Classifying Pseudoknots

Classifying Pseudoknots
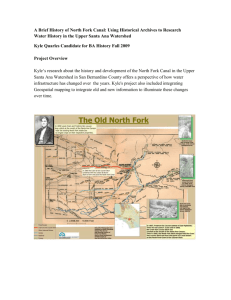
Kyle L. Spafford
Recap – What’s a pseudoknot again?
• Substructure with nonnested base pairings
• Makes RNA secondary structure prediction NPcomplete
• Looks pretty simple…
Classifying Pseudoknots -- Kyle Spafford 2
Not so simple...
• Quite a complex space
• Some simpler examples
• Should they be treated as equals?
Classifying Pseudoknots -- Kyle Spafford 3
Biological Motivation
• In nature, things get complicated…
A) Hepatitis Delta B) Diels-Alder Ribozyme C) Human telomerase D) Mouse mammary tumor virus E) pea
4
Biological Motivation
• Function of these pseudoknots?
– Viral Frameshifting
• SARS, Hep C, MMTV, some HIV
– Catalytically Active
• Genome replication
• Self-cleaving ribozymes
• Break down C-C bonds
– Some things remain a mystery
• Telomeres, aging, and cancer
Classifying Pseudoknots -- Kyle Spafford 5
My Project
• Examined 3 approaches to classifying pseudoknots
• Looked at prevalence results for what’s been found in nature
• Formed an argument which explains which approach should be used in different scenarios
Classifying Pseudoknots -- Kyle Spafford 6
Patterns (from Condon et al)
• A pattern is a string P over some alphabet
A, s.t. every element of A appears exactly twice, or not at all in P.
Classifying Pseudoknots -- Kyle Spafford 7
Patterns
• Classification idea - Sort pseudoknots by the algorithm(s) that can predict them
• Algorithms from: Uemura & Akutsu, Rivas
& Eddy, Lyngso & Pederson, Dirks &
Pierce
• Also, have a pseudoknot-free class
Classifying Pseudoknots -- Kyle Spafford 8
Patterns
• Pros
– O(n) existence test and classification
– Really easy to implement
– Given a pseudoknot, if is in one of the categories (with high probability)
• Cons
– Not very useful for biologists
Classifying Pseudoknots -- Kyle Spafford 9
Dual Graphs (from Gan et al)
• Represent RNA SS’s as dual graphs
– Vertex - stem
– Edge – single strand that may occur in segments, connects other secondary elements
Classifying Pseudoknots -- Kyle Spafford 10
Dual Graphs
• Classification idea – work with topological characteristics from dual graphs
Classifying Pseudoknots -- Kyle Spafford 11
Dual Graphs
• Pros
– Very useful for biologists
– Topological qualities are easy to compute
• Cons
– Hard to specify in words
– Not efficient to store
– Problems with accuracy
Classifying Pseudoknots -- Kyle Spafford 12
Knot-Components (from Rodland)
• Simplify the complex space
• Find “building blocks” of pseudoknots
• Describe structure in a short, precise method
• Ignore nested substructures which complicate things
Classifying Pseudoknots -- Kyle Spafford 13
Bottom-Up
• Start basic – bonds
– Orthodox or knotted
• Hairpin – P 2
• The notation
– Superscript: Number of stems involved in the pseudoknot
– Subscript (used when not reduced): number of stem components replacing a single stem in reduced form
Classifying Pseudoknots -- Kyle Spafford 14
Knot-Components
Optional second superscript when non-unique (double hairpin vs. pseudotrefoil
Classifying Pseudoknots -- Kyle Spafford 15
Top-Down
Classifying Pseudoknots -- Kyle Spafford 16
Knot-component
• Pros
– Precise biological information
– No overlap (like Condon’s system)
– Mapping to Condon’s categories
• Cons
– High learning curve
– Not so easy to implement
– Mapping has loss of specificity
Classifying Pseudoknots -- Kyle Spafford 17
A Brief Word on Prevalence
• Most pseudoknots in nature are P 5 and below
• Probability of finding more complex pseudoknots drops almost exponentially as superscript grows
– Exception: Group II introns
Classifying Pseudoknots -- Kyle Spafford 18
When to use each system
• Condon’s Patterns – Large scale analysis
• Gan’s Dual Graphs – When you need a lot of biological information (including substructures)
• Rodland’s Knot-Components – Any other time
Classifying Pseudoknots -- Kyle Spafford 19
Summary
• Pseudoknots range from trivially simple to extraordinarily complex.
• They perform a myriad of exciting biological roles.
• Classifying them is important in determining those roles.
• Almost always, stick with Rodland’s knotcomponent system
Classifying Pseudoknots -- Kyle Spafford 20
Thank You
• Questions?
• Dying to read the paper?
• kls@gatech.edu
Classifying Pseudoknots -- Kyle Spafford 21