Warfarin
advertisement

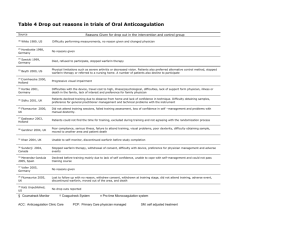
Recent Genetic Advances in Cardiovascular Disease Linnea M. Baudhuin, Ph.D. Mayo Clinic Dec. 6, 2008 DISCLOSURE Information Relevant Financial Relationships None Off Label Usage None Recent Developments in CV Genetics Mendelian genetics Autosomal Dominant Hypercholesterolemia Familial Thoracic Aortic Aneurysm and Dissection syndromes: LDLR, APOB, PCSK9 FBN1, TGFBR1, TGFBR2, ACTA2, MYH11, NOTCH Genetic markers for CAD Chromosome 9p21/ANRIL KIF6 Pharmacogenetics Warfarin sensitivity Statin efficacy Coronary Artery Disease Unable to predict significant portion of CAD “CAD could be eliminated in 21st century by treating all risk factors” 35% of CAD occurs in population with TC<200 mg/dL R. Roberts, Genetics of Premature Myocardial Infarction, Curr Ather Reports, 2008, 10: 186-193 Atherosclerotic CVD has strong heritable component Estimates as high as >50% Families with early CAD (14% of population) account for 72% of early CAD cases Most of the risk factors have predominant genetic component Need for identifying genetic modifiers and their function Complex Genetics of CAD Minority are single-gene disorders FH, FDB, ARH, Tangier, Sitosterolemia, etc. What about the majority of CAD? What’s going on genetically? Early stages of atherosclerosis Late stages of atherosclerosis Lusis et al., Ann Rev Gen Hum Gen, 2004, 5:189-218 Genes contributing to CAD susceptibility Lusis A, Trends Cardio Med, 2003; 13:309-16. How can we identify genetic markers for complex disease? Family studies Genetic variants that segregate with disease in affected vs. non-affected family members Linkage analysis Population studies Genetic variants that segregate with disease in cases vs. controls Candidate genes Localized genomic regions Genome-wide Association studies CAD Linkage Studies At least 7 family studies Linkage: Chr 1, 2, 3, 13, 14, 15, 16, X Occurrence of MI or subclinical atherosclerosis Lack of replication Genes identified: MEF2A (Chr 15q26) ALOX5AP LTA4 Candidate Gene Association Analyses in CAD Generally single gene/single polymorphism Small cohorts Inability to replicate findings in additional studies Genetic analysis of 103 candidate genes (n=1400, with follow-up n= 806) unable to replicate findings (Pare et al, Am Hum Genet, 2007, 80:673-682) Similar results in n=811 ACS patients, 70 genes (Morgan et al, JAMA, 2007, 297: 1551-1561 Why are the results of Linkage and Candidate Gene studies so disappointing? Inadequately powered Phenotypic heterogeneity and imprecise definition of phenotypes Bias (e.g. population stratification) Multiple genes each contribute to small percentage of phenotype (<5-10%) A single gene is insufficient to induce disease Information from multiple polymorphisms should be integrated to become clinically useful Multiple possible combinations of genotypes Population and disease heterogeneity What about Genome Wide Association Studies? Genome-wide association studies Previously: single SNP/single cohort associations with little replication Now: large international consortium studies of tens of thousands of individuals and hundreds of thousands of SNPs Technology Ultra high-throughput genotyping with ability to analyze 500K to >1M SNPs in each individual Topol, EJ et al., JAMA, 2007; 298:218-221 Multiple GWASs confirming 9p21 in Caucasian populations McPherson et al, Science (316), June 8 2007, 1488-91 Initial study, 322 cases vs 312 controls; 73K SNPs Confirmed in 5 independent Caucasian populations (n=23,000) Helgadottir et al, Science (316), June 8 2007, 1491-3 Initial study, 1607 cases vs 6728 controls; 306K SNPs Replicated in 665 cases and 3533 controls Three other case control populations Philadelphia, Atlanta, & Durham, NC 4587 cases, 12,767 controls Wellcome Trust Case Consortium, Nature, 447:661-678, 2007 Samani et al, NEJM (357), Aug 2 2007, 443-53 PROCARDIS Consortium (Broadbent et al, Hum Mol Gen, 2008, 17) Chromosome 9p21 9p21 confined to 58K bp region Risk associated with MI and CAD independent of known risk factors 50% of individuals heterozygous for allele 25% of individuals homozygous for allele 15-20% increased risk of CAD 30-40% increased risk of CAD Odds ratio Heterozygous: 1.3-1.5 Homozygous: 1.8-2.1 9p21 and ANRIL High-risk CAD 9p21 haplotype overlaps with exons 13-19 of ANRIL Newly annotated gene, encodes large antisense non-coding RNA (ncRNA) Identified through deletion analysis of French family with hereditary melanoma-neural system tumors Expressed in atheromatous human vessels Comparable cell type profiles to atherosclerotic arteries Expressed in vascular endothelial cells, monocyte-derived macrophages, and coronary smooth muscle cells Very little known about function Gene class is thought to be involved in transcriptional control Broadbent, et al, Human Mol Gen, 2008, 17: 806-14 KIF6 Variant and CAD CARE: Cholesterol and Recurrent Events WOSCOPS: West of Scotland Coronary Prevention Study Association of KIF6 with CAD 2 Prospective Trials of Pravastatin CARE: Cholesterol and Recurrent Events Secondary prevention in patients with a prior MI 40 mg pravastatin vs placebo 87% men and 13% women LDL-C 115 to 174 mg/dL at baseline 2,697 participants studied WOSCOPS: West of Scotland Coronary Prevention Study Primary prevention 40 mg pravastatin vs placebo Men aged 45 to 64 LDL-C 174 to 232 mg/dL at baseline 1,527 participants studied Risk of CAD in CARE Placebo Arm KIF6 Variant and Traditional Risk Factors Smoking Diabetes KIF6 Variant Age ≥ 55 HDL-C < 40 Hypertension LDL-C ≥130 0.5 1 1.5 2 2.5 3 Adjusted Hazard Ratio Magnitude of risk predicted by KIF6 variant was: Similar to that of traditional risk factors Independent of traditional risk factors Risk of CAD in WOSCOPS Placebo Arm KIF6 Variant and Traditional Risk Factors Diabetes KIF6 Variant HDL-C < 40 LDL-C ≥ 189* Hypertension 0.5 1 1.5 2 2.5 3 Adjusted Risk Ratio Magnitude of risk predicted by KIF6 variant was: Similar to that of traditional risk factors Independent of traditional risk factors Case and control patients were matched for age and smoking; all were men * Median level in placebo arm KIF6 Variant is Associated with CAD 5 Prospective Studies CARE Untreated patients WOSCOPS CHS Some treatment ARIC WHS 0.5 1 1.5 2 2.5 Hazard Ratio* KIF6 variant predicts risk of CAD Up to 55% increased risk in untreated populations *Adjusted for traditional risk factors Recent Developments in CV Genetics Mendelian genetics Autosomal Dominant Hypercholesterolemia Familial Thoracic Aortic Aneurysm and Dissection syndromes: LDLR, APOB, PCSK9 FBN1, TGFBR1, TGFBR2, ACTA2, MYH11, NOTCH Genetic markers for CAD Chromosome 9p21/ANRIL KIF6 Pharmacogenetics Warfarin sensitivity Statin efficacy PROVE IT - TIMI 22 study Pravastatin or Atorvastatin Evaluation and Infection Therapy, Thrombolysis in MI Patients hospitalized within 10 days after an acute coronary syndrome Prevention of CHD Events by Statins Carriers of the KIF6 Variant Benefit the Most WOSCOPS PROVE IT Pravastatin 40 mg Atorvastatin 80 mg All Carriers Noncarriers All NonCarriers carriers 0 2 4 Absolute Risk Reduction (%) 6 8 10 12 14 3.5% 5.5% 0.1% 6.2% 10.0% 0.8% KIF6 carriers received significant risk reduction from atorvastatin in PROVE IT, suggesting KIF6 may also predict response with other statins KIF6 carriers also received significantly greater benefit than noncarriers (p=0.003 for WOSCOPS and 0.018 for PROVE IT) Statin Intensity and CHD Event Reduction According to KIF6 719Arg Carrier Status Noncarriers Death or major CV events KIF6 Carriers 40 40 Pravastatin 30 30 p≤0.001 20 20 Atorvastatin 10 Pravastatin 10 0 0 0 3 6 9 12 15 18 21 Months of follow-up 24 27 30 0 3 6 Inclusion Hospit Total c Stabili P=1.0 Major Exc Co-mo Atorvastatin Curren Need f 9 12 15 18 21 24 CABG 27 30 Months of follow-up Liver d Strong KIF6 carriers received greater benefit from 80 mg atorvastatin, compared with 40 mg pravastatin, than did noncarriers Number Needed to Treat with atorvastatin for 2 years to prevent 1 event was 10 for KIF6 carriers and 125 for noncarriers KIF6 Summary Risk for CAD and Risk Reduction from Statins KIF6 carriers, ~60% of the population, have greater risk for CAD Up to 55% increased risk of CAD Independent of traditional risk factors, but similar in magnitude of risk Associated with CAD risk in men (WOSCOPS), women (WHS), the middle aged (ARIC) and the elderly (CHS) KIF6 carriers treated with pravastatin or atorvastatin had a greater reduction of coronary events than noncarriers Warfarin PGx Warfarin is commonly prescribed drug Variants in CYP2C9 and VKORC1 account for high percentage of variability of warfarin response FDA relabeled warfarin in Aug. 2007 to encourage pharmacogenetic testing Utility still remains low Controversial topic Unanswered questions Warfarin PGx: Ready for Prime Time? How is the information applied to clinical practice? Can genetic testing be used to determine the right warfarin dosage? Does it add significantly to the information discerned by careful INR monitoring and other factors (e.g. age, underlying disease state, drug-drug interactions)? Will it really reduce clinical complications? Will it result in shorter time to stable INR? Is it cost-effective? Should genotyping be ordered on all patients taking Warfarin? Can the information be obtained in a timely fashion? Warfarin Widely prescribed anticoagulant 12th most commonly prescribed drug Annual Number of Outpatient Warfarin Prescriptions, 1998-2004 35000000 45% increase from 1998 to 2004 30000000 Annual Number of Prescriptions 25000000 20000000 15000000 10000000 5000000 0 1998 1999 2000 2001 2002 2003 2004 Year Wysowski, D. K. et al. Arch Intern Med 2007;167:1414-1419. Warfarin Challenges in regulating warfarin dosing Prothrombin time (INR) must remain in narrow therapeutic range Elevated INR: risk for major bleeding complications Risk for serious bleeding increases with INR > 4.0 Most likely to occur within first few weeks of initiating treatment Subtherapeutic INR: thrombotic complications Warfarin is the Most Commonly Implicated Medication in U.S. ED Visits Furosemide Bactrim Lisinopril Levofloxacin Glipizide Phenytoin Vicodin Glyburide Metformin Digoxin Clopodigrel Aspirin Insulin Warfarin 0 5 10 Frequency 15 20 Budnitz et al, Ann Intern Med 2007;147:755-765. Wide Range in Warfarin Dose Requirements Variability in Warfarin Dose Requirements, N = 2305 800 780 Number of Patients 750 700 600 500 Warfarin 400Sensitivity 390 300 200 Warfarin Resistance 180 180 100 70 30 0 25 Daily Dose, mg James AH. J Clin Path 1992;45:704-06 12 Sources of variability in Warfarin dosing VKORC1 25% Other 28% CYP2C9 17% Drugs 12% CYP4F2 2% Age 7% Weight 9% Warfarin Pharmacology Racemic mixture of R- and S-enantiomers S-Warfarin approx. 7-10X as potent as RMajority of in vivo activity of warfarin resides in S-warfarin Metabolized mainly through CYP2C9 Targets VKORC1 (Vitamin K epoxide reductase complex subunit 1) Interferes with recycling of Vit K in the liver Reduced activation of clotting factors Pharmacodynamics Pharmacokinetics (hepatocyte) R-Warfarin S-Warfarin CYP2C19 CYP3A4 CYP1A1 CYP1A2 CYP2C8 CYP2C9 (hepatocyte) R-Warfarin S-Warfarin CYP2C9 VKOR inactive 4-hydroxywarfarin 6-hydroxywarfarin 6-hydroxywarfarin 7-hydroxywarfarin Vitamin K (oxidized) Vitamin K (reduced) 7-hydroxywarfarin 8-hydroxywarfarin GGCX 10-hydroxywarfarin Elimination Inactive Vitamin Kdependent clotting factors Active Vitamin Kdependent clotting factors Warfarin PGx CYP2C9 CYP2C9*2 variant CYP2C9*3 variant Arg144Cys 30% decrease in enzymatic activity Ile359Leu 70%-95% decreased enzymatic activity VKORC1 Promoter SNP -1639G>A 44% decrease in promoter activity Allele Frequencies CYP2C9 CYP2C9 VKORC1 *2 *3 -1639G>A Caucasian 8-13% 6-10% 42% Asian 2-5% <1% 89% African American <1% 1-4% 8% Genotype-guided Warfarin Dosing CYP2C9 Genotype Rapid Intermediate *1/*1 *1/*2 *1/*3 *2/*2 *2/*3 *3/*3 Low Sensitivity (GG) 140% 113% 94% 92% 75% 63% Med Sensitivity (AG) 100% 81% 67% 67% 54% 46% High Sensitivity (AA) 73% 58% 48% 48% 40% 33% VKORC1 Genotype Slow McClain, et al., Gen Med, 2008, 10(2): 89-98 www.warfarindosing.org Very Slow Can Pharmacogenomics Assist Warfarin Dosing? Warfarin Dosing 185 Patients Initiation Stabilization PGxInformed (CYP2C9 only) Algorithm Guided (Ageno et al, 2000) Caraco, Clin Pharm Ther, 2008 CYP2C9 Genotype-guided Warfarin Prescribing Efficacy and Safety Time to First Therapeutic INR P<0.001* Proportion of patients with INR>2 1.0 0.8 PGx-informed Controls 0.6 0.4 0.2 *Time to therapeutic INR 2.73 days earlier 0.0 0 5 10 15 20 25 Days Caraco, Clin Pharm Ther, 2008 CYP2C9 Genotype-guided Warfarin Prescribing Efficacy and Safety Proportion of patients at stable anticoagulation Time to First Stable Anticoagulation 1.0 P<0.001* 0.8 PGx-informed Controls 0.6 0.4 *Time to stable anticoagulation 18 days earlier (22+7 vs 40+21 days) 0.2 0.0 0 25 50 Days 75 100 125 Caraco, Clin Pharm Ther, 2008 CYP2C9 Genotype-guided Warfarin Prescribing Efficacy and Safety Time spent in the therapeutic range was higher (80.4 vs. 63.4%) Bleeding incidence was lower (3.2 vs. 12.5%) Patients with 1 or 2 variant alleles required 77% and 52% of dose used by *1/*1, respectively Caraco, Clin Pharm Ther, 2008 Using genotyping results to make Warfarin dosing decisions No clinically validated algorithm A “work in progress” Several large multi-center studies being undertaken to address this question www.warfarindosing.org Warfarin PGx: Summary Some studies suggest that warfarin PGx testing is beneficial Large multi-center studies currently underway Reduced time to stable INR Reduced adverse events Clinical outcomes Validated dosing algorithms New technology allowing for more rapid results Genetic variants explain 40-45% of variability in response to warfarin Important to realize the impact of the compound effects of polymorphisms in CYP2C9 AND VKORC1 Drug-drug interactions, co-morbidities, age, BMI Recent Developments in CV Genetics Mendelian genetics Autosomal Dominant Hypercholesterolemia Familial Thoracic Aortic Aneurysm and Dissection syndromes: FBN1, TGFBR1, TGFBR2, ACTA2, MYH11, NOTCH Genetic markers for CAD LDLR, APOB, PCSK9 Multiple genes contribute to small percentage of phenotype Chromosome 9p21/ANRIL KIF6 Pharmacogenetics Controversial, but gathering speed Warfarin sensitivity Statin efficacy Thank You!!