(LRG) DNA sequence format for LSDBs
advertisement
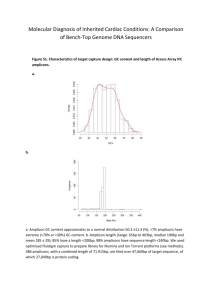
Locus Reference Genomic (LRG) Sequences Raymond Dalgleish Department of Genetics University of Leicester Background • Descriptions of sequence variants should use HGVS nomenclature • Variants should be described with respect to a reference DNA sequence specified by an accession number and a version e.g. NM_000088.3:c.2362G>T • Mostly works well, but three key issues frequently cause problems for LSDB curators and for diagnostic laboratories Issue 1: Version not specified • The autosomal dominant RP10 form of retinitis pigmentosa is caused by variants in the IMPDH1 gene • Variants for this gene are described with respect to NM_000883.1, but the version is rarely mentioned in the literature • The current version (NM_000883.3) records a shorter mRNA & protein which could lead to confusion and delay Issue 2: Alternative splicing • ~93% of genes have alternatively spliced transcripts & may yield several proteins • The CDKN2A locus encodes the tumour suppressor proteins p16INK4a and p14ARF • The mRNAs for the two proteins share exon 2 in common but in different reading frames, due to different upstream exons • Separate RefSeq records for the mRNAs CDKN2A alternate splicing Issue 3: Legacy numbering (1) • The “sickle cell” variant of β-globin is due to the substitution of glutamic acid by valine at amino acid 6 • Determined by amino acid sequencing prior to completion of the genetic code • HGVS protein-level description is p.Glu7Val counting from the start codon Issue 3: Legacy numbering (2) • Type I & III collagen variants were originally numbered from the start of the Gly-X-Y triple-helical repeat region • Legacy and HGVS descriptions still run in parallel: e.g. Gly610Cys & p.Gly788Cys • The exons of these genes were originally numbered in a 3´ to 5´ direction Issue 3: Legacy numbering (3) • New exons are often discovered in genes long after their initial characterisation • This interferes with simple sequential numbering of exons from 5´ to 3´ • Non-simple numbering is well-established: – COL1A1: 33/34 – CFTR: 6a, 6b,14a, 14b, 17a, 17b – OPRM: O, X, Y – CDKN2A: 1B, 1A So what is the solution? • An ideal reference sequence would: – be stable over periods as long as 25 years – be free of version confusion – comprise an “idealised” genomic DNA sequence haplotype providing a practical working framework – contain comprehensive information about the transcripts and proteins encoded by the gene (including alternative numbering schemes) – be mapped to the current genome assembly Primary design decisions • LRGs will be a working representation of a gene with a permanent ID: i.e. no versions • Based on any existing RefSeqGene record • 5 kb upstream and 2 kb downstream • There can be more than one LRG for a given region of the genome • LRGs will have both fixed and updatable feature annotations Primary fixed annotations • Coding sequence coordinates • Transcripts essential to the reporting of sequence variants • The conceptual translated protein(s) • Non-coding transcripts Primary updatable annotations • • • • Mapping to current genome assembly Chromosome number Any alternative IDs Cross references to other reference sequences • “Legacy” exon and amino acid numbering systems • Links to LSDBs • Overlapping genes Variant reporting with LRGs • The calcitonin gene (CALCA) encodes the peptide hormones calcitonin and calcitonin gene related peptide (CGRP) by alternative splicing • A SNP in the first base of exon 4 affects the transcript (t2) and the resulting precursor protein (p2) for calcitonin • The variant can be reported at gene, mRNA and protein level with reference just to LRG_13 (CALCA) Description Level RefSeqGene or RefSeq LRG gene NG_015960.1:g.8290C>A LRG_13:g.8290C>A mRNA NM_001033952.2:c.228C>A LRG_13t2:c.228C>A protein NP_001029124.1:p.Ser76Arg LRG_13p2:p.Ser76Arg Progress • LRGs can be viewed at the LRG web site: http://www.lrg-sequence.org • The first 10 LRGs have been finalised: – COL1A1, COL1A2, COL3A1, CRTAP, ATP1A2, CACNA1A, SCN1A, PPIB, FKBP10, CALCA • Another 4 await final approval: – LEPRE1, CDKN2A, L1CAM, UBE3A • Requests have been received for around 100 others Other tools to view LRGs • Ensembl, NCBI Genome Workbench, NCBI Sequence Viewer will soon provide support for LRGs • NGRL Universal Browser displays LRGs with links through to LSDBs and dbSNP • Mutalyzer will be updated to parse LRGs to support their use in LOVD • Alamut will probably be the first commercial software support for LRGs How do I learn more? • Dalgleish et al., 2010, Genome Medicine, in press • LRG web site: http://www.lrg-sequence.org • LRG specification document: http://www.lrg-sequence.org/docs/LRG.pdf • The LRG XML schema is available for download • E-mail addresses: – Request help: help@lrg-sequence.org – Provide feedback: feedback@lrg-sequence.org – Request a new LRG: request@lrg-sequence.org Acknowledgements Raymond Dalgleish Tony Brookes University of Leicester, Leicester, UK Donna Maglott Alex Astashyn Ray Tully NCBI, Bethesda, USA Fiona Cunningham Paul Flicek Ewan Birney Yuan Chen Pontus Larsson Will McLaren Glenn Proctor Brendon Vaughan EBI, Hinxton, UK Andrew Devereau Glen Dobson Christophe Béroud INSERM, Montpellier, France NGRL, Manchester, UK Peter Taschner Johan den Dunnen LUMC, Leiden, Netherlands Heikki Lehväslaiho CBRC, Jeddah, Saudi Arabia Coordination and funding • LRGs were devised by the GEN2PHEN project: http://www.gen2phen.org • The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement number 200754 — the GEN2PHEN project