Molecular Descriptors
advertisement
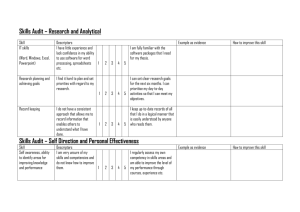
Molecular Descriptors C371 Fall 2004 INTRODUCTION • Molecular descriptors are numerical values that characterize properties of molecules • Examples: – Physicochemical properties (empirical) – Values from algorithms, such as 2D fingerprints • Vary in complexity of encoded information and in compute time Descriptors for Large Data Sets • Descriptors representing properties of complete molecules – Examples: LogP, Molar Refractivity • Descriptors calculated from 2D graphs – Examples: Topological Indexes, 2D fingerprints • Descriptors requiring 3D representations • Example: Pharmacophore descriptors DESCRIPTORS CALCULATED FROM 2D STRUCTURES • Simple counts of features – Lipinski Rule of Five (H bonds, MW, etc.) – Number of ring systems – Number of rotatable bonds • Not likely to discriminate sufficiently when used alone • Combined with other descriptors for best effect Physicochemical Properties • Hydrophobicity – LogP – the logarithm of the partition coefficient between n-octanol and water • ClogP (Leo and Hansch) – based on small set of values from a small set of simple molecules – BioByte: http://www.biobyte.com/ – Daylight’s MedChem Help page – http://www.daylight.com/dayhtml/databases/medchem/m edchem-help.html – Isolating carbon: one not doubly or triply bonded to a heteroatom ACD Labs Calculated Properties • http://www.acdlabs.com • ACD Labs values now incorporated into the CAS Registry File for millions of compounds • I-Lab: http://ilab.acdlabs.com/ – Name generation – NMR prediction – Physical property prediction Molar Refractivity • MR = n2 – 1 MW -------- ----n2 + 2 d where n is the refractive index, d is density, and MW is molecular weight. • Measures the steric bulk of a molecule. Topological Indexes • Single-valued descriptors calculated from the 2D graph of the molecule • Characterize structures according to size, degree of branching, and overall shape • Example: Wiener Index – counts the number of bonds between pairs of atoms and sums the distances between all pairs Topological Indexes: Others • Molecular Connectivity Indexes – Randić (et al.) branching index • Defines a “degree” of an atom as the number of adjacent non-hydrogen atoms • Bond connectivity value is the reciprocal of the square root of the product of the degree of the two atoms in the bond. • Branching index is the sum of the bond connectivities over all bonds in the molecule. – Chi indexes – introduces valence values to encode sigma, pi, and lone pair electrons Kappa Shape Indexes • Characterize aspects of molecular shape – Compare the molecule with the “extreme shapes” possible for that number of atoms • Range from linear molecules to completely connected graph 2D Fingerprints • Two types: – One based on a fragment dictionary • Each bit position corresponds to a specific substructure fragment • Fragments that occur infrequently may be more useful – Another based on hashed methods • Not dependent on a pre-defined dictionary • Any fragment can be encoded • Originally designed for substructure searching, not for molecular descriptors Atom-Pair Descriptors • Encode all pairs of atoms in a molecule • Include the length of the shortest bond-bybond path between them • Elemental type plus the number of nonhydrogen atoms and the number of πbonding electrons BCUT Descriptors • Designed to encode atomic properties that govern intermolecular interactions • Used in diversity analysis • Encode atomic charge, atomic polarizability, and atomic hydrogen bonding ability DESCRIPTORS BASED ON 3D REPRESENTATIONS • Require the generation of 3D conformations – Can be computationally time consuming with large data sets – Usually must take into account conformational flexibility – 3D fragment screens encode spatial relationships between atoms, ring centroids, and planes Pharmacophore Keys & Other 3D Descriptors • Based on atoms or substructures thought to be relevant for receptor binding • Typically include hydrogen bond donors and acceptors, charged centers, aromatic ring centers and hydrophobic centers • Others: 3D topographical indexes, geometric atom pairs, quantum mechanical calculations for HUMO and LUMO DATA VERIFICATION AND MANIPULATION • Data spread and distribution – Coefficient of variation (standard deviation divided by the mean) • Scaling (standardization): making sure that each descriptor has an equal chance of contributing to the overall analysis • Correlations • Reducing the dimensionality of a data set: Principal Components Analysis