (% of all proteins) CL500 5% 10% 25% DE 5% effect size (q-value) 10% 25% DE Energy production and conversion 104 (4.5 %) 16 (14.5 %) 30 (13.3 %) 62 (10.6 %) 9 (11.4 %) 10.07 (<0.01) 8.8 (<0.01) 6.09 (<0.01) 6.92 (0.02) Posttranslational modification, protein turnover, chaperones 47 (2 %) 11 (10 %) 18 (8 %) 25 (4.3 %) 9 (11.4 %) 7.98 (<0.01) 5.94 (<0.01) 2.24 (0.01) 9.37 (<0.01) 21 (9.3 %) 14 (6.2 %) 9 (2.6 %) 10 (4.4 %) 24 (10.6 %) 4 (1.8 %) 59 (10.1 %) 24 (4.1 %) 9 (1.1 %) 25 (4.3 %) 55 (9.4 %) 10 (1.7 %) 8 (10.1 %) 3 (3.8 %) 6.2 (0.15) 0.94 (1) 1.7 (0.35) 1.78 (1) 1.04 (1) 0.6 (1) -0.47 (1) 0 (0 %) 1.37 (0.9) 0 (0 %) 1.09 (0.67) 3.44 (0.02) 2.09 (0.21) 2.27 (0.14) 5.63 (<0.01) 1.04 (0.21) 1.33 (0.3) 2 (2.5 %) 1.79 (0.78) 4.73 (<0.01) 1.48 (0.65) 17 (0.7 %) 16 (14.5 %) 5 (4.5 %) 9 (5.2 %) 4 (3.6 %) 7 (6.4 %) 2 (1.8 %) Coenzyme transport and metabolism 64 (2.8 %) 4 (3.6 %) 7 (3.1 %) 16 (2.7 %) 7 (8.9 %) 0.88 (0.95) 0.34 (1) -0.03 (1) 6.11 (0.01) Cell motility 29 (1.2 %) 2 (1.8 %) 6 (2.7 %) 16 (2.7 %) 0 (0 %) 0.57 (0.92) 1.41 (0.21) 1.48 (0.04) -1.25 (0.83) Defense mechanisms 33 (1.4 %) 2 (1.8 %) 3 (1.3 %) 5 (0.9 %) 6 (7.6 %) 0.4 (1) -0.09 (1) -0.57 (0.45) 6.17 (<0.01) Secondary metabolites biosynthesis, transport and catabolism 40 (1.7 %) 0 (0 %) 0 (0 %) 5 (0.9 %) 3 (3.8 %) -1.72 (0.72) -1.72 (0.15) -0.87 (0.33) 2.08 (0.64) 169 (7.3 %) 885 (38.1 %) 412 (14.9 %) 5 (4.5 %) 21 (19.1 %) 4 (2.3 %) 8 (3.5 %) 41 (18.1 %) 33 (5.6 %) 122 (20.8 %) 38 (4.5 %) 3 (3.8 %) 16 (20.3 %) -2.73 (0.66) -18.99 (<0.01) -12.59 (<0.01) -3.73 (0.13) -19.94 (<0.01) -12.92 (<0.01) -1.65 (0.38) -17.3 (<0.01) -10.38 (<0.01) -3.47 (0.69) -17.83 (0.01) -12.89 (0.07) Amino acid transport and metabolism Inorganic ion transport and metabolism pfam00496 - ABC transporter (di/oligopeptides) Transcription Translation, ribosomal structure and biogenesis Cell cycle control, cell division, chromosome partitioning General function prediction only None No pfam 194 (8.3 %) 64 (2.8 %) 13 (0.5 %) 50 (2.2 %) 116 (5 %) 7 (2 %) 0 (0 %) 2 (2 %) 2.11 (0.02) 4.38 (<0.01) 0.97 (0.1) 0.38 (1) -4.99 (0.18) -0.73 (0.97) Table S12: Differentially expressed genes. Genes identified as differentially expressed using DEseq2 (Benjamini-Hochberg false-discovery rate adjusted p < 0.05) between spring surface and all seasons deep samples. The fold change is the ratio of the average number of reads mapped for the deep samples and the average number of reads mapped to the same gene for the surface samples. log2 Fold p Change value Higher transcript levels at 108 m depth (All seasons) 3.96EGa0063436_10363 282.27 1.19 04 1.55EGa0063436_10608 236.12 1.04 04 Locus Avg Ga0063436_106012 262.90 1.18 Ga0063436_12044 740.20 0.90 Ga0063436_12195 309.82 0.89 Ga0063436_12953 481.56 1.21 Ga0063436_13261 58.46 1.73 Ga0063436_13843 97.67 1.37 Ga0063436_13845 534.87 1.14 Ga0063436_13847 63.88 2.04 Ga0063436_13848 21.31 2.16 Ga0063436_138411 18.79 1.71 Ga0063436_138412 74.03 2.58 Ga0063436_14013 46.72 1.25 Ga0063436_10022 179.82 -1.27 Ga0063436_10023 735.39 -1.58 Ga0063436_10024 68.19 -1.38 Ga0063436_10061 41.13 -2.23 Ga0063436_10062 91.88 -3.05 Ga0063436_10064 58.21 -1.97 Ga0063436_101425 118.36 -1.34 Ga0063436_10223 444.08 -1.16 Ga0063436_10225 379.23 -0.84 Ga0063436_10226 122.41 -1.06 Ga0063436_10411 49.56 -1.77 Ga0063436_10412 88.58 -1.74 Ga0063436_10421 112.06 -1.39 Ga0063436_10571 245.46 -0.99 2.36E05 5.27E04 5.64E04 9.91E04 6.28E05 1.21E04 3.09E04 1.86E05 2.16E05 8.27E04 3.00E08 1.57E03 p adjusted Annotation COG 1.22E-02 Ferrous iron transport protein B C terminus/Nucleoside recognition None 5.97E-03 acetyl-coenzyme A synthetase (EC 6.2.1.1) I 1.29E-03 1.54E-02 1.62E-02 2.43E-02 2.79E-03 4.76E-03 1.02E-02 1.12E-03 1.22E-03 2.15E-02 Aerobic-type carbon monoxide dehydrogenase, small subunit CoxS/CutS homologs carbohydrate ABC transporter ATP-binding protein, CUT1 family (TC 3.A.1.1.-) pfam pfam07664 pfam07670 pfam00501 pfam13193 C pfam00111 pfam01799 G pfam00005 pfam08402 Predicted acetamidase/formamidase C pfam03069 glutamine synthetase, type III E pfam00120 FR pfam07969 Cytosine deaminase and related metaldependent hydrolases electron transfer flavoprotein alpha subunit apoprotein Glycine cleavage system T protein (aminomethyltransferase) anthranilate synthase, component I (EC 4.1.3.27) anthranilate synthase, component II (EC 4.1.3.27) phosphoribosylanthranilate isomerase (EC 5.3.1.24) C E EH pfam00766 pfam01012 pfam01266 pfam01571 pfam08669 pfam00425 pfam04715 EH pfam00117 E pfam00697 6.04E-06 tryptophan synthase, beta subunit E pfam00291 3.70E-02 condensin subunit ScpA S pfam02616 1.01E-02 Adenylate cyclase, family 3 (some proteins contain HAMP domain) T pfam00211 pfam00672 1.46E-05 Bacteriorhodopsin R pfam01036 2.33E-02 Phytoene dehydrogenase and related proteins Q pfam01266 pfam13450 6.44E-05 AhpC/TSA antioxidant enzyme None pfam13911 1.28E-15 Arabinose efflux permease G pfam07690 8.88E-04 Phytoene dehydrogenase and related proteins Q pfam13450 3.62E-03 Geranylgeranyl pyrophosphate synthase H pfam00348 2.30E-03 FeS assembly ATPase SufC O pfam00005 3.90E-02 FeS assembly protein SufD O pfam01458 1.99E-02 cysteine desulfurases, SufSfamily E pfam00266 8.08E-04 Phytoene/squalene synthetase I pfam00494 1.20E-04 phytoene desaturase Q pfam01593 3.03E-03 Molybdopterin-binding domain of aldehyde dehydrogenase None 1.01E-02 cytochrome c oxidase, subunit II C Higher transcript levels at 5 m depth (Spring) 2.73E04 8.64E08 9.37E04 6.46E07 3.01E18 1.41E05 8.98E05 4.89E05 1.68E03 7.54E04 1.19E05 1.41E06 7.35E05 2.80E04 pfam01315 pfam02738 pfam00034 pfam00116 pfam02790 Ga0063436_10572 408.22 -1.07 Ga0063436_10676 76.74 -1.72 Ga0063436_10806 87.19 -1.47 Ga0063436_11352 38.64 -1.55 Ga0063436_11364 164.56 -1.02 Ga0063436_113726 177.60 -0.95 Ga0063436_11381 668.46 -1.07 Ga0063436_11533 48.50 -1.33 Ga0063436_11563 43.24 -2.32 Ga0063436_11564 19.40 -1.63 Ga0063436_11565 73.83 -2.48 Ga0063436_11566 27.94 -1.70 Ga0063436_116710 210.83 -1.76 Ga0063436_11851 949.46 -1.04 Ga0063436_12065 132.06 -1.56 Ga0063436_12101 43.87 -1.52 Ga0063436_12103 51.52 -1.61 Ga0063436_121110 279.81 -1.08 Ga0063436_121210 453.68 -1.24 Ga0063436_121211 949.67 -1.23 Ga0063436_12128 371.85 -0.87 Ga0063436_12193 83.72 -1.39 Ga0063436_122617 215.88 -1.12 Ga0063436_12264 765.81 -1.82 Ga0063436_12281 54.43 -1.59 Ga0063436_12282 68.55 -1.46 Ga0063436_12312 162.32 -2.41 Ga0063436_123211 305.25 -0.93 Ga0063436_12375 18.39 -2.63 Ga0063436_12432 413.33 -2.53 Ga0063436_12433 282.91 -2.53 Ga0063436_12892 367.02 -1.04 Ga0063436_12981 43.53 -2.10 Ga0063436_13092 299.91 -1.14 Ga0063436_13093 56.92 -1.26 Ga0063436_13218 558.44 -1.15 Ga0063436_13221 3265.88 -2.42 Ga0063436_13373 44.01 -1.31 4.09E05 9.93E07 3.26E04 3.15E04 3.09E04 1.18E03 2.45E05 7.22E04 1.16E08 5.77E04 4.18E11 3.09E04 3.21E08 1.42E05 4.06E07 3.14E04 3.25E04 6.59E05 3.05E07 2.72E07 4.51E04 3.67E05 2.03E-03 9.34E-05 1.02E-02 1.02E-02 1.02E-02 Heme/copper-type cytochrome/quinol oxidases, subunit 1 ABC-type multidrug transport system, ATPase component PUCC protein ABC-type antimicrobial peptide transport system, ATPase component Dipeptidyl peptidase IV (DPP IV) N-terminal region C pfam00115 V pfam00005 None pfam03209 V pfam00005 None pfam00930 2.85E-02 Excinuclease ABC subunit A L pfam00005 1.29E-03 ATP-dependent Zn proteases O pfam00004 pfam01434 1.94E-02 short chain dehydrogenase None pfam00106 2.81E-06 Predicted secreted hydrolase R pfam07143 1.63E-02 FtsX-like permease family None pfam02687 M pfam02687 pfam12704 V pfam00005 1.18E-08 1.02E-02 6.04E-06 8.88E-04 4.29E-05 1.02E-02 1.02E-02 2.79E-03 ABC-type transport system, involved in lipoprotein release, permease component ABC-type antimicrobial peptide transport system, ATPase component Trypsin-like serine proteases, typically periplasmic, contain C-terminal PDZ domain O pfam13180 pfam13365 pfam00004 pfam01434 ATP-dependent metalloprotease FtsH O Cytochrome c biogenesis factor O pfam01578 P pfam01497 P pfam01032 C pfam00180 ABC-type Fe3+-hydroxamate transport system, periplasmic component ABC-type Fe3+-siderophore transport system, permease component isocitrate dehydrogenase (NADP) (EC 1.1.1.42) 3.68E-05 Cytochrome c553 C pfam00034 3.55E-05 Cytochrome b subunit of the bc complex C pfam00032 pfam13631 1.36E-02 pyridoxal phosphate synthase yaaD subunit H pfam01680 1.88E-03 cysteine synthase (EC 2.5.1.47) E pfam00291 5.47E05 2.50E-03 Dinucleotide-utilizing enzymes involved in molybdopterin and thiamine biosynthesis family 2 H 3.99E07 4.29E-05 RNA polymerase sigma factor, sigma-70 family K 2.10E-03 Protein of unknown function (DUF3090) None pfam11290 1.64E-02 Fructose-2,6-bisphosphatase G pfam00300 7.49E-10 Cupredoxin-like domain None pfam13473 3.70E-02 Protease subunit of ATP-dependent Clp proteases OU pfam00574 hypothetical protein None None V pfam02687 pfam12704 V pfam00005 Glutamate-1-semialdehyde aminotransferase H pfam00202 1.72E-04 MerR HTH family regulatory protein None pfam13411 1.15E-03 2-oxoacid:acceptor oxidoreductase, alpha subunit C 2.17E-02 Transcriptional regulators K 4.34E05 5.91E04 2.21E12 1.57E03 2.18E07 6.83E22 6.35E21 1.78E03 2.24E06 1.97E05 8.44E04 6.21E06 2.12E06 9.24E- 3.08E-05 5.78E-19 3.58E-18 4.03E-02 4.38E-04 1.71E-04 2.33E-02 ABC-type antimicrobial peptide transport system, permease component ABC-type antimicrobial peptide transport system, ATPase component Membrane protease subunits, stomatin/prohibitin homologs Secreted and surface protein containing fasciclin-like repeats Cupin domain O M None pfam00581 pfam00899 pfam05237 pfam04539 pfam04542 pfam04545 pfam01558 pfam01855 pfam00392 pfam07729 pfam01145 pfam02469 pfam13345 pfam07883 04 Ga0063436_13382 236.67 -0.95 Ga0063436_13493 131.78 -1.60 Ga0063436_13571 104.17 -1.81 Ga0063436_13651 265.62 -1.10 Ga0063436_13811 38.02 -1.63 Ga0063436_13876 114.87 -1.18 Ga0063436_13981 651.51 -2.66 Ga0063436_14441 25.40 -1.82 Ga0063436_14491 36.06 -2.19 6.34E04 2.17E07 1.23E06 2.76E04 4.81E04 1.79E03 2.05E29 6.50E05 4.85E06 1.73E-02 thioredoxin None pfam00085 3.08E-05 hypothetical protein None None 1.10E-04 ABC-type nitrate/sulfonate/bicarbonate transport systems, periplasmic components P pfam09084 1.01E-02 chaperone protein DnaK O pfam00012 1.43E-02 MacB-like periplasmic core domain None pfam12704 4.03E-02 LysM domain None pfam01476 3.47E-26 RND family efflux transporter, MFP subunit V pfam12700 2.79E-03 Molybdopterin converting factor, small subunit H pfam02597 3.57E-04 Ribonucleotide reductase, beta subunit F pfam00268 Fig. S1: Microscopic measurements of CL500-11-LM. Average cell sizes measured for cells hybridizing to the CL500-11-specific probe in offshore spring surface (n = 48), summer deep (n = 48), and fall deep (n = 57) samples. For cells occurring in pairs, measurements of both the cell pair (both) as well as individual cells (single, single small (smallest cell of the pair), single large (largest cell of the pair)) were included. Error bars indicate 90% confidence interval and asterisk indicates significantly different means (t-test, p < 0.10). Figure S2: Extraction of the Chloroflexi sequence bin from the gamNGS merged assembly based on TETRA-eSOM analysis. Colors indicate taxonomic assignment of the contig, based on RAPsearch2 best hits. The inset provides a magnified view of the Chloroflexi bin. The two data points in the top of the inset that are highlighted with red boxes and white arrows are the two 3-kb sequence windows of the contig that contains a bacteriorhodopsin gene. Figure S3: Differential sequence coverage analysis of the Chloroflexi sequence bin. (A) Sequence read coverage resulting from bowtie2 alignment of three metagenomic datasets (10 million paired end read pairs from each) to the three Chloroflexi bins identified by MaxBin analysis of the TETRA-esom Chloroflexi bin (Fig. S2). Data was scaled to 100 % to normalize absolute read coverage differences, which shows that relative sequence coverage in MaxBin2 corresponded to iTag-based differences in relative abundance of CL500-11-LM (Fig. 2) patterns, MaxBin3 was a different population, and MaxBin1 was a mixture of CL500-11-LM and other populations. (B) Read recruitment data for the contig that contains the bacteriorhodopsin gene. Figure S4: Extraction of the refined Chloroflexi CL500-11-LM bin using the mmetagenome package. Each contig is represented by a contig, scaled by contig size, colored by Chloroflexi MaxBin 1-3, and plotted on the graph based on sequence read coverage determined by bowtie2 recruitment of ten million paired end reads from spring surface and summer deep water metagenomic datasets generated from the off-shore station.