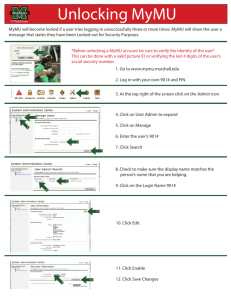
Sec7-1 - Personal.psu.edu
advertisement

STAT 250 Dr. Kari Lock Morgan Testing Goodness-ofFit for a Single Categorical Variable SECTION 7.1 • Testing the distribution of a single categorical variable : 2 goodness of fit (7.1) Statistics: Unlocking the Power of Data Lock5 Statistics! Statistics might be the most important class you take in college http://college.usatoday.com/2015/04/08/voices- statistics-might-be-the-most-important-class-youtake-in-college/ (4/8/15) Why you need to study statistics https://www.youtube.com/watch?v=wV0Ks7aS7YI (4/2/15) Statistics: Unlocking the Power of Data Lock5 Multiple Categories • So far, we’ve learned how to do inference for categorical variables with only two categories • Today, we’ll learn how to do hypothesis tests for categorical variables with multiple categories Statistics: Unlocking the Power of Data Lock5 Genetic Variants for Fast-Twitch Muscles A gene called ACTN3 encodes a protein which functions in fast twitch muscles Three different variants of the gene: RR, RX, and XX In a sample, we observe 130 RR, 226 RX, and 80 XX. If both R and X are equally likely, then by the Hardy- Weinberg principle about 50% of the population should be heterozygotes (RX) and about 25% should be each of the homozygotes (25% RR, 25% XX) Do our data contradict these hypothesized proportions? Yang, N. et. al. (2003). “ACTN3 genotype is associated with human elite athletic performance,” American Journal of Human Genetics, 73: 627-631. Statistics: Unlocking the Power of Data Lock5 Hypothesis Testing 1. State Hypotheses 2. Calculate a statistic, based on your sample data 1. Create a distribution of this statistic, as it would be observed if the null hypothesis were true 2. Measure how extreme your test statistic from (2) is, as compared to the distribution generated in (3) Statistics: Unlocking the Power of Data Lock5 Hypotheses Define the null hypothesized proportions in each category: H0 : pRR = 0.25, pRX = 0.5, pxx = 0.25 Ha : At least one pi is not as specified in H0 Statistics: Unlocking the Power of Data Lock5 Observed Counts • The observed counts are the actual counts observed in the study Observed RR 130 Statistics: Unlocking the Power of Data RX 226 XX 80 Lock5 Test Statistic Why can’t we use the familiar formula sample statistic null value SE to get the test statistic? We need something a bit more complicated… Statistics: Unlocking the Power of Data Lock5 Expected Counts • The expected counts are the expected counts if the null hypothesis were true • For each cell, the expected count is the sample size (n) times the null proportion, pi expected = npi Statistics: Unlocking the Power of Data Lock5 Expected Counts n = 436 Null Proportion Expected RR 0.25 Statistics: Unlocking the Power of Data RX 0.5 XX 0.25 Lock5 Chi-Square Statistic Observed Expected RR 130 109 RX 226 218 XX 80 109 Need a way to measure how far the observed counts are from the expected counts… Use the chi-square statistic : 2 observed - expected Statistics: Unlocking the Power of Data 2 expected Lock5 Chi-Square Statistic Observed Expected RR 130 109 Statistics: Unlocking the Power of Data RX 226 218 XX 80 109 Lock5 What Next? We have a test statistic. What else do we need to perform the hypothesis test? How do we get this? Two options: 1) Simulation 2) Distributional Theory Statistics: Unlocking the Power of Data Lock5 Upper-Tail p-value To calculate the p-value for a chi-square test, we always look in the upper tail Why? Values of the χ2 are always positive The higher the χ2 statistic is, the farther the observed counts are from the expected counts, and the stronger the evidence against the null Statistics: Unlocking the Power of Data Lock5 Simulation p-value Statistics: Unlocking the Power of Data Lock5 Chi-Square (χ2) Distribution • If each of the expected counts are at least 5, AND if the null hypothesis is true, then the χ2 statistic follows a χ2 –distribution, with degrees of freedom equal to df = number of categories – 1 • Gene variants: df = 3 – 1 = 2 Statistics: Unlocking the Power of Data Lock5 Chi-Square Distribution Statistics: Unlocking the Power of Data Lock5 p-value using χ2 distribution Statistics: Unlocking the Power of Data Lock5 Conclusion Do our data provide evidence that the population proportions differ from 25% RR, 50% RX, and 25% XX? a) Yes b) No Statistics: Unlocking the Power of Data Lock5 Chi-Square Test for Goodness of Fit 1. State null hypothesized proportions for each category, pi. Alternative is that at least one of the proportions is different than specified in the null. 2. Calculate the expected counts for each cell as npi . 3. Calculate the χ2 statistic: observed - expected 2 expected 2 4. Compute the p-value as the proportion above the χ2 statistic for either a randomization distribution or a χ2 distribution with df = (# of categories – 1) if expected counts all > 5 5. Interpret the p-value in context. Statistics: Unlocking the Power of Data Lock5 Mendel’s Pea Experiment In 1866, Gregor Mendel, the “father of genetics” published the results of his experiments on peas • • He found that his experimental distribution of peas closely matched the theoretical distribution predicted by his theory of genetics (involving alleles, and dominant and recessive genes) Source: Mendel, Gregor. (1866). Versuche über Pflanzen-Hybriden. Verh. Naturforsch. Ver. Brünn 4: 3–47 (in English in 1901, Experiments in Plant Hybridization, J. R. Hortic. Soc. 26: 1–32) Statistics: Unlocking the Power of Data Lock5 Mendel’s Pea Experiment Mate SSYY with ssyy: 1st Generation: all Ss Yy Mate 1st Generation: => 2nd Generation Second Generation S, Y: Dominant s, y: Recessive Statistics: Unlocking the Power of Data Phenotype Theoretical Proportion Round, Yellow 9/16 Round, Green 3/16 Wrinkled, Yellow 3/16 Wrinkled, Green 1/16 Lock5 Mendel’s Pea Experiment Phenotype Theoretical Observed Proportion Counts Round, Yellow Round, Green Wrinkled, Yellow 9/16 3/16 3/16 315 101 108 Wrinkled, Green 1/16 32 Let’s test this data against the null hypothesis of each pi equal to the theoretical value, based on genetics H 0 : p1 9 /16, p2 3 /16, p3 3 /16, p4 1/16 H a : At least one pi is not as specified in H 0 Statistics: Unlocking the Power of Data Lock5 Mendel’s Pea Experiment Phenotype Round, Yellow Null pi Observed Counts Wrinkled, Yellow 9/16 3/16 3/16 315 101 108 Wrinkled, Green 1/16 32 Round, Green Expected Counts The expected count for the round, yellow phenotype is a)177.2 b)310.5 c) 312.75 d)318.25 Statistics: Unlocking the Power of Data Lock5 Mendel’s Pea Experiment Phenotype Round, Yellow Null pi Observed Counts Expected Counts Contribution to χ2 Wrinkled, Yellow 9/16 3/16 3/16 315 101 108 312.75 104.25 104.25 0.101 0.135 Wrinkled, Green 1/16 32 34.75 0.1218 Round, Green The contribution to the χ2 statistic for the round, yellow phenotype is a)0.012 b)0.014 c) 0.016 d)0.018 Statistics: Unlocking the Power of Data Lock5 Mendel’s Pea Experiment • χ2 = 0.47 • Two options: o Simulate a randomization distribution o Compare to a χ2 distribution with 4 – 1 = 3 df Statistics: Unlocking the Power of Data Lock5 Mendel’s Pea Experiment p-value = 0.925 Does this prove Mendel’s theory of genetics? Or at least prove that his theoretical proportions for pea phenotypes were correct? a) Yes b) No Statistics: Unlocking the Power of Data Lock5 To Do Read Section 7.1 Do HW 7.1 (due Friday, 4/17) Statistics: Unlocking the Power of Data Lock5