Ch 17 - Concept Check Questions
advertisement

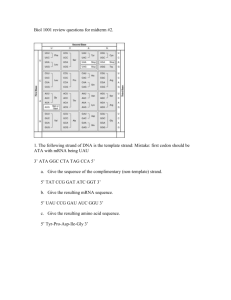
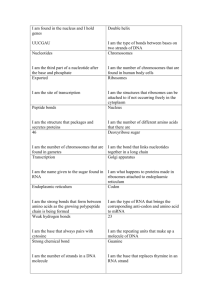
17.1 1. Draw the nontemplate strand of DNA for the template shown below. Compare and contrast its base sequence with the mRNA molecule. DNA A C C A A A C C G A G T mRNA U G G U U U G G C U C A 17.1 1. Draw the nontemplate strand of DNA for the template shown below. Compare and contrast its base sequence with the mRNA molecule. DNA A C C A A A C C G A G T T G G T T T G G C T C A mRNA U G G U U U G G C U C A 17.1 1. Draw the nontemplate strand of DNA for the template shown below. Compare and contrast its base sequence with the mRNA molecule. DNA A C C A A A C C G A G T T G G T T T G G C T C A mRNA U G G U U U G G C U C A 17.1 2. What protein product would you expect from a poly-G mRNA that is 30 nucleotides long? 5’-GGGGGGGGGGGGGGGGGGGGGGGGGGGGGG-3’ 17.1 2. What protein product would you expect from a poly-G mRNA that is 30 nucleotides long? 5’-GGGGGGGGGGGGGGGGGGGGGGGGGGGGGG-3’ Gly-Gly-Gly-Gly-Gly-Gly-Gly-Gly-Gly-Gly 17.2 1. Compare and contrast the functioning of DNA polymerase and RNA polymerase. DNA polymerase RNA polymerase DNA polymerase RNA polymerase •Assembles chains •Assembles chains from monomers from monomers DNA polymerase RNA polymerase •Assembles chains •Assembles chains from monomers from monomers •Complementary base pairing •Complementary base pairing DNA polymerase RNA polymerase •Assembles chains •Assembles chains from monomers from monomers •Complementary base pairing •Complementary base pairing •Reads 3’→5’ •Reads 3’→5’ •Assembles 5’ →3’ •Assembles 5’ →3’ DNA polymerase RNA polymerase •Needs a primer •Can start from scratch DNA polymerase RNA polymerase •Needs a primer to •Can start from start scratch •Uses A, T, G & C •Uses A, U, G & C •Uses nucleotides containing deoxyribose •Uses nucleotides containing ribose 17.2 2. Is the promoter at the upstream or downstream end of a transcription unit? 17.2 2. Is the promoter at the upstream or downstream end of a transcription unit? Upstream 17.2 3. In a prokaryote, how does RNA polymerase “know” where to start transcribing a gene? In a eukaryote? 17.2 3. In a prokaryote, how does RNA polymerase “know” where to start transcribing a gene? In a eukaryote? Prokaryote: RNA polymerase recognizes promoter Eukaryote: Transcription factors mediate binding 17.2 4. How is the primary transcript produced by a prokaryotic cell different from that produced by a eukaryotic cell? 17.2 4. How is the primary transcript produced by a prokaryotic cell different from that produced by a eukaryotic cell? Prokaryote: used immediately as mRNA Eukaryote: Must be modified before being used as mRNA 17.3 1. How does the alteration of the 5’ and 3’ ends of pre-mRNA affect the mRNA that exists in the nucleus? 17.3 1. How does the alteration of the 5’ and 3’ ends of pre-mRNA affect the mRNA that exists in the nucleus? •Facilitates transportation •Prevents degradation •Facilitates ribosomal attachment 17.3 2. Describe the role of snRNPs in RNA splicing. 17.3 2. Describe the role of snRNPs in RNA splicing. •Joins with other proteins to form spliceosomes. •Removes introns, splices exons together. 17.3 3. How can alternative RNA splicing generate a greater number of polypeptide products than there are genes? 17.3 3. How can alternative RNA splicing generate a greater number of polypeptide products than there are genes? THE CAT ATE THE RBAT 17.3 3. How can alternative RNA splicing generate a greater number of polypeptide products than there are genes? THE CAT ATE THE RBAT 17.3 3. How can alternative RNA splicing generate a greater number of polypeptide products than there are genes? THE CAT ATE THE RBAT 17.4 1. Which two processes ensure that the correct amino acid is added to a growing polypeptide chain? 17.4 1. Which two processes ensure that the correct amino acid is added to a growing polypeptide chain? •Aminoacyl-tRNA synthase •tRNA codon 17.4 2. Describe how the formation of polyribosomes can benefit the cell. 17.4 2. Describe how the formation of polyribosomes can benefit the cell. Multiple copies of a protein in a short time. 17.4 3. Describe how a polypeptide to be secreted is transported to the endomembrane system. 17.4 3. Describe how a polypeptide to be secreted is transported to the endomembrane system. •Signal peptide is recognized by SRP. •SPR brings polypeptide to ER lumen. 17.5 1. Describe three properties of RNA that allow it to perform diverse roles in the cell. 17.5 1. Describe three properties of RNA that allow it to perform diverse roles in the cell. •Hydrogen bonds with DNA or RNA •Specific 3-D shape •Catalize chemical reactions. 17.6 1. In figure 17.22 (orange book and green book) number the RNA polymerases in order of their initiation of transcription. Then number each mRNA’s ribosomes in order of their initiation of translation. 17.6 2. Would the arrangement shown in Figure 17.22 be found in a eukaryotic cell? Explain. 17.7 1. What happens when one nucleotide pair is lost from the middle of the coding sequence of a gene? 17.7 1. What happens when one nucleotide pair is lost from the middle of the coding sequence of a gene? •Frame shift mutation •Nonfunctional protein 17.7 2. The template strand of a gene contains the sequence 3’-TACTTGTCCGATATC5’. Draw a double strand of DNA and the resulting strand of mRNA, labeling all 5’ and 3’ ends. Determine the amino acid sequence. Then show the same after a mutation changes the template DNA sequence to 3’TACTTGTCCAATATC-5’. What is the effect on the amino acid sequence? 17.7 2. Template strand: 3’-TACTTGTCCGATATC-5’ Draw a double strand of DNA. 17.7 2. Double strand: 3’-TACTTGTCCGATATC-5’ 5’-ATGAACAGGCTATAG-3’ Draw the resulting strand of mRNA, labeling all 5’ and 3’ ends 17.7 2. Double strand: 3’-TACTTGTCCGATATC-5’ 5’-ATGAACAGGCTATAG-3’ mRNA: 5’-AUGAACAGGCUAUAG-3’ 17.7 2. mRNA: 5’-AUGAACAGGCUAUAG-3’ Determine the amino acid sequence. 17.7 2. mRNA: AUG AAC AGG CUA UAG Determine the amino acid sequence. 17.7 2. mRNA: AUG AAC AGG CUA UAG Polypeptide: Met-Asn-Arg-Leu 17.7 2. Then show the same after a mutation changes the template DNA sequence to 3’-TACTTGTCCAATATC-5’. 17.7 2. Template strand: 3’-TACTTGTCCAATATC-5’ Draw a double strand of DNA. 17.7 2. Double strand: 3’-TACTTGTCCAATATC-5’ 5’-ATGAACAGGTTATAG-3’ Draw the resulting strand of mRNA, labeling all 5’ and 3’ ends 17.7 2. Double strand: 3’-TACTTGTCCAATATC-5’ 5’-ATGAACAGGTTATAG-3’ mRNA: 5’-AUGAACAGGUUAUAG-3’ 17.7 2. mRNA: 5’-AUGAACAGGUUAUAG-3’ Determine the amino acid sequence. 17.7 2. mRNA: AUG AAC AGG UUA UAG Determine the amino acid sequence. 17.7 2. mRNA: AUG AAC AGG UUA UAG Polypeptide: Met-Asn-Arg-Leu 17.7 2. mRNA: AUG AAC AGG UUA UAG Polypeptide: Met-Asn-Arg-Leu The resulting polypeptide is the same. .