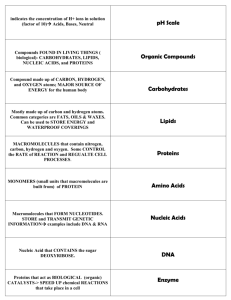
Genetic Variability
advertisement
Genetic Variability 1 Genetic Variability A population is monomorphic at a locus if there exists only one allele at the locus. A population is polymorphic at a locus if two or more alleles coexist in the population. 2 Genetic Variability At a polymorphic locus, if one allele has a very high frequency (> 99%), then the other alleles are unlikely to be observed, unless the sample size is very large. Thus, a locus is commonly defined as polymorphic only if the frequency of its most common allele is < 99%. This definition is arbitrary, and other criteria may be found in the literature. 3 4 5 Influenced by mating system 6 Gene Diversity (Mean Expected Heterozygosity) Gene diversity at a locus (single-locus expected heterozygosity) is defined as: m h 1 x 2 i i 1 where xi = frequency of allele i and m = total number of alleles at the locus. h = the probability that two alleles chosen at random from the population are different from 7 each other. Gene Diversity (Mean Expected Heterozygosity) The average of the h values over all the loci studied, H, can be used as an estimate of the extent of genetic variability within the population. That is, 1 n H hi ni 1 where hi is the gene diversity at locus i, and n is the number of loci. 8 Gene Diversity (Mean Expected Heterozygosity) •H does not depend on an arbitrary definition of polymorphism •H can be computed directly from knowledge of the allele frequencies •H is not affected by sampling effects or mating systems. 9 Random genetic drift is an anti-polymorphic force. Gene diversity is expected to decrease under random genetic drift. In the absence of mutational input, gene diversity will decrease by a fraction of 1/2Ne each generation (Ne = effective population size). 10 11 h and H are unsuitable for DNA data, since the extent of genetic variation at the DNA level in nature is extensive, and both h and H will be ~1 in most cases. Thus, h and H will not be informative measures of polymorphism. The values of h and H is the same for both groups of sequences above. 12 Nucleotide Diversity ( Nucleotide Diversity = Average number of nucleotide differences per site between two randomly chosen sequences. P xi x j pij ij where xi and xj are the frequencies of the ith and jth type of DNA sequences, respectively, and ij is the proportion of different nucleotides between the ith and jth types. 13 14 The alcohol dehydrogenase locus in Drosophila melanogaster Total number of compared sites = 2,379. S = slow migrating electrophoretic allele. F = fast migrating electrophoretic allele. 15 Pairwise percent nucleotide differences among 11 alleles of the alcohol dehydrogenase locus in Drosophila melanogaster. Allele 1 S 1 S 2 S 3 S 4 S 5 S 6 S 7 F 8 F 9 F 10 F 2S 3S 4S 0.13 0.59 0.67 0.55 0.63 0.25 5 S 6 S 0.80 0.80 0.84 0.67 0.55 0.38 0.46 0.46 0.59 7F 8F 0.84 1.13 0.71 1.10 0.50 0.88 0.59 0.97 0.63 0.59 0.21 0.59 0.38 9 F 1.13 10 F 1.13 11 F 1.22 1.10 1.10 1.18 0.88 0.88 0.97 0.97 0.97 1.05 0.59 0.59 0.84 0.59 0.59 0.67 0.38 0.38 0.46 0.00 0.00 0.42 0.00 0.42 0.42 16 17 Types of Genetic Variation: Single nucleotide polymorphisms (SNPs) due to point mutations. Structural variation due to deletions, duplications, insertions, inversions, and translocations. 18 Types of Structural Genetic variation: Submicroscopic variation (less than 3Mb). Microscopic variation (more than 3Mb). Copy number variants (CNVs) are submicroscopic structural variations that are due to deletion, duplication, and replicative transposition. If the variation in copy number occurs in tandem, it is referred to as variable number of tandem repeats (VNTRs). 19 Types of Structural Genetic variation: Inversion. A segment of DNA that is reversed in orientation with respect to the rest of the chromosome. Pericentric inversions include the centromere, whereas paracentric inversions do not. Translocation. A change in position of a chromosomal segment within a genome that involves no change in total DNA content. Translocations can be intra- or interchromosomal. Segmental uniparental disomy. Uniparental disomy describes the phenomenon in which a pair of homologous chromosomes or portions of a chromosome in a diploid 20 individual is derived from a single parent. Which mammal has the most size variation? 21 Which mammal has the most size variation? 22 Human Genetic Variation 23 With the exception of monozygotic twins, which are NEARLY identical genetically every one of us is genetically different from every other human who ever lived. 24 Geographic distribution of skin and hair color Distribution of Human Skin Color Clinal distribution of hair color among Australian Aborigines Discontinuous distribution of red hair in Britain 25 Genetic variation may be important from a medical point of view For example, because of genetic differences, different people may respond differently to the same drug In the 1950s, anesthesiologists began using the muscle relaxant succinylcholine Succinylcholine is normally metabolized by cholinesterase One out of 2,500 people are heterozygous for a variant of cholinesterase that does not metabolize succinylcholine These people are OK unless exposed to succinylcholine, in which case they go into breathing arrest 26 How are genomes of individuals different from one another? More than 90% of the differences are single base substitutions. These are called single nucleotide polymorphisms (SNPs) Nature 409, 822 - 823 (2001) SNP Any two human genomes are roughly 99.9% identical chr - chromosome n - Number of samples examined S - Number of polymorphic sites - Nucleotide divergence Mean = ~ 0.1% 28 Przeworski, M., et al. (2000) Trends Genet 16, 296-302. If (1) two genomes are roughly 99.9% identical to each other, and (2) a haploid genome is 3.2 billion base-pairs in length, then, there are 3.2 million differences (SNPs) between any two genomes. (remember that each diploid individual has two genomes) 29 Kruglyak and Nickerson Nature Genetics (2001) 27 234 How many SNPs have been identified in humans? Build 135, November 3, 2011 30 Where are the SNPs found? protein-coding exon Total: 60,480,978 SNPs In protein-coding exons: 862,465 SNPs (1.4%) 31 http://www.ncbi.nlm.nih.gov/SNP/snp_summary.cgi How many SNPs are “born” in each generation? Number of genomes: N = 14×109 (twice the number of people) Mutation rate: m = ~2×10-8 per base-pair per generation New mutations = Nm = 280 per base-pair per generation Each nucleotide in the genome gets mutated on average in 280 individuals in each generation The overwhelming majority of these will never attain polymorphic status (arbitrarily set at 1% of the population) 32 Kruglyak and Nickerson Nature Genetics (2001) 27 234 How are the frequencies of the SNPs distributed? 35,989 SNPs in a sample of 20 chromosomes 21. 33 Patil et al. Science (2001) 294:1719-1723 Percentage of human genetic variation within and between populations. 34 R.A.Brown,G.J.Armelagos,Evol.Anthropol.10 ,34 (2001). Owens and King Science (1999) 286: 451-453. Percentage of human genetic variation within and between populations. An average population from anywhere in the world contains 85% of all human variation at autosomal loci and 81% of all human variation in mtDNA sequences. Differences among populations from the same continent contribute another 6% of variation; only 9-13% of 35 genetic variation differentiates populations from different continents. Most alleles are geographically widespread 377 autosomal microsallelite loci 1056 individuals from 52 populations from seven regions 36 Rosenberg et al. Science Dec 20 2002: 2381-2385. (supplement) There are non major genetic differences across ‘races’ = NO ‘races’ “The possibility that human history has been characterized by genetically relatively homogeneous groups (‘races’), distinguished by major biological differences, is not consistent with genetic evidence.” 37 Owens and King. Science (1999) 286: 451-453. Copy number variation (CNV) of DNA sequences in 270 individuals from four populations with ancestry in Europe, Africa or Asia. A total of 1,447 copy number variable regions (CNVRs), covering 360 megabases (12% of the genome) were identified in these populations. These CNVRs encompassed more nucleotide content per genome than SNPs, underscoring the importance of CNV in genetic diversity 38 and evolution.