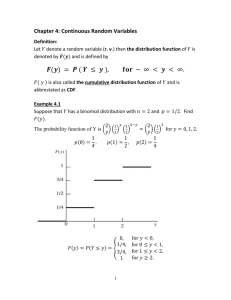
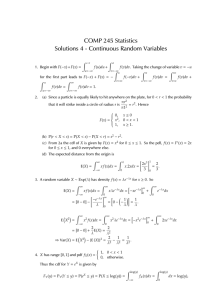
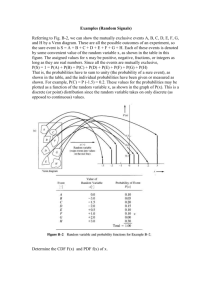
CDF thesis: lab meeting, Mar 17, 2004
advertisement
Computational analysis of membrane proteins implicated in metal transport in Arabidopsis thaliana CIAVVLCLVFMSVEVVGGIKANSLAILTDAAHLLSDVAAFAISLFSLWAAGWEATPRQTYGFFRIEILGALVSIQLIWLLT ALFLLINTAYMVVEFVAGFMSNSLGLISDACHMLFDCAALAIGLYASYISRLPANHQYNYGRGRFEVLSGYVNAVFLVLVG CFVVVLCLLFMSIEVVCGIKANSLAILADAAHLLTDVGAFAISMLSLWASSWEANPRQSYGFFRIEILGTLVSIQLIWLLT LIAVLLCAIFIVVEVVGGIKANSLAILTDAAHLLSDVAAFAISLFSLWASGWKANPQQSYGFFRIEILGALVSIQMIWLLA ---IFLYLIVMSVQIVGGFKANSLAVMTDAAHLLSDVAGLCVSLLAIKVSSWEANPRNSFGFKRLEVLAAFLSVQLIWLVS Stefanie Hartmann Max Planck Institute for Molecular Plant Physiology Supervisors: Joachim Selbig, Ute Krämer QuickTime™ and a TIFF (Uncompress ed) decompress or are needed to s ee this picture. Qu ic kTi me™ a nd a TIFF (U nc omp res se d) de co mpre ss or are n ee de d to se e thi s p i cture . Quick Time™a nd a TIFF ( Uncomp res sed) deco mpre ssor are n eede d to s ee this picture . Qu i ck Ti me ™a nd a TIFF (Unc om pres se d) de co mp re ss or are n ee de d to s ee th is pi ctu re . Q ui ck Ti m e ™ an d a T I FF ( U nc om p r es se d) de co m pr e ss or ar e n ee de d t o se e t hi s p i ct ur e . Q ui ck Ti m e ™ an d a T I FF ( U nc om p r es se d) de co m pr e ss or ar e n ee de d t o se e t hi s p i ct ur e . Q ui ck Ti m e ™ an d a T I FF ( U nc om p r es se d) de co m pr e ss or ar e n ee de d t o se e t hi s p i ct u re . QuickTi me™ and a T IFF (Uncom pressed) decom pressor are needed to see t his pict ure. QuickTime™ and a TIFF (Uncompress ed) decompress or are needed to s ee this picture. Qu ic kTi me™ a nd a TIFF (U nc omp res se d) de co mpre ss or are n ee de d to se e thi s p i cture . Quick Time™a nd a TIFF ( Uncomp res sed) deco mpre ssor are n eede d to s ee this picture . Qu i ck Ti me ™a nd a TIFF (Unc om pres se d) de co mp re ss or are n ee de d to s ee th is pi ctu re . 12 membrane proteins involved in metal transport in Arabidopsis Q ui ck Ti m e ™ an d a T I FF ( U nc om p r es se d) de co m pr e ss or ar e n ee de d t o se e t hi s p i ct ur e . Q ui ck Ti m e ™ an d a T I FF ( U nc om p r es se d) de co m pr e ss or ar e n ee de d t o se e t hi s p i ct ur e . Q ui ck Ti m e ™ an d a T I FF ( U nc om p r es se d) de co m pr e ss or ar e n ee de d t o se e t hi s p i ct u re . QuickTi me™ and a T IFF (Uncom pressed) decom pressor are needed to see t his pict ure. Metal transporters are of great importance because… …they provide an adequate supply of essential trace metals …they prevent an excess of these potentially toxic ions in silico analyses may help design further experiments on • basic research on metal homeostasis • development of new ways of phytoremediation Cation Diffusion Facilitator (CDF) proteins also referred to as cation efflux (CE) proteins • occur in archaea, bacteria, eukaryotes • are involved in transporting heavy metals (Co2+, Cd2+, Zn2+, Ni2+) • the CDF family of proteins had 13 members in 1997 • the CE Pfam family today has 348 members (July 2003) 426 (Jan 2004) CDF signature sequence: S X (ASG) (LIVMT)2 (SAT) (DA) (SGAL) (LIVFYA) (HDN) X3 D X2 (AS) The Arabidopsis thaliana CDF protein family CDF1: At2g46800 S LAILTDAAHLLS D VAA CDF2: At3g61940 CDF3: At3g58810 CDF4: At2g29410 CDF5: At2g04620 CDF6: At2g47830 CDF7: At2g39450 CDF8: At1g16310 CDF9: At1g79520 CDF10: At3g58060 CDF11: At3g12100 CDF12: At1g51610 S LAILADAAHLLT D VGA exact match S LAILTDAAHLLS D VAA S LAVMTDAAHLLS D VAG S LGLISDACHMLF D CAA 1 mismatch S TAIIADAAHSVS D VVL S LAIIASTLDSLL D LLS S MAVIASTLDSLL D LLS 2 mismatches S MAVIASTLDSLL D LLS S IAIAASTLDSLL D LMA R VGLVSDAFHLTF G CGL S HVIMAEVVHSVA D FAN 3 mismatches 4 mismatches Research questions: Can all 12 proteins be classified as CDF proteins? i.e., are there predicted structural and functional similarities of these 12 Arabidopsis proteins? secondary structure prediction, inclusion in membrane- and transporter databases, evaluation of common motifs, etc Research questions: Can all 12 proteins be classified as CDF proteins? i.e., are there predicted structural and functional similarities of these 12 Arabidopsis proteins? secondary structure prediction, inclusion in membrane- and transporter databases, evaluation of common motifs, etc What are the relationships of the 12 Arabidopsis proteins among each other and to other published sequences? intron/exon structure, phylogenetic reconstructions Research questions: Can all 12 proteins be classified as CDF proteins? i.e., are there predicted structural and functional similarities of these 12 Arabidopsis proteins? secondary structure prediction, inclusion in membrane- and transporter databases, evaluation of common motifs, etc What are the relationships of the 12 Arabidopsis proteins among each other and to other published sequences? intron/exon structure, phylogenetic reconstructions Is it possible to predict the 3D structure of these proteins? fold recognition by threading Sequence retrieval - four ambiguous sequences TIGR Arabidopsis thaliana database TAIR: The Arabidopsis Information Resource MIPS Arabidopsis thaliana genome database • different assignment of introns, use of alternative start codons Sequence analysis - three additional ambiguous sequences SWALL Pfam vs. TIGR/TAIR/MIPS • insertions and deletions, different amino acid sequence Cloning and RT-PCR revealed correct sequences for six of the seven ambiguous CDFs Inclusion in membrane and transport databases cation efflux, Pfam entry PF01545 Membrane Protein Library (AMPL) CDF1 CDF2 Arabidopsis ARAMEMNON Transport Protein Database PlantsT CDF3 CDF4 CDF5 () - CDF6 CDF7 - CDF8 - - CDF9 - - CDF10 - CDF11 CDF12 - Inclusion in membrane and transport databases cation efflux, Pfam entry PF01545 Membrane Protein Library (AMPL) CDF1 CDF2 Arabidopsis ARAMEMNON Transport Protein Database PlantsT CDF3 CDF4 CDF5 () () - CDF6 () () CDF7 () () - CDF8 () () - - CDF9 () () - - CDF10 () - CDF11 () CDF12 () - Inclusion in membrane and transport databases cation efflux, Pfam entry PF01545 Membrane Protein Library (AMPL) CDF1 CDF2 Arabidopsis ARAMEMNON Transport Protein Database PlantsT CDF3 CDF4 CDF5 () () – CDF6 () () CDF7 () () – CDF8 () () – – CDF9 () () – – CDF10 () – CDF11 () CDF12 () – Hidden Markov models used for secondary structure prediction cytoplasmic side membrane non-cytoplasmic side • states (loops, transmembrane domains, etc) are defined • states are connected in a biologically reasonable way (transitions) • each state has a specific probability distribution over the 20 amino acids • each transition has a specific transition probability • amino acid probabilities and transition probabilities are learned • models are first taught using a training set, the trained model is then used for the prediction Results of secondary structure predictions number of TMD N-terminus within cytoplasm CDF1 6 2/3 CDF2 6 3/3 CDF3 6 2/3 CDF4 5-6 2/3 CDF5 6 CDF6 0-6 1/3 CDF7 4-6 2/3 CDF8 5-6 3/3 CDF9 5-6 3/3 CDF10 4-6 2/3 CDF11 6 3/3 CDF12 4-6 3/3 TMHMM v2 HMMTOP v2 Memsat2 (14) (Tusnady and Simon, 1998, 2001) (Sonnhammer et al. 1998) (Jones et al. 1994, McGuffin et al. 2000) 3/3 Results of secondary structure predictions number of TMD N-terminus within cytoplasm CDF1 6 2/3 CDF2 6 3/3 CDF3 6 2/3 CDF4 5-6 2/3 CDF5 6 CDF6 0-6 1/3 CDF7 4-6 2/3 CDF8 5-6 3/3 CDF9 5-6 3/3 CDF10 4-6 2/3 CDF11 6 3/3 CDF12 4-6 3/3 TMHMM v2 HMMTOP v2 Memsat2 (14) (Tusnady and Simon, 1998, 2001) (Sonnhammer et al. 1998) (Jones et al. 1994, McGuffin et al. 2000) 3/3 CDF signature CE signature Prediction of subcellular localization mTP: mitochondrial targeting peptide cTP: chloroplast transit peptide SP: signal peptide (ER/secretory pathway) Prediction of subcellular localization - methods • N-terminal sorting signals display characteristic amino acid compositions • sequence-based methods predicting N-terminal sorting signals are based on this observation TargetP mTP, cTP, SP neural network-based iPSORT mTP, cTP, SP decision list Predotar mTP, cTP neural network-based SignalP NN SignalP HMM mTP: mitochondrial targeting peptide SP neural network-based SP based on hidden Markov models cTP: chloroplast transit peptide SP: signal peptide (ER/secretory pathway) Prediction of subcellular localization - results TargetP iPSORT Predotar SignalP NN HMM CDF1 CDF2 3/4 CDF3 CDF4 CDF5 CDF6 mTP cTP cTP cTP mTP cTP* mTP* 1/4 CDF7 CDF8 2/4* Y* CDF9 CDF10 CDF11 CDF12 mTP: mitochondrial targeting peptide mTP mTP cTP: chloroplast transit peptide SP: signal peptide (ER/secretory pathway) Exon structure of the CDF proteins # of exons CDF1 1 CDF2 1 CDF3 1 CDF4 1 CDF5 1 CDF11 9 CDF6 12 CDF12 13 CDF7 6 CDF8 6 CDF9 7 CDF10 5 Gene organization of the CDF proteins CDF1 CDF1 CDF2 CDF2 CDF3 CDF3 CDF4 CDF4 CDF5 CDF5 CDF11 CDF10 CDF6 CDF6 CDF12 CDF12 CDF7 CDF7 CDF8 CDF8 CDF9 CDF9 CDF11 CDF10 Phylogenetic Relationships within Cation Transporter Families of Arabidopsis Plant Physiology 2001; 126 (4): 1646–1667 omitted: CDFs 5, 7, 8, 9 CDF6 CDF11 CDF4 CDF3 CDF2 CDF10 CDF12 CDF1 Phylogenetic analysis of the Arabidopsis CDF proteins AtCDF4 100 AtCDF3 group I 100 AtCDF1 98/94/99 AtCDF2 AtCDF12 100 AtCDF6 AtCDF10 67/–/69 100 AtCDF7 group II 86/100/95 –/9479 AtCDF9 100 AtCDF8 AtCDF5 100/ 73/68 8 AtCDF11 RmCzcD Phylogenetic analysis of sequences containing the CE signature Escherischia coli ZITB Ralstonia metallidurans CZCD Ralstonia metallidurans CZCD Mus musculus ZNT4 Rattus norvegigus ZNT2 Mus musculus ZNT3 Arabidopsis thaliana CDF4 Arabidopsis thaliana CDF2 Eucalyptus grandis Arabidopsis thaliana CDF3 Arabidopsis thaliana CDF1 Thlaspi caerulescens ZTP1 Thlaspi goesingense MTP1 Thlaspi goesingense MTP1 Thlaspi goesingense MTP1 Oryza sativa Zea mays Lotus japonicus Medicago trunculata Oryza sativa Triticum aestivum Caenorhabditis elegans CDF1 Rattus norvegicus ZNT1 Schizosachharomyces pombe ZHF Saccharomyces cerevisiae COT1 Saccharomyces cerevisiae ZRC1 Oryza sativa Stylosanthes hamata MTP1 Oryza sativa Arabidopsis thaliana CDF10 Arabidopsis thaliana CDF7 Stylosanthes hamata MTP4 Stylosanthes hamata MTP2 Stylosanthes hamata MTP3 Oryza sativa Arabidopsis thaliana CDF8 Arabidopsis thaliana CDF9 Arabidopsis thaliana CDF6 Saccharomyces cerevisiae Mmt2 Saccharomyces cerevisiae Mmt1 T. thermophilus czrB Oryza sativa Arabidopsis thaliana CDF12 Homo sapiens ZNT5 Homo sapiens ZTL1 Homo sapiens ZNT7 Homo sapiens ZNT6 Arabidopsis thaliana CDF11 S. cerevisiae MSC2 Oryza sativa Arabidopsis thaliana CDF5 Staphylococcus aureus Staphylococcus aureus Bacillus stearothermophilus Bacillus stearothermophilus Arabidopsis group I sequences, monocot and dicot sequences, mammalian metal transporters Arabidopsis group II sequences, monocot and dicot sequences, prokaryotic and eukaryotic seqs several two-domain proteins outgroup working model: topology of Arabidopsis CDF proteins CDF signature sequence cell exterior/organelle cytoplasm N C Information derived from the 3D structure of a protein assignment of function guide mutagenesisexperiments ligand and functional sites evolutionary relationships residue solvent exposure putative interaction sites Structure determination 1. Classical approaches • X-ray crystallography • NMR spectroscopy 2. Computational approaches • comparative (“homology”) modeling • fold recognition (“threading”) • ab initio methods The basis of fold recognition (“threading”) The number of folds occurring in nature is limited: QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. PDB statistics: http://www.rcsb.org/pdb/holdings.html There are many sequences with no significant sequence identity but with the same or similar folds …HEAIDHKPKLTGMKTGRVVSSMKSNFFADLP… …HDGRSSMTRFSRYFRKTGRVSEYYKKQERLLE… Fold recognition methods aim: to find an optimal sequence-structure alignment 1. “threading” of an unknown target sequence into the backbone structure of template proteins of known structure ………CLVFMSVEVVGGIKANSLAILTD……… Fold recognition methods 2. evaluation of the compatibility between target sequence and proposed 3D structure using environment-based mean force potentials or using knowledge-based mean force potentials 3. Output: 4.99 Å a list of folds (sorted or unsorted), their “compatibility score”, sometimes other information such as SCOP descriptors, alignment, rudimentary 3D model of the query protein, raw scores, solvation energy for the model, links No new insights regarding the structure of CDF proteins Membrane proteins are significantly under-represented in structural databases – and therefore also in fold libraries If there is no fold similar to the native fold of the target protein, this approach cannot succed. Threading methods cannot be used for modeling of transmembrane proteins Will the 3D structure of CDFs be available soon? • for fold recognition methods to be used successfully: significantly more 3D structures of membrane proteins are needed • fold recognition methods specifically for integral membrane proteins may eventually be developed • cyrystallization of bacterial homologs and subsequent extraploation of structural features as an alternative? • approach for globular proteins: predicting a protein’s solubility and propensity to crystallize, based on results from high-throughput structure determination Can threading results be used as an independent way to verify group assignment? Were some structural hits specific for any of the CDF groups? 1. Which hits were common to which of the CDF sequences? 1 • • • • • • • 2 • • • • • • 3 • • • • • • 4 • • • • • • 5 • • • • • • • • • • • 1 • • • • • • 2 • • • 3 • • • • 2. “Phylothreading” • • • • 4 • • 5 Can threading results be used as an independent way to verify group assignment? Were some structural hits specific for any of the CDF groups? 1. Which hits were common to which of the CDF sequences? 1 • • • • • • • 2 • • • • • • 3 • • • • • • 4 • • • • • • 5 • • • • • • • • • • • 1 • • • • • • 2 • • • 3 • • • • 2. “Phylothreading” • • • • 4 • • 5 Which hits were common to which of the CDF sequences? Structural hits predicted • for most CDF sequences • for group I sequences • for group II sequences • for CDF5 and CDF11 • for CDF6 and CDF12 1… …170 1 • • • • • • 2 • • • • • • … • • • • • • 11 • • • • • • 12 • • • • • • • • • • • • • • • • • • • • • • Results were unable to provide evidence to verify group assignments based on other methods • • • • • • • • • “Phylothreading” CDF1 CDF2 CDF3 CDF4 CDF5 CDF6 78 CDF7 99 68 CDF8l CDF8s CDF9 CDF10 CDF11 CDF12 Phylothreading results can neither verify nor refute group assignments based on other methods Threading: non-transmembrane CDF fragments cell exterior/organelle cytoplasm N C N-terminus histidine-rich loop between TMD 4 and 5 C-terminus “Phylothreading”: CDF C-terminal fragments CDF1 CDF2 group I CDF3 “phylothreading” results confirm the assignment of CDF sequences to groups that were based on independent methods CDF4 CDF5 67 CDF11 CDF6 69 CDF7 68 79 85 CDF8 group II CDF9 CDF10 CDF12 Conclusions • The 12 Arabidopsis protein sequences reveal structural and therefore probably functional conservation • My results support the classification of these proteins as CDF metal transporters • I propose that the CDF protein family of A. thaliana contains two groups, each containing at least four proteins that are structurally and functionally closely related • Threading methods cannot be used for transmembrane proteins or for their non-transmembrane domains (yet) • Threading results for multiple sequences may be used to confirm (or find?) relationships among these sequences (“phylothreading”) • I was able to evaluate and compare a number of online tools that are available for the analysis of sequence data Conclusions 1. Sequence retrieval revealed conflicting information for 7 of the 12 proteins 2. The 12 Arabidopsis protein sequences reveal striking structural and therefore probably functional conservation 3. My results support the classification of these proteins as CDF metal transporters 4. I propose that the CDF protein family of A. thaliana contains two groups, each containing four proteins that are structurally and functionally closely related 5. I was able to evaluate and compare a variety of online tools available for the analysis of sequence data Conclusions 1. Sequence retrieval revealed conflicting information for 7 of the 12 proteins 2. The 12 Arabidopsis protein sequences reveal striking structural and therefore probably functional conservation 3. My results support the classification of these proteins as CDF metal transporters 4. I propose that the CDF protein family of A. thaliana contains two groups, each containing four proteins that are structurally and functionally closely related 5. I was able to evaluate and compare a variety of online tools available for the analysis of sequence data 6. Threading methods cannot be used for transmembrane proteins or for their non-transmembrane domains (yet) 7. Threading results for multiple sequences can be used to confirm (or find?) relationships among these sequences (“phylothreading”) METHODS Phylogenetic analysis: tree-building methods • distance-based methods overall distance between all pairs of sequences are calculated and then used to calculate a tree (Neighbor Joining) • character-based methods the individual substitutions among the sequences are used to determine the most likely ancestral relationships (Maximum Parsimony, Maximum Likelihood) • Bayesian inference of phylogenies ...CLVFMSVEVVGGIKANSLAILTD... ...NTAYMVVEFVAGFMSNSLGLISD... ...CLLFMSIEVVCGIKANSLAILAD... ...CAIFIVVEVVGGIKANSLAILTD... ...YLIVMSVQIVGGFKANSLAVMTD... Phylogenetic analysis: statistical evaluation of trees • bootstrap analysis how much support exists for particular branches in a phylogeny? 1. 2. 3. 4. 5. 6. tree construction, determination of the “best” tree bootstrap datasets (pseudosamples) are created from the original dataset by random sampling with replacement tree construction using the bootstrap datasets comparison of the bootstrap tree with the inferred tree this is repeated several hundred times bootstrap value: percentage of times an interior branch in the bootstrap tree was the same as the one in the inferred tree ...CLVFMSVEVVGGIKANSLAILTD... ...NTAYMVVEFVAGFMSNSLGLISD... ...CLLFMSIEVVCGIKANSLAILAD... ...CAIFIVVEVVGGIKANSLAILTD... ...YLIVMSVQIVGGFKANSLAVMTD... Fold recognition methods 2. evaluation of the compatibility between target sequence and proposed 3D structure • using environment-based mean force potentials (Bowie, Fischer, Eisenberg: 1991-1996) - residue positions are categorized into environment classes - the 3D protein structure is converted into a 1D sequence - generate alignment of this 1D string to target sequence • using knowledge-based mean force potentials (Sippl: 1990-1995) - information is automatically learned from databases of protein structures - pairwise interactions between structurally adjacent residues are calculated - transformation of mean force potentials as a function of distance Fold recognition methods aim: to find an optimal sequence-structure alignment 1. “threading” of an unknown target sequence into the backbone structure of template proteins of known structure ………CLVFMSVEVVGGIKANSLAILTD……… fold library query sequence Fold recognition methods 2. evaluation of the compatibility between target sequence and proposed 3D structure using environment-based mean force potentials or using knowledge-based mean force potentials 4.99 Å Fold recognition methods 2. evaluation of the compatibility between target sequence and proposed 3D structure using environment-based mean force potentials* or using knowledge-based mean force potentials* 4.99 Å * distant-dependent forces that act between atoms/residues (electrostatic and van der Waals interactions, influences on the surrounding medium on these interactions, contacts between two or three amino acids, angles between residue pairs, …) Fold recognition methods 2. evaluation of the compatibility between target sequence and proposed 3D structure using environment-based mean force potentials or using knowledge-based mean force potentials 3. Output: 4.99 Å a list of folds (sorted or unsorted), their “compatibility score”, sometimes other information such as SCOP descriptors, alignment, rudimentary 3D model of the query protein, raw scores, solvation energy for the model, links Threading methods used Qu i ck Ti me ™a nd a TIFF (Unc om pres se d) de co mp re ss or are n ee de d to s ee th is pi ctu re . QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Quick Time™a nd a TIFF ( Uncomp res sed) deco mpre ssor are n eede d to s ee this picture . Qu i ck Ti me ™a nd a TIFF (Unc om pres se d) de co mp re ss or are n ee de d to s ee th is pi ctu re . UCLA-DOE Fold Server P. Mallick et al., 2002 (BLAST, PSI-BLAST, SDP, DASEY) Threader D.T. Jones et al., 1992 mGenThreader L.J. McGuffin & D.T. Jones 2003 3D-PSSM L.A. Kelley et al., 2000 Arby I. Sommer et al., unpublished (PSI-BLAST, 123D, Jprop) Selection of structural hits for further analysis UCLA-DOE: top 10 structural hits are returned, all were kept Threader: compatibility of target sequence and all 2000 available templates is evaluated; lists were sorted by Z-value, approximately 10-20 best hits were kept mGenThreader: top 20 structural hits are returned, all were kept 3D-PSSM: top 20 structural hits are returned, all were kept Arby: a list of the 10-20 best scores is returned; the corresponding hits were extracted from a large table Evaluation of the top score for each CDF sequence UCLA 350 Threader 300 0 very poor score 250 1 200 2 150 100 50 0 3 borderline significant 4 significant very significant scores: no guidelines mGenThreader 3D-PSSM 1.0 0.8 guess 0.6 0.4 2.5 2.0 1.5 low confidence 0.2 0.0 poor score 1.0 worthy of attention 0.5 medium confidence high confidence certain 0.0 highly confident There is no consensus of top fold predicted by different methods example: top two structural hits for CDF1 Threader: 1ONE 1C3Q phosphopyruvate hydrolase thiazole kinase mGenThreader: 1L8M 1QGR his-rich protein (model) importin beta UCLA-DOE: 1B8F 1HFA histidine ammonia-lyase clathrin assembly protein 3D-PSSM: 1PW4 1KPW glycerol-3-phosphate transporter green cone pigment Arby: 1HZX 1EZV bovine rhodopsin yeast cytochrome bc1 No new insights regarding the structure of CDF proteins Membrane proteins are significantly under-represented in structural databases – and therefore also in fold libraries If there is no fold similar to the native fold of the target protein, this approach cannot succed. Threading methods cannot be used for modeling approaches Threading results: C-termini 1. Structural information no information of domains for metal transport available. BUT: several of the returned hits are proteins in which bound metals have structural or catalytic roles 2. Verification of group assignment i. Hits predicted for more than one C-terminus: specific for group I: specific for group II: specific for CDF5 and CDF11: ii. “Phylothreading” 48 folds 3 2 2 Positions of conserved domains and signature sequences 1 2 3 4 5 11 6 12 7 8 9 10 TMD I II III IV V VI CDF signature Pfam CE signature BLOCKS (eMOTIF) 10, 11 11, 12 6-12 Arabidopsis CDF proteins AtCDF1 AtCDF2 AtCDF3 AtCDF4 AtCDF5 AtCDF11 AtCDF6 AtCDF12 AtCDF9 AtCDF8 AtCDF10 AtCDF7 outgroup group I: - contain his-rich region between TMD 4 and 5 - one member is confirmed to transport Zn ions - genome structure conserved (no introns) no group assignment: - CDF6, CDF12: possibly distant common ancestry and mitochondrial localization - CDF5, CDF11: close relationship also in PFAM tree group II: - lack the his-rich region between TMD 4 and 5 - proteins may transport Mn ions - C-terminal regions differ from group I sequences working model: topology of Arabidopsis CDF proteins CDF signature sequence cell exterior/organelle cytoplasm N C Gene organization of the CDF proteins CDF1 CDF2 CDF3 CDF4 CDF5 CDF10 CDF6 CDF12 CDF7 CDF8 CDF9 CDF11 Phylogenetic analysis of sequences containing the CE signature 100/52/83 100/100/70 100/98/97 100/100/69 100/ 99/82 100/99/88 99/ 94/ 72 100/100/63 100/100/74 100/100/– 71/55 100/99/95 99/–/61 -/75 1000/100/94 100/ 100/ 53 62 100/100/82 100/100 /95 97/–/– 68/–/83 100/–/63 83/–/67 100/100/71 99/65/– 100/100/– 100/99/75 96/96/78 100/100/63 100/100/69 74/–/– 100/100/92 100 /100 /85 5 100 /100 /84 Escherischia coli ZITB Ralstonia metallidurans CZCD Ralstonia metallidurans CZCD Mus musculus ZNT4 Rattus norvegigus ZNT2 Mus musculus ZNT3 Arabidopsis thaliana CDF4 Arabidopsis thaliana CDF2 Eucalyptus grandis Arabidopsis thaliana CDF3 Arabidopsis thaliana CDF1 Thlaspi caerulescens ZTP1 Thlaspi goesingense MTP1 Thlaspi goesingense MTP1 Thlaspi goesingense MTP1 Oryza sativa Zea mays Lotus japonicus Medicago trunculata Oryza sativa Triticum aestivum Caenorhabditis elegans CDF1 Rattus norvegicus ZNT1 Schizosachharomyces pombe ZHF Saccharomyces cerevisiae COT1 Saccharomyces cerevisiae ZRC1 Oryza sativa Stylosanthes hamata MTP1 Oryza sativa Arabidopsis thaliana CDF10 Arabidopsis thaliana CDF7 Stylosanthes hamata MTP4 Stylosanthes hamata MTP2 Stylosanthes hamata MTP3 Oryza sativa Arabidopsis thaliana CDF8 Arabidopsis thaliana CDF9 Arabidopsis thaliana CDF6 Saccharomyces cerevisiae Mmt2 Saccharomyces cerevisiae Mmt1 T. thermophilus czrB Oryza sativa Arabidopsis thaliana CDF12 Homo sapiens ZNT5 Homo sapiens ZTL1 Homo sapiens ZNT7 Homo sapiens ZNT6 Arabidopsis thaliana CDF11 S. cerevisiae MSC2 Oryza sativa Arabidopsis thaliana CDF5 Staphylococcus aureus Staphylococcus aureus Bacillus stearothermophilus Bacillus stearothermophilus IV I V II III Phylogenetic analysis: tree-building methods • maximum parsimony methods the best tree topology minimizes the total amount of evolutionary change that has occurred • distance methods the best tree topology minimizes the the total distance among taxa • maximum likelihood methods given a particular substitution model and given a particular tree, how likely is the observed data? ...CLVFMSVEVVGGIKANSLAILTD... ...NTAYMVVEFVAGFMSNSLGLISD... ...CLLFMSIEVVCGIKANSLAILAD... ...CAIFIVVEVVGGIKANSLAILTD... ...YLIVMSVQIVGGFKANSLAVMTD... Inclusion in membrane and transport databases cation efflux, Pfam entry PF01545 Membrane Protein Library (AMPL) CDF1 CDF2 Arabidopsis ARAMEMNON Transport Protein Database PlantsT CDF zinc transporter CDF CDF CDF putative MTP CDF CDF CDF3 CDF putative MTP CDF CDF CDF4 CDF putative MTP CDF CDF CDF5 singleton (CDF related) putative cation transporter CDF - CDF6 singleton unknown protein CDF CDF CDF7 family unknown protein CDF - CDF8 family hypothetical protein - - CDF9 family unknown protein - - CDF10 family putative MTP - CDF CDF11 singleton putative MTP CDF CDF CDF12 singleton putative MTP - CDF Inclusion in membrane and transport databases cation efflux, Pfam entry PF01545 Membrane Protein Library (AMPL) CDF1 CDF2 Arabidopsis ARAMEMNON Transport Protein Database PlantsT CDF zinc transporter CDF CDF CDF putative MTP CDF CDF CDF3 CDF putative MTP CDF CDF CDF4 CDF putative MTP CDF CDF CDF5 singleton (CDF related) putative cation transporter CDF - CDF6 singleton unknown protein CDF CDF CDF7 family unknown protein CDF - CDF8 family hypothetical protein - - CDF9 family unknown protein - - CDF10 family putative MTP - CDF CDF11 singleton putative MTP CDF CDF CDF12 singleton putative MTP - CDF Inclusion in membrane and transport databases cation efflux, Pfam entry PF01545 Membrane Protein Library (AMPL) CDF1 CDF2 Arabidopsis ARAMEMNON Transport Protein Database PlantsT CDF zinc transporter CDF CDF CDF putative MTP CDF CDF CDF3 CDF putative MTP CDF CDF CDF4 CDF putative MTP CDF CDF CDF5 singleton (CDF related) putative cation transporter CDF - CDF6 singleton unknown protein CDF CDF CDF7 family unknown protein CDF - CDF8 family hypothetical protein - - CDF9 family unknown protein - - CDF10 family putative MTP - CDF CDF11 singleton putative MTP CDF CDF CDF12 singleton putative MTP - CDF Inclusion in membrane and transport databases cation efflux, Pfam entry PF01545 Membrane Protein Library (AMPL) CDF1 CDF2 Arabidopsis ARAMEMNON Transport Protein Database PlantsT CDF zinc transporter CDF CDF CDF putative MTP CDF CDF CDF3 CDF putative MTP CDF CDF CDF4 CDF putative MTP CDF CDF CDF5 singleton (CDF related) putative cation transporter CDF - CDF6 singleton unknown protein CDF CDF CDF7 family unknown protein CDF - CDF8 family hypothetical protein - - CDF9 family unknown protein - - CDF10 family putative MTP - CDF CDF11 singleton putative MTP CDF CDF CDF12 singleton putative MTP - CDF