Continued
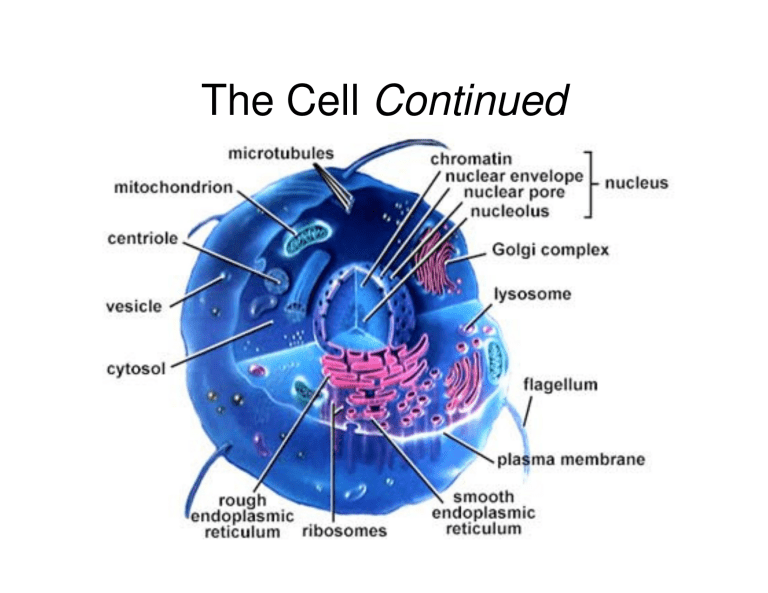
The Cell Continued
Golgi Apparatus
• Directs the trafficking of synthesized proteins
• Functions in modification, concentration, and packaging of proteins made in the rough ER
• Transport vessels from the ER fuse with the cis face of the Golgi apparatus
• Modifications involve the addition or removal of sugars and phosphates
Golgi Apparatus
• Proteins then pass through the Golgi apparatus to the trans face
• Secretory vesicles leave the trans face of the Golgi stack and move to designated parts of the cell
– Three paths can be take:
– Exocytosis
– Plasma membrane
– lysosome
Golgi Apparatus
Figure 3.20a
Role of the Golgi Apparatus
Cisterna
Rough ER
Proteins in cisterna
Phagosome
Membrane
Vesicle
Lysosomes containing acid hydrolase enzymes
Vesicle incorporated
Pathway 3 into plasma membrane
Coatomer coat
Golgi apparatus
Pathway 1
Proteins
Pathway 2
Secretory vesicles
Plasma membrane
Secretion by exocytosis
Extracellular fluid
Figure 3.21
Lysosomes
• Spherical membranous bags containing digestive enzymes
– Low pH in lysosome
• Abundant in phagocytes
• Digest ingested bacteria, viruses, and toxins
• Degrade nonfunctional organelles
• Breakdown glycogen and release thyroid hormone
Lysosomes
• Breakdown nonuseful tissue (during development, termed autolysis) and organelles
(lysosomal rupture yields cell death via autolysis)
– Digest particles taken in by endocytosis
– Breakdown glycogen for energy
– Breakdown bone to release Ca2 +
• Secretory lysosomes are found in white blood cells, immune cells, and melanocytes
Endomembrane System
• System of organelles that function to:
– Produce, store, and export biological molecules
– Degrade potentially harmful substances
• System includes:
– Nuclear envelope, smooth and rough ER, lysosomes, vacuoles, transport vesicles, Golgi apparatus, and the plasma membrane
Endomembrane System
Figure 3.23
Peroxisomes
• Membranous sacs containing oxidases and catalases
• Detoxify harmful or toxic substances
• Neutralize dangerous free radicals
– Free radicals – highly reactive chemicals with unpaired electrons (i.e., O
2
– )
– Oxidases + free radicals H
2
O
2
+ catalase
H
2
O
Cytoskeleton
• The “skeleton” of the cell
• Consists of microtubules, microfilaments, and intermediate filaments running through the cytosol to give structure and to support the organelles
Cytoskeleton
Figure 3.24a-b
Microtubules
• Dynamic, hollow tubes made of the spherical protein tubulin
• Determine the overall shape of the cell and distribution of organelles
Microfilaments
• Dynamic strands of the protein actin
• Attached to the cytoplasmic side of the plasma membrane
• Braces and strengthens the cell surface
• Attach to CAMs (cell adhesion molecule) and function in endocytosis and exocytosis, as well as the
“crawling” motion associated with amoeba
• Involved in the formation of cleavage furrows during cell division
Microfilaments
Intermediate Filaments
• Tough, insoluble protein fibers woven like rope and exhibit high tensile strength
• Stable, permanent cytoskeletal elements
• Serve as tracks for motility
• Attach to desmosomes and act as guy wires to resist pulling forces exerted on the cell
Intermediate Filaments
Cilia
• Whip-like, motile cellular extensions on exposed surfaces of certain cells
• Move substances in one direction across cell surfaces
• E.g. in the respiratory tract, cilia move dust laden mucous away from the lungs
Cilia
Figure 3.27a
Flagella
• Substantially longer cilia
• Only example in human is sperm
• Self propulsion
• Microtubules = 9 triplets
• Cilia/flagella = 9 doublets surrounding a central pair
Nucleus
• Contains nuclear envelope, nucleoli, chromatin, and distinct compartments rich in specific protein sets
• Gene-containing control center of the cell
• Contains the genetic library with blueprints for nearly all cellular proteins
• Dictates the kinds and amounts of proteins to be synthesized
Nucleus
Figure 3.28a
Nucleus
• Most cells have a single nucleus, but some are multinucleated and are often associated with large cytoplasmic mass and high protein production
• Mature red blood cells (RBCs) are anuclated and do not divide (why?)
• Largest of all cytoplasmic organelles
• Divided into three regions:
–Nuclear envelope
–Nucleoli
–Chromatin
Nuclear Envelope
• Selectively permeable double membrane barrier containing pores
• Encloses jellylike nucleoplasm, which contains essential solutes
• Outer membrane is continuous with the rough
ER and is studded with ribosomes on its external face
• Inner membrane is lined with the nuclear lamina, which maintains the shape of the nucleus
• Pore complex regulates transport of large molecules into and out of the nucleus
Nucleolus
• Dark-staining spherical bodies within the nucleus
• Non-membrane bound structure
• Site of ribosome subunit production
Chromatin
• Threadlike strands of DNA and histones
• Arranged in fundamental units called nucleosomes (8 histones wrapped twice w/ DNA)
• Histones help pack the long strands of DNA and are involved in gene regulation
• Form condensed, barlike bodies of chromosomes when the nucleus starts to divide
Figure 3.29
End of The Cell
• Let’s see that Harvard movie again and see what we’ve learned http://multimedia.mcb.harvard.edu/media.html
Cell Cycle
• Two major periods:
• Interphase
– Growth (G
1
), synthesis (S), growth (G
2
)
• Mitotic phase (Cell
Division
– Mitosis and cytokinesis
Figure 3.30
Interphase
• Period from cell formation to cell division
• “Resting phase”: period between divisions
• During interphase, cell carries out all its routine activities
• G
1
(gap 1) – metabolic activity and vigorous growth
• G
0
– cells that permanently cease dividing
• S (synthetic) – DNA replication
• G
2
(gap 2) – preparation for division
DNA Replication
• Replication begins simultaneously on several chromatin threads & continues until all DNA has been replicated
Steps in DNA Replication
• 1) DNA helices unwind from the nucleosomes
• 2) Helicase untwists the double helix into 2 complementary nucleotide chains exposing the nitrogenous bases
• 3) Each nucleotide strand serves as a template for building a new complementary strand.
Occurs at the replication fork
Steps in DNA Replication
• 4) The replisome uses RNA primers to begin
DNA synthesis of 2 nd strand
• 5) DNA polymerase III continues from the primer and covalently adds complementary nucleotides to the template
• 6) DNA ligase splices the short segments together
Steps in DNA Replication (cont.)
• 7) After replication:
• histones associate w/ the DNA
• chromatin strands condense forming chromatids and are held together by the centromere
• at anaphase, chromatids are distributed to each daughter cell
Steps in DNA Replication (cont.)
• Since DNA polymerase only works in one direction:
– A continuous leading strand is synthesized
– A discontinuous lagging strand is synthesized
– DNA ligase splices together the short segments of the discontinuous strand
• This process is called semiconservative replication
• Two new telomeres are also synthesized
DNA Replication
Figure 3.31
http://video.yahoo.com/watch/648006/30174
82
• Video of DNA Replication
Protein Synthesis
• Many genes contain exons, regions encoding for a polypeptide sequence, and
• Introns, noncoding intervening sequences.
We are still not sure why introns exist
From DNA to Protein
Nuclear envelope
Transcription
RNA Processing mRNA
DNA
Pre-mRNA
Ribosome
Translation
Polypeptide
Figure 3.33
Roles of the Three Types of
RNA
• Messenger RNA (mRNA) – carries the genetic information from DNA in the nucleus to the ribosomes in the cytoplasm
• Transfer RNAs (tRNAs) – bound to amino acids base pair with the codons of mRNA at the ribosome to begin the process of protein synthesis
• Ribosomal RNA (rRNA) – a structural component of ribosomes
Genetic Code
• Rules by which base sequences of a gene are translated into an amino acid sequence.
• 2 Major steps: Transcription
Translation
Transcription
• Transfer of information from the sense strand of DNA to RNA
• Transcription factor
– Loosens histones from DNA in the area to be transcribed
– Binds to promoter, a DNA sequence specifying the start site of RNA synthesis
– Mediates the binding of RNA polymerase to promoter
Transcription: RNA Polymerase
• An enzyme that oversees the synthesis of
RNA
• Unwinds the DNA template
• Adds complementary ribonucleoside triphosphates on the DNA template
• (5’ to 3’ direction)
• Joins these RNA nucleotides together
• Encodes a termination signal to stop transcription
Coding strand Promoter
Template strand
Unwound DNA
Termination signal
RNA polymerase
Transcription unit
In a process mediated by a transcription factor, RNA polymerase binds to promoter and unwinds 16–18 base pairs of the DNA template strand
RNA polymerase bound to promoter
RNA nucleotides mRNA synthesis begins mRNA RNA nucleotides RNA polymerase moves down DNA; mRNA elongates
(b)
RNA polymerase mRNA synthesis is terminated
DNA
(a)
Rewinding of DNA
Coding strand mRNA transcript
RNA polymerase
Unwinding of DNA
Template strand mRNA
RNA nucleotides
RNA-DNA hybrid region
Figure 3.34
Transcription
• Codon: 3 base sequence on mRNA corresponding to a specific amino acid
• 4 nucleotides (A,C,G,U) in RNA so there are:
• 4 3 = 64 codons
• 3 are stop codons (termination of the polypeptide chain)
• 61 code for amino acids
• There are only 20 amino acids, so more than 1 codon codes for a specific amino acid
Genetic Code
• RNA codons code for amino acids according to a genetic code
Figure 3.35
Transcription
• Pre mRNA contains introns and exons
• Pre mRNA is processed whereby the introns are spliced out and the exons are spliced together.
• This is done by the splicesome
• mRNA complex proteins then associate w/ the processed mature mRNA and guide its export from the nucleus
Transcription
Translation
• Nucleic acid sequences are “translated” into amino acid sequences (polypeptides)
• Occurs in the cytoplasm and involves mRNA, tRNA, rRNA
• A leader sequence on mRNA attaches to the small subunit of the ribosome by base pairing to rRNA (ribosomal RNA)
• tRNA (transfer RNA) loads a single amino acid, migrates to the ribosome, and maneuvers the amino acid into position as specified by the mRNA
• tRNA has 2 active sites: 1) binding of amino acid at one end, and 2) a 3-base complementary to the mRNA codon
(anticodon) calling for the amino acid carried by the particular tRNA
Translation
• Anticodons form hydrogen bonds w/ complementary codons (base pairing)
• tRNA is the link (translator) between nucleic acids and amino acids
• There are 45 different tRNAs each capable of binding to a specific amino acid
Information Transfer from DNA to RNA
• DNA triplets are transcribed into mRNA codons by RNA polymerase
• Codons base pair with tRNA anticodons at the ribosomes
• Amino acids are peptide bonded at the ribosomes to form polypeptide chains
• Start and stop codons are used in initiating and ending translation
Nuclear membrane
Nuclear pore
RNA polymerase
Nucleus mRNA
Template strand of DNA
Released mRNA
1 After mRNA processing, mRNA leaves nucleus and attaches to ribosome, and translation begins.
Codon 15
Small ribosomal subunit
Codon 16 Codon 17 Direction of ribosome advance
Amino acids tRNA
Aminoacyl-tRNA synthetase
Portion of mRNA already translated
4 Once its amino acid is released, tRNA is ratcheted to the E site and then released to reenter the cytoplasmic pool, ready to be recharged with a new amino acid.
tRNA “head” bearing anticodon
Large ribosomal subunit
3 As the ribosome moves along the mRNA, a new amino acid is added to the growing protein chain and the tRNA in the A site is translocated to the P site.
2 Incoming aminoacyltRNA hydrogen bonds via its anticodon to complementary mRNA sequence (codon) at the A site on the ribosome.
Energized by ATP, the correct amino acid is attached to each species of tRNA by aminoacyl-tRNA synthetase enzyme.
Figure 3.36
Information Transfer from DNA to RNA
Figure 3.38