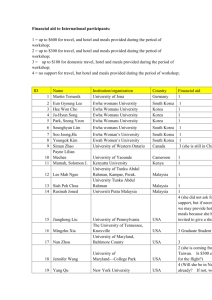
II. Topological Nature of Metabolic Networks at Peripheral Nodes
advertisement
Toward Automatically Drawn Metabolic
Pathway Atlas with Peripheral Node
Abstraction Algorithm
Myungha Jang, Arang Rhie, and Hyun-Seok Park*
Bioinformatics Laboratory, School of Engineering
Ewha Womans University
Seoul, Korea
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
Table of Contents
I.
Introduction
II.
Topological Nature of Metabolic Networks at Peripheral Nodes
III. Node Abstraction Featured Scale-free Algorithm
IV. Experimental Results
V. Discussion and Future Work
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
I. INTRODUCTION
Automatic graph layout algorithms in systems biology
• Abstract graph structure ⇒ visual representation
• Graphical diagrams are intuitively helpful to understand
biochemical reaction networks
- Node : compound, Edge : reactions
•Optimal solutions : NP-hard problems
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
I. INTRODUCTION
Focusing on Global Metabolic Pathway
•
•
•
•
•
A complete metabolic network indicates all the metabolic potential and capacity.
The shift of research focus: single pathways to multiple pathways.
Visualization serves an important role in understanding large scale metabolic
network.
KEGG Atlas(http://www.genome.ad.jp/kegg), 2008
Terms : Global (metabolic) pathway, Multiple pathway, Atlas
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
I. INTRODUCTION
Our Efforts Toward Automatic Global Layout
• Not enough to deal with the global pathway!
• How can we obtain a complete view?
• No attempts for automatic visualization for Atlas
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
I. INTRODUCTION
How To Deal With Large-scale Metabolic Pathway?
Related work: KEGG Atlas
• The map integration process is carried out manually by curators.
• Based on curator’s experience
• However, that metabolic networks are dynamic in nature should not be
disregarded Systematic approach is necessary
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
INTRODUCTION
How To Deal With Large-scale Metabolic Pathway? (con’d)
Our Strategy
We provide a novel algorithmic approach in drawing multiple metabolic
pathways by considering two properties:
1. Automatic abstraction criteria: by analyzing a topological
nature of metabolic networks based on the graphical property of relation
distance, linear reactions were abstracted as a unit reaction.
2. the consistency of highly connected nodes
II. TOPOLOGICAL NATURE OF METABOLIC NETWORKS AT PERIPHERAL NODES
• We obtained 255 map data by parsing KEGG XML (KGML) documents of
version 0.6 using our KGML Parser.
KG
ML
Two terms were defined:
+
1. Relation degree
the number of edges branching from a node
2. Relation distance
a factor to measure the length between any two compounds
encompassing nodes which all have relation degrees less than or equal
to p (p = 2)
• A dedicated analysis on peripheral nodes with low connectivity was performed.
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
II. TOPOLOGICAL NATURE OF METABOLIC NETWORKS AT PERIPHERAL NODES
Relation Distance Term Clarification
• Definition: The length between any two compounds encompassing nodes
which all have relation degrees equal to p
• Here, p = 2
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
II. TOPOLOGICAL NATURE OF METABOLIC NETWORKS AT PERIPHERAL NODES
Relation Distance Example in Map
RD(C01290, C00369) = 7
cpd:C01291
cpd:C01290
cpd:C16466
cpd:C16475
cpd:C16468
cpd:C16470
cpd:C16471
cpd:C16469
cpd:C00369
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
III. NODE ABSTRACTION FEATURED SCALE-FREE ALGORITHM
Basic Motivation
• Observation: 66.83% of the total compounds within the complete metabolic
pathways were of low connectivity, with less than relation degree of 3.
• The number of compounds with higher relation degree, i.e. more than 6 edges,
was much less.
Abstracting Compounds With
Linear Interaction
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Layout Components according
to High Connectivity
Ewha Womans University
III. NODE ABSTRACTION FEATURED SCALE-FREE ALGORITHM
A. Abstracting Compounds With Linear Interaction
• We abstracted and hid all those
compounds that appear within these
linear interactions.
• This approach could be called “chain
reduction”(M. Chimani et al)
• All green compounds in the figure
will be hidden in the graph layout
according to this approach.
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
III. NODE ABSTRACTION FEATURED SCALE-FREE ALGORITHM
B. Layout Components according to High Connectivity
• Highly Connected Nodes: Nodes with relation degree
bigger than 6
Input : Metabolic Pathway Graph
Output : coordinates of each node
void LayoutPathway (Pathway graph)
{
IF highly connected nodes (Nd) exist in graph
LayoutHighlyConnectedNode (graph, Nd);
• LayoutHighConnectedNode() Algorithm Steps
1.
Find a highly Connected node Nd
2.
Each component connected to Nd is decomposed
into sub-graph
3.
Each decomposed sub-graph is treated as a super
node to apply the spring-embedding algorithm
ELSE IF any cycle(Nc) exists in graph
AND size of cycle ≥ 6
LayoutCircular (graph, Nc);
ELSE LayoutHierarchic (graph);
}
IEEE BIBM, 18-21 Dec 2010, Hong Kong
3
Ewha Womans University
6
IV. EXPERIMENTAL RESULTS
Experiments : To compare compression rate of compounds, we obtained the number of
abstracted compounds and edge crossings by applying two different layout algorithms:
Result 1
…
• Scope
1. 84 single metabolic pathways
2. 8 major categorized metabolic pathways
3. the global pathway
single pathways
Result 2
• The number of edge crossing comparison between by
1. Conventional algorithm
2. Our Node abstraction featured scale-free layout algorithm
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
…
• Node compression rate performance
Categorized
pathways
Global
pathway
Peripheral path as supplementary nodes
III. EXPERIMENTAL RESULTS
Result 1B
The Number of Nodes Before and After Applying Node Abstraction
Number of Nodes
Before Abstraction
Number of Nodes After
Abstraction
Abstraction Rate
Carbohydrate Metabolism
1235
972
21.2%
Lipid Metabolism
1043
805
22.8%
Nucleotide Metabolism
424
351
17.21%
Amino Acid Metabolism
1327
980
26.14%
Metabolism of Other Amino Acid
332
262
21.08%
Metabolism of Cofactor and
Vitamins
Biosynthesis of Secondary
Metabolism
Xenobiotics Biodegradation
250
175
30%
800
536
33%
542
348
35.79%
Global Pathway (Atlas)
5675
4371
22.98%
Pathway
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
III. EXPERIMENTAL RESULTS
Peripheral path as super edges
Result 1A
Original Network
Abstracted Network
Results drawn with Cytoscape, using conventional spring embedding
The red-colored edges represent the abstracted edges. (abstraction rate : 70%)
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
III. EXPERIMENTAL RESULTS
Result 2 : Edge Crossing Reduction
• In single metabolic pathways, the node abstraction featured algorithm
reduced edge crossings by 63.31%.
• In a global metabolic pathway, the number of edge crossings has reached a
reduction of 58.08% in total.
• Our proposed algorithm with node abstraction resulted in 86,067 edge
crossings, whereas the one without node abstraction resulted in 205,316 edge
crossings.
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
IV. DISCUSSION
• Two approaches were used:
1. Abstracting compound pairs according to a consistent criteria
2. Layout components according to high connectivity
• Our experimental results show that node abstraction feature reduced the
number of compounds by approximately 23% in global pathway.
•
Further discussion is necessary regarding enzyme reactions
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University
IV. WHY IS OUR WORK IMPORTANT?
•
The first systematic approach for Atlas visualization focusing on
peripheral nodes
•
Fundamental to building a hierarchical structure of Atlas
•
Our approach is flexible upon pathway database change that frequently
updates
•
It is a crucial preliminary step toward automatically drawn
metabolic pathway
•
Future research on individual biological meaning of each peripheral nodes
and abstracted path
IEEE BIBM, 18-21 Dec 2010, Hong Kong
Ewha Womans University