Lac Operon
advertisement
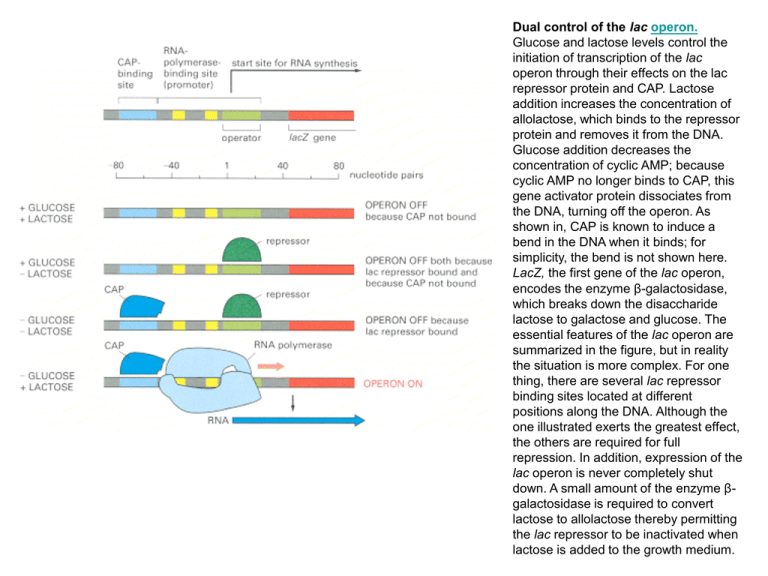
Dual control of the lac operon. Glucose and lactose levels control the initiation of transcription of the lac operon through their effects on the lac repressor protein and CAP. Lactose addition increases the concentration of allolactose, which binds to the repressor protein and removes it from the DNA. Glucose addition decreases the concentration of cyclic AMP; because cyclic AMP no longer binds to CAP, this gene activator protein dissociates from the DNA, turning off the operon. As shown in, CAP is known to induce a bend in the DNA when it binds; for simplicity, the bend is not shown here. LacZ, the first gene of the lac operon, encodes the enzyme β-galactosidase, which breaks down the disaccharide lactose to galactose and glucose. The essential features of the lac operon are summarized in the figure, but in reality the situation is more complex. For one thing, there are several lac repressor binding sites located at different positions along the DNA. Although the one illustrated exerts the greatest effect, the others are required for full repression. In addition, expression of the lac operon is never completely shut down. A small amount of the enzyme βgalactosidase is required to convert lactose to allolactose thereby permitting the lac repressor to be inactivated when lactose is added to the growth medium. Induction of the LAC Operon. (A) In the absence of lactose, the lac repressor binds DNA and represses transcription from the lac operon. (B) Allolactose or another inducer binds to the lac repressor, leading to its dissociation from DNA and to the production of lac mRNA. Positive control of the lac operon by glucose Low levels of glucose activate adenylyl cyclase, which converts ATP to cyclic AMP (cAMP). Cyclic AMP then binds to the catabolite activator protein (CAP) and stimulates its binding to regulatory sequences of various operons concerned with the metabolism of alternative sugars, such as lactose. CAP interacts with the α subunit of RNA polymerase to activate transcription Negative control of the lac operon The i gene encodes a repressor which, in the absence of lactose (top), binds to the operator (o) and blocks transcription of the three structural genes (z, β-galactosidase; y, permease; and a, transacetylase). Lactose induces expression of the operon by binding to the repressor (bottom), which prevents the repressor from binding to the operator. P = promoter; Pol = polymerase Catabolite control of the lac operon. The operon is inducible by lactose to the maximal levels when cAMP and CAP form a complex. (a) Under conditions of high glucose, a glucose breakdown product inhibits the enzyme adenylate cyclase, preventing the conversion of ATP into cAMP. (b) Under conditions of low glucose, there is no breakdown product, and therefore adenylate cyclase is active and cAMP is formed. (c) When cAMP is present, it acts as an allosteric effector, complexing with CAP. (d) The cAMP–CAP complex acts as an activator of lac operon transcription by binding to a region within the lac promoter. (CAP = catabolite activator protein; cAMP = cyclic adenosine monophosphate.) Negative and positive control of the lac operon by the Lac repressor and catabolite activator protein (CAP), respec-tively. (a) In the absence of lactose to serve as an inducer, the Lac repressor is able to bind the operator; regardless of the levels of cAMP and the presence of CAP, mRNA production is repressed. (b) With lactose present to bind the repressor, the repressor is unable to bind the operator; however, only small amounts of mRNA are produced because the presence of glucose keeps the levels of cAMP low, and thus the cAMP–CAP complex does not form and bind the promoter. (c) With the repressor inactivated by lactose and with high levels of cAMP present (owing to the absence of glucose), cAMP binds CAP. The cAMP–CAP complex is then able to bind the promoter; the lac operon is thus activated, and large amounts of mRNA are produced. (d) When CAP binds the promoter, it creates a bend greater than 90° in the DNA. Apparently, RNA polymerase binds more effectively when the promoter is in this bent configuration. (e) CAP bound to its DNA recognition site. This part is derived from the structural analysis of the CAP–DNA complex Regulation of the lac operon. The I gene continually makes repressor. The repressor binds to the O (operator) region, blocking the RNA polymerase bound to P (the promoter region) from transcribing the adjacent structural genes. When lactose is present, it binds to the repressor and changes its shape so that the repressor no longer binds to O. The RNA polymerase is then able to transcribe the Z, Y, and A structural genes, so the three enzymes are produced. A simplified lac operon model. The three genes Z, Y, and A are coordinately expressed. The product of the I gene, the repressor, blocks the expression of the Z, Y, and A genes by interacting with the operator (O). The inducer can inactivate the repressor, thereby preventing interaction with the operator. When this happens, the operon is fully expressed. The base sequence and the genetic boundaries of the control region of the lac operon, with partial sequences for the structural genes. Structure of the inducer of the lac operon, IPTG. The β-d-thiogalactoside linkage is not cleaved by β-galactosidase, allowing manipulation of the intracellular concentration of this inducer Heterodimerization of leucine zipper proteins can alter their DNA-binding specificity. Leucine zipper homodimers bind to symmetric DNA sequences, as shown in the left-hand and center drawings. These two proteins recognize different DNA sequences, as indicated by the red and blue regions in the DNA. The two different monomers can combine to form a heterodimer, which now recognizes a hybrid DNA sequence, composed from one red and one blue region A leucine zipper dimer bound to DNA. Two α-helical DNA-binding domains (bottom) dimerize through their α-helical leucine zipper region (top) to form an inverted Y-shaped structure. Each arm of the Y is formed by a single α helix, one from each monomer, that mediates binding to a specific DNA sequence in the major groove of DNA. Each α helix binds to one-half of a symmetric DNA structure. The structure shown is of the yeast Gcn4 protein, which regulates transcription in response to the availability of amino acids in the environment. One type of zinc finger protein. This protein belongs to the Cys-Cys-His-His family of zinc finger proteins, named after the amino acids that grasp the zinc. (A) Schematic drawing of the amino acid sequence of a zinc finger from a frog protein of this class. (B) The three-dimensional structure of this type of zinc finger is constructed from an antiparallel β sheet (amino acids 1 to 10) followed by an α helix (amino acids 12 to 24). The four amino acids that bind the zinc (Cys 3, Cys 6, His 19, and His 23) hold one end of the α helix firmly to one end of the β sheet. Summary of sequence-specific interactions between different six zinc fingers and their DNA recognition sequences. Even though all six Zn fingers have the same overall structure (see Figure 7-17), each binds to a different DNA sequence. The numbered amino acids form the α helix that recognizes DNA (Figures 7-17 and 7-18), and those that make sequence-specific DNA contacts are colored green. Bases contacted by protein are orange. Although arginine-guanine contacts are common (see Figure 7-27), guanine can also be recognized by serine, histidine, and lysine, as shown. Moreover, the same amino acid (serine, in this example) can recognize more than one base. Two of the Zn fingers depicted are from the TTK protein (a Drosophila protein that functions in development); two are from the mouse protein (Zif268) that was shown in Figure 7-18; and two are from a human protein (GL1), whose aberrant forms can cause certain types of cancers DNA binding by a zinc finger protein. (A) The structure of a fragment of a mouse gene regulatory protein bound to a specific DNA site. This protein recognizes DNA using three zinc fingers of the Cys-Cys-His-His type (see Figure 7-17) arranged as direct repeats. (B) The three fingers have similar amino acid sequences and contact the DNA in similar ways. In both (A) and (B) the zinc atom in each finger is represented by a small sphere A dimer of the zinc finger domain of the intracellular receptor family bound to its specific DNA sequence. Each zinc finger domain contains two atoms of Zn (indicated by the small gray spheres); one stabilizes the DNA recognition helix (shown in brown in one subunit and red in the other), and one stabilizes a loop (shown in purple) involved in dimer formation. Each Zn atom is coordinated by four appropriately spaced cysteine residues. Like the helix-turn-helix proteins shown in Figure 7-14, the two recognition helices of the dimer are held apart by a distance corresponding to one turn of the DNA double helix. The specific example shown is a fragment of the glucocorticoid receptor. This is the protein through which cells detect and respond transcriptionally to the glucocorticoid hormones produced in the adrenal gland in response to stress Families of DNA-binding domains (A) Zinc finger domains consist of loops in which an α helix and a β sheet coordinately bind a zinc ion. (B) Helix-turn-helix domains consist of three (or in some cases four) helical regions. One helix (helix 3) makes most of the contacts with DNA, while helices 1 and 2 lie on top and stabilize the interaction. (C) The DNA-binding domains of leucine zipper proteins are formed from two distinct polypeptide chains. Interactions between the hydrophobic side chains of leucine residues exposed on one side of a helical region (the leucine zipper) are responsible for dimerization. Immediately following the leucine zipper is a DNA-binding helix, which is rich in basic amino acids. (D) Helix-loop-helix domains are similar to leucine zippers, except that the dimerization domains of these proteins each consist of two helical regions separated by a loop.