AOV Assumption Checking and §8.4, 8.5) Transformations (
advertisement

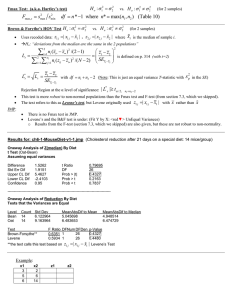
AOV Assumption Checking and Transformations (§8.4, 8.5) • How do we check the Normality assumption in AOV? • How do we check the Homogeneity of variances assumption in AOV? (§7.4) • What to do if these assumptions are not met? Model Assumptions • Homoscedasticity (common group variances). • Normality of responses (or of residuals). • Independence of responses (or of residuals). (Hopefully achieved through randomization…) • Effect additivity. (Only an issue in multi-way AOV; later). Checking the Equal Variance Assumption H0 : 12 22 2t HA: some of the variances are different from each other Little work but little power Hartley’s Test: A logical extension of the F test for t=2. Requires equal replication, n, among groups. Requires normality. Fmax 2 smax 2 smin Reject if Fmax > Fa,t,n-1, tabulated in Table 12. Bartlett’s Test More work but better power Bartlett’s Test: Allows unequal replication, but requires normality. T.S. t t 2 2 C (ni 1) loge s (ni 1) loge si i1 i1 If C > c2 (t-1),a then apply the correction term 1 t 1 1 t CF 1 3( t 1) i1 (ni 1) (ni 1) i1 R.R. Reject if C/CF > c2(t-1),a si2 s i1 t t 2 Levene’s Test More work but powerful result. Let zij yij yi median of yi = sample i-th group t T.S. L 2 n ( z z ) i i /(t 1) i 1 ni t 2 n ( z z ) i ij i /( nT t ) t nT ni i 1 i 1 j 1 R.R. Reject H0 if L Fa,df1 ,df2 Essentially an AOV on the zij df1 = t -1 df2 = nT - t Test for Equal Variances Response Resist Factors Sand ConfLvl 95.0000 Minitab Bonferroni confidence intervals for standard deviations Lower Sigma Upper N Factor Levels 1.70502 3.28634 14.4467 5 15 1.89209 3.64692 16.0318 5 20 1.07585 2.07364 9.1157 5 25 1.07585 2.07364 9.1157 5 30 1.48567 2.86356 12.5882 5 35 Bartlett's Test (normal distribution) Test Statistic: 1.890 P-Value : 0.756 Levene's Test (any continuous distribution) Test Statistic: 0.463 P-Value : 0.762 Stat > ANOVA > Test for Equal Variances Minitab Help Use Bartlett’s test when the data come from normal distributions; Bartlett’s test is not robust to departures from normality. Use Levene’s test when the data come from continuous, but not necessarily normal, distributions. The computational method for Levene’s Test is a modification of Levene’s procedure [10] developed by [2]. This method considers the distances of the observations from their sample median rather than their sample mean. Using the sample median rather than the sample mean makes the test more robust for smaller samples. Do not reject H0 since p-value > 0.05 (traditional a) SAS Program proc glm data=stress; class sand; model resistance = sand / solution; means sand / hovtest=bartlett; means sand / hovtest=levene(type=abs); means sand / hovtest=levene(type=square); means sand / hovtest=bf; /* Brown and Forsythe mod of Levene */ title1 'Compression resistance in concrete beams as'; title2 ' a function of percent sand in the mix'; run; Hovtest only works when one factor in (right hand side) model. hovtest=bartlett; Bartlett's Test for Homogeneity of resistance Variance Source sand DF Chi-Square Pr > ChiSq 4 1.8901 0.7560 SAS Levene's Test for Homogeneity of resistance Variance ANOVA of Absolute Deviations from Group Means Source sand Error hovtest=levene(type=abs); Sum of Mean DF Squares Square F Value Pr > F 4 8.8320 2.2080 0.95 0.4573 20 46.6080 2.3304 Levene's Test for Homogeneity of resistance Variance ANOVA of Squared Deviations from Group Means Source sand Error hovtest=levene(type=square); Sum of Mean DF Squares Square F Value Pr > F 4 202.2 50.5504 0.85 0.5076 20 1182.8 59.1400 Brown and Forsythe's Test for Homogeneity of resistance Variance ANOVA of Absolute Deviations from Group Medians Source sand Error hovtest=bf; Sum of Mean DF Squares Square F Value Pr > F 4 7.4400 1.8600 0.46 0.7623 20 80.4000 4.0200 SPSS RESIST Levene Statistic .947 Test of Homogeneity of Variances df1 4 df2 20 Sig. .457 Since the p-value (0.457) is greater than our (typical) a =0.05 Type I error risk level, we do not reject the null hypothesis. This is Levene’s original test in which the zij are centered on group means and not medians. R Tests of Homogeneity of Variances bartlett.test(): Bartlett’s Test. fligner.test(): Fligner-Killeen Test (nonparametric). Checking for Normality Reminder: Normality of the RESIDUALS is assumed. The original data are assumed normal also, but each group may have a different mean if HA is true. Practice is to first fit the model, THEN output the residuals, then test for normality of the residuals. This APPROACH is always correct. TOOLS 1. Histogram and/or boxplot of all residuals (eij). 2. Normal probability (Q-Q) plot. 3. Formal test for normality. Histogram of Residuals proc glm data=stress; class sand; model resistance = sand / solution; output out=resid r=r_resis p=p_resis ; title1 'Compression resistance in concrete beams as'; title2 ' a function of percent sand in the mix'; run; proc capability data=resid; histogram r_resis / normal; ppplot r_resis / normal square ; run; Probability Plots (QQ-Plots) A scatter plot of the percentiles of the residuals against the percentiles of a standard normal distribution. The basic idea is that if the residuals came from a normal distribution, values for these percentiles should lie on a straight line. • Compute and sort the residuals e(1), e(2),…, e(n). • Associate with each residual a standard normal percentile: z(i) = F-1((i-.5)/n). • Plot z(i) versus e(i). Compare to straight line (don’t care so much about which line). Percentile Residual Normal Percentile 0.02 -4.4 -2.0537 0.06 -3.8 -1.5548 0.10 -3.6 -1.2816 0.14 -3.4 -1.0803 0.18 -2.6 -0.9154 0.22 -2.6 -0.7722 0.26 -2.6 -0.6433 0.30 -1.6 -0.5244 0.34 -0.8 -0.4125 0.38 -0.6 -0.3055 0.42 0.2 -0.2019 0.46 0.2 -0.1004 0.50 0.4 0.0000 0.54 0.4 0.1004 0.58 0.4 0.2019 0.62 0.4 0.3055 0.66 1.4 0.4125 0.70 1.4 0.5244 0.74 1.4 0.6433 0.78 1.6 0.7722 0.82 2.4 0.9154 0.86 2.6 1.0803 0.90 3.6 1.2816 0.94 4.2 1.5548 0.98 5.4 2.0537 Software EXCEL: Use AddLine option. Percentile pi = (i-0.5)/n Normal percentile =NORMSINV(pi) MTB: Graph -> Probability Plot R: with residuals in “y” qqnorm(y) qqline(y) Excel Probability Plot Probability Plot - Percent Sand Data 2.500 2.000 Normal Percentiles 1.500 1.000 0.500 0.000 -6 -4 -2 -0.500 0 -1.000 -1.500 -2.000 -2.500 Data Percentiles 2 4 6 Probability Plot Minitab SAS (note axes changed) These look normal! Formal Tests of Normality Many, many tests (a favorite pass-time of statisticians is developing new tests for normality.) • Kolmogorov-Smirnov test; Anderson-Darling test (both based on the empirical CDF). • Shapiro-Wilk’s test; Ryan-Joiner test (both are correlation based tests applicable for n < 50). • D’Agostino’s test (n>=50). All quite conservative – they fail to reject the null hypothesis of normality more often than they should. Shapiro-Wilk’s W test e1, e2, …, en represent data ranked from smallest to largest. H0: The population has a normal distribution. HA: The population does not have a normal distribution. 1 W d a i (en j1 e j ) j1 k T.S. 2 Coefficients ai come from a table. R.R. Reject H0 if W < W0.05 n d (e i e ) 2 i 1 n If n is even 2 (n 1) k If n is odd. 2 k Critical values of Wa come from a table. Shapiro-Wilk Coefficients Shapiro-Wilk Coefficients Shapiro-Wilk W Table D’Agostino’s Test e1, e2, …, en represent data ranked from smallest to largest. H0: The population has a normal distribution. HA: The population does not have a normal distribution. T.S. (D 0.28209479) n Y 0.02998598 1 2 s n (e j e ) j1 n n R.R. (two sided test) Reject H0 if Y Y0.025 or Y Y0.975 D [ j j1 1 2 1 2 (n 1)]e j n 2s Y0.025 and Y0.975 come from a table of percentiles of the Y statistic. Transformations to Achieve Homoscedasticity What can we do if the homoscedasticity (equal variances) assumption is rejected? 1. Declare that the AOV model is not an adequate model for the data. Look for alternative models. (Later.) 2. Try to “cheat” by forcing the data to be homoscedastic through a transformation of the response variable Y. (Variance Stabilizing Transformations.) Square Root Transformation Response is positive and continuous. zi yi This transformation works when we notice the variance changes as a linear function of the mean. k i 2 i • Useful for count data (Poisson Distributed). • For small values of Y, use Y+.5. 35.00 30.00 Sample Variance Typical use: Counts of items when counts are between 0 and 10. k>0 25.00 20.00 15.00 10.00 5.00 0.00 0 10 20 Sample Mean 30 40 Logarithmic Transformation Response is positive and continuous. Z ln(Y ) Typical use: Growth over time. Concentrations. Counts of times when counts are greater than 10. k 2 i 2 i k>0 200 180 160 Sample Variance This transformation tends to work when the variance is a linear function of the square of the mean • Replace Y by Y+1 if zero occurs. • Useful if effects are multiplicative (later). • Useful If there is considerable heterogeneity in the data. 140 120 100 80 60 40 20 0 0 10 20 Sample Mean 30 40 ARCSINE SQUARE ROOT Response is a proportion. 1 Z sin Y arcsin Y With proportions, the variance is a linear function of the mean times (1-mean) where the sample mean is the expected proportion. i2 k i 1 i • Y is a proportion (decimal between 0 and 1). • Zero counts should be replaced by 1/4, and N by N-1/4 before converting to percentages Typical use: Proportion of seeds germinating. Proportion responding. Reciprocal Transformation Response is positive and continuous. 1 Z Y This transformation works when the variance is a linear function of the fourth power of the mean. • Use Y+1 if zero occurs. • Useful if the reciprocal of the original scale has meaning. Typical use: Survival time. i2 k i4 Power Family of Transformations (1) Suppose we apply the power transformation: Suppose the true situation is that the variance is proportional to the th power of the mean. In the transformed variable we will have: zy p i ki 2 i 2 If p is taken as 1-, then the variance of Z will not depend on the mean, i.e. the variance will be constant. This is a Variance stabilizing transformation. p 1 i Power Family of Transformations (2) With replicated data, can sometimes be found empirically by fitting: Estimate: ln( ˆ i ) C ln( ˆ i ) 2 1 ni 2 ˆi ( y y ) ij i ni 1 j 1 ˆ i yi can be estimated by least squares (regression – Next Unit). pˆ 1 ˆ If p̂ is zero use the logarithmic transformation. Box/Cox Transformations (advanced) suggested transformation geometric mean of the original data. y i 1 0 1 zi ln y 0 i 1 n exp lny i n i 1 Exponent, , is unknown. Hence the model can be viewed as having an additional parameter which must be estimated (choose the value of that minimizes the residual sum of squares). Handling Heterogeneity no Regression? yes ANOVA Fit Effect Model Fit linear model Test for Homoscedasticity Plot residuals reject Transform Not OK OK accep t Traditional Box/Cox Family Power Family Transformed Data OK Transformations to Achieve Normality Regression? no ANOVA yes Fit linear model Estimate group means Probability plot Formal Tests yes Residuals Normal? no Transform Different Model OK