poster - The Department of Ecology and Evolutionary Biology
advertisement
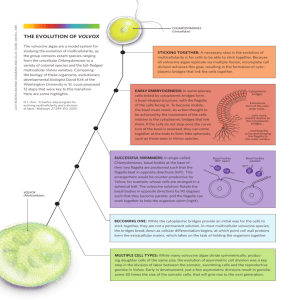
Estimation of divergence times in the volvocine algae Matthew D. Herron, Jeremiah D. Hackett, Frank Aylward, and Richard E. Michod Department of Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ, USA INTRODUCTION MATERIALS AND METHODS Phylogenetic reconstruction of red algae, green algae, and land plants using Bayesian Markov Chain Monte Carlo methods in MrBayes. The volvocine green algae (Volvox and its close relatives) are an important model system for the evolution of multicellularity and cellular differentiation. Three chloroplast genes: P700 chlorophyll a-apoprotein A1 (psaA), P700 chlorophyll aapoprotein A2 (psaB), and photosystem II CP43 apoprotein (psbC) partitioned by codon. The evolutionary transition from single cells to completely differentiated multicellular individuals can be describe as a series of small steps, many represented in extant volvocine algae (Kirk 2005). One nuclear gene: small ribosomal subunit (SSU). Divergence times estimated using Langley-Fitch (local molecular clock method; LF), nonparametric rate smoothing (NPRS), and penalized likelihood (PL) in r8s (Figure 1). Reconstructing the history of these changes allows us to describe an increase in the level of complexity within a gradualist framework. Sample of 300 posterior trees. Multiple fossil calibrations (see inset, Figure 1). Rausch et al. (1989) estimated the divergence of colonial volvocine algae from unicellular relatives at 50-75 MYA using nuclear genes; these estimates assume a constant substitution rate across lineages. Analyses evaluated by fossil cross-validation. Divergences within the volvocine algae estimated using LF (Figure 2). Sample of 300 posterior trees from the five chloroplast gene analysis of Herron & Michod (2008). We used multiple genes with multiple fossil calibrations to estimate divergence times among unicellular and colonial volvocine algae. Beta subunit of ATP synthase (atpB), large subunit of Rubisco (rbcL), psaA, psaB, and psbC, partitioned by codon. RESULTS Compsopogon coeruleus 1 Rhodella violacea Porphyra yezoensis Gracilaria tenuistipitata Rhodophyta Porphyridium aeruginium Tetrabaenaceae 0.97 8 9 Palmaria palmata 2 5 6 Chaetosphaeridium globosum Psilotum nudum 3 Pinus 4 Nymphaea / Cabomba 5 Streptophyta Marchantia polymorpha 7 2 0.84 1 3 4 Arabidopsis thaliana 6 0.96 Goniacaceae 0.88 Mesostigma viride 9 10 Euvolvox 8 0.88 Triticum aestivum 0.67 Ostreococcus tauri Chlorella vulgaris 7 8 Oltmannsiellopsis viridis 8 Stigeoclonium helveticum Paulschulzia pseudovolvox Fossil calibrations from Yoon et al. (2004), Berney & Pawlowski (2006): 1. 1174-1222 MYA for Bangiomorpha (red alga) 2. 596-603 MYA for Florideophyte red algae 3. 432-476 MYA for first land plants 4. 355-370 MYA for first seed plants 5. 290-320 MYA for angiosperm vs. gymnosperm split 6. 115 MYA min. for oldest known Nymphaeales 7. 90-130 MYA for monocot vs. dicot split 8. 750 MYA min. for Proterocladus (Cladophoraceae) 1800 1600 1400 1200 1000 Chlamydomonas debaryana 8 9 Chlamydomonas reinhardtii 0.99 800 600 400 9 Tetrabaena socialis Volvox tertius # Gains and Volvox carteri # losses of characters defined by Kirk (2005): 200 MYA Figure 1. Chronogram of red algae, green algae and land plants based on three chloroplast genes (psaA, psaB, and psbC) and one nuclear gene (SSU) using LF. Gray boxes are ranges of estimates across the sample of 300 posterior trees. Outlined boxes are fossil calibrations from Yoon et al. (2004); gray boxes for these nodes are ranges inferred during fossil cross-validation. Bayesian posterior probabilities below 1.0 are shown. Volvocaceae 0.97 8 Chlorophyta Pseudendoclonium akinetum 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 300 Incomplete cytokinesis Partial inversion Rotation of basal bodies Organismal polarity Transformation of cell wall into ECM Genetic control of cell number Complete inversion Increased volume of ECM Sterile somatic cells Specialized germ cells * Asymmetric division Bifurcated cell division program 250 200 0.94 9 10 0.98 0.94 8 0.97 10* 0.99 11 12 0.59 150 Lobomonas monstruosa Paulschulzia pseudovolvox Chlamydomonas reinhardtii Vitreochlamys pinguis Vitreochlamys aulata Vitreochlamys aulata Vitreochlamys ordinata Chlamydomonas debaryana Basichlamys sacculifera Tetrabaena socialis Astrephomene perforata Astrephomene gubernaculifera Astrephomene gubernaculifera Gonium pectorale Gonium viridistellatum Gonium viridistellatum Gonium viridistellatum Gonium quadratum Gonium octonarium Gonium multicoccum Gonium multicoccum Volvox globator Volvox barberi Volvox rousseletii Platydorina caudata Volvulina compacta Volvulina pringsheimii Volvulina steinii Volvulina steinii Volvulina steinii Pandorina morum Pandorina morum Pandorina morum Pandorina colemaniae Pandorina morum Pandorina morum Volvulina boldii Yamagishiella unicocca Yamagishiella unicocca Yamagishiella unicocca Eudorina elegans Eudorina elegans Eudorina unicocca Eudorina unicocca Eudorina elegans Volvox gigas Pleodorina indica Pleodorina illinoisensis Eudorina cylindrica Pleodorina californica Pleodorina japonica Volvox aureus Volvox aureus Volvox aureus Volvox africanus Volvox dissipatrix Volvox tertius Volvox tertius Volvox obversus Volvox carteri f. nagariensis Volvox carteri f. kawasakiensis Volvox carteri f. weismannia 100 50 MYA CONCLUSIONS The colonial volvocine algae (Tetrabaenaceae, Goniaceae, and Volvocaceae) diverged from unicellular relatives at least 200 MYA. An early, rapid radiation established all of the major colonial lineages (Tetrabaenaceae; Astrephomene; Gonium; Euvolvox; Pandorina + Volvulina; Yamagishiella; Eudorina + Pleodorina + remaining Volvox) by 150 MYA. Most colonial body plans are not recent innovations but ancient adaptations that have been stable over a time scale of tens of millions of years. Several nominal species include divergences over 50 MYA (Astrephomene gubernaculifera, Gonium multicoccum, Volvulina steinii, Pandorina morum, Yamagishiella unicocca, Eudorina elegans) A body plan similar to that of modern Eudorina, with complete inversion and a large volume of extracellular matrix (ECM) was present at least 180 MYA. Sterile somatic cells originated at least 30 MYA in Euvolvox (V. globator + V. barberi + V. rousseletii), at least 80 MYA in the clade including the remaining Volvox and Pleodorina, and at least 125 MYA in Astrephomene. Figure 2. Chronogram of volvocine algae based on five chloroplast genes using LF, calibrated with the divergence between Paulschulzia (Tetrasporales) and Volvox carteri derived from analysis in Figure 1. Gray boxes are ranges of estimates across the sample of 300 posterior trees. Bayesian posterior probabilities below 1.0 are shown. * It is unclear if the MRCA of V. aureus and V. carteri had specialized germ cells. BIBLIOGRAPHY Berney, C. and Pawlowski, J. 2006. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc. R. Soc. B 273:1867-1872. Herron, M. D. and R. E. Michod. 2008. Evolution of complexity in the volvocine algae: transitions in individuality through Darwin's eye. Evolution 62:436-451. Kirk, D. L. 2005. A twelve-step program for evolving multicellularity and a division of labor. BioEssays 27:299-310. Rausch, H., N. Larsen, and R. Schmitt. 1989. Phylogenetic relationships of the green alga Volvox carteri deduced from small-subunit ribosomal RNA comparisons. J. Mol. Evol. 29:255-265. Sanderson M. J. 1999 r8s ("rates"), version 0.9. Software for the analysis of rates of evolution and miscellaneous other stuff. University of California, Davis, California, USA Yoon, H. S., J. D. Hackett, C. Ciniglia, G. Pinto, and D. Bhattacharya. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21:809-818.